Abstract
Interspecific or intergeneric hybridization, followed by chromosome doubling, can lead to the formation of new allopolyploid species. Recent studies indicate that allopolyploid formation is associated with genetic and epigenetic changes, although little is known about the type of changes that occur, how rapidly they occur, and the type of sequences involved. To address these matters, we have surveyed F1 hybrids between diploid species from the wheat (Aegilops and Triticum) group and their derived allotetraploids by screening a large number of loci using amplified fragment length polymorphism and DNA gel blot analysis and by assaying the extent of cytosine methylation. We found that sequence elimination is one of the major and immediate responses of the wheat genome to wide hybridization or allopolyploidy, that it affects a large fraction of the genome, and that it is reproducible. In one cross between Ae. sharonensis × Ae. umbellulata, 14% of the loci from Ae. sharonensis were eliminated compared with only 0.5% from Ae. umbellulata, with most changes occurring in the F1 hybrid. In contrast, crosses between Ae. longissima × T. urartu showed that sequence elimination was more frequent after chromosome doubling. Alterations in cytosine methylation occurred in ∼13% of the loci, either in the F1 hybrid or in the allopolyploid. For eight of nine bands that were isolated, the sequences that underwent elimination corresponded to low-copy DNA, whereas alterations in methylation patterns affected both repetitive DNA sequences, such as retrotransposons, and low-copy DNA in approximately equal proportions.
INTRODUCTION
The duplication of genomes (polyploidy), either of the same genome (autopolyploidy) or of diverged genomes (allopolyploidy or amphiploidy), is a major force of evolution. It is thought that most living eukaryotes have undergone one or more polyploidization events during their history (Spring, 1997; Wolfe and Shields, 1997). One interpretation of the success of polyploidy is that it releases one set of alleles from selective pressure, thus facilitating the acquisition of new functions and buffering the mutation load. In the case of allopolyploidy, the fixation of permanent heterozygosity (Stebbins, 1971; Allard et al., 1993) has the potential to offer a substantial heterozygote advantage. Despite these potential benefits, polyploidy is an enormous challenge with regard to the orchestration of gene expression, DNA replication, and chromosome pairing. How the new polyploid genome deals with such genomic stress is a significant, albeit poorly understood, question.
In plants, it is estimated that polyploidy occurred in the history of ∼70% of flowering species (Masterson, 1994). Genome sequencing data may provide further evidence for polyploidy, as suggested by the recent finding that Arabidopsis, whose genome is among the smallest in angiosperms, is probably an ancient tetraploid (The Arabidopsis Genome Initiative, 2000). Allopolyploidy, brought about by interspecific or intergeneric hybridization followed by chromosome doubling, was found to be associated with genomic alterations, such as sequence elimination, activation of transposons, intergenomic coconversion, methylation, gene silencing, or subtler changes in gene expression (for reviews, see Matzke et al., 1999; Comai, 2000; Wendel, 2000).
Previous works showing allopolyploidy-induced genetic and epigenetic alterations have focused on selected sequences such as rDNA genes (Sasakuma et al., 1995), cDNAs (Song et al., 1995), restriction fragment length polymorphisms (RFLPs), or low-copy sequences from chromosome- or genome-specific DNA (Feldman et al., 1997; Liu et al., 1998a, 1998b) or transgenes (Scheid et al., 1996). In this work, we have analyzed an unbiased set of wheat DNA loci in diploid wheat species, the interspecific F1 offspring, and their derived synthetic allotetraploids using amplified fragment length polymorphism (AFLP) (Vos et al., 1995) and methylation-sensitive amplification polymorphism (MSAP) (Xiong et al., 1999) to obtain a quantitative estimate of the timing and frequency of allopolyploidy-associated genetic and epigenetic alterations and to determine what type of sequences are affected. The wheat (Aegilops and Triticum) group offers an ideal system in which to study the evolution of polyploids because several species are young, such as the hexaploid bread wheat, which is only ∼85,000 years old (Feldman, 2001), most diploid progenitors are known, and allopolyploids can be synthesized easily.
RESULTS
Frequency of Allopolyploidy-Associated Genetic Alterations as Estimated by AFLP
To assess the frequency of allopolyploidy-associated genetic changes in a quantitative and unbiased manner, two different crosses were studied: Ae. sharonensis (SlSl) × Ae. umbellulata (UU) (cross A) and Ae. longissima (SlSl) × T. urartu (AA) (cross B). The F1 hybrids and the allopolyploids derived from each cross were analyzed by AFLP. The banding pattern of the hybrids is expected to yield a pattern that is additive of both parents because parents derive from inbred lines (confirmed in this study) and because AFLP markers are dominant. All of the cases of deviation from such additivity scored in this study were reproducible (see Methods). Deviation was observed for bands present in only one of the parents (polymorphic bands) and absent in the F1 hybrid and/or the allotetraploid. Another, albeit more rare, type of deviation from additivity was the appearance of new bands that were not present in the parents. Using 24 different primer pairs, 1829 bands were obtained in cross A and 1832 bands were obtained in cross B (Table 1). Of these, the number of polymorphic bands was 373 in cross A and 346 in cross B. In cross A, 20 bands were absent in both F1 and the allotetraploid, suggesting that these bands disappeared at the hybrid level (Figure 1). Similarly, in cross B, 13 bands disappeared in both F1 and allotetraploid plants. In another group of loci, bands that were present in the parents and in the F1 hybrid disappeared in the allotetraploid plants: there were five such bands in cross A and 28 in cross B. Interestingly, band disappearance was not random: all 20 bands that had disappeared in F1 in cross A originated from Ae. sharonensis, and in cross B, of 13 bands that disappeared in F1, 12 were from Ae. longissima. For bands that were eliminated only in the allopolyploid, there was also a preferential elimination of bands from Ae. sharonensis in cross A (four of five bands that disappeared originated from Ae. sharonensis), whereas in cross B, there was no strong bias for elimination (10 bands in Ae. longissima and 18 in T. urartu). Overall, the percentage of bands that disappeared can be estimated only out of the polymorphic bands because of the dominant nature of AFLP. In cross A, 14% of the bands (24 of 171) from Ae. sharonensis disappeared compared with 0.5% (one of 202) from Ae. umbellulata. Most of the bands had disappeared in the F1 hybrid (20 versus five in the allopolyploid). In cross B, 12.2% of the bands (22 of 180) from Ae. longissima disappeared compared with 11.4% (19 of 166) from T. urartu, and the timing of band disappearance was later than in cross A (i.e., more events occurred in the allopolyploid [28 events] than in the F1 hybrid [13 events]).
Table 1.
AFLP Banding Pattern in the Parental Plants of One Interspecific Cross (Ae. sharonensis [SlSl] × Ae. umbellulata [UU]) and One Intergeneric Cross (Ae. longissima [SlSl] × T. urartu [AA]) in Their F1 Hybrids and in the First Generation of the Allotetraploids [SlSlUU and SlSlAA]
| Bands in Parents
|
Bands That Disappeared
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Polymorphic
|
In F1 and Allotetraploid
|
In Allotetraploid Only
|
||||||
| Crossa |  |
 |
Totalb |  |
 |
 |
 |
Total |
| SlSl × UU (cross A) | 171 | 202 | 1829 | 20 | 0 | 4 | 1 | 25 |
| SlSl × AA (cross B) | 80 | 166 | 1832 | 12 | 1 | 10 | 18 | 41 |
In cross A, the maternal genome (SlSl) was from Ae. sharonensis, whereas in cross B, it was from Ae. longissima (SlSl).
The total number of bands for both parents. Monomorphic bands were scored only once.
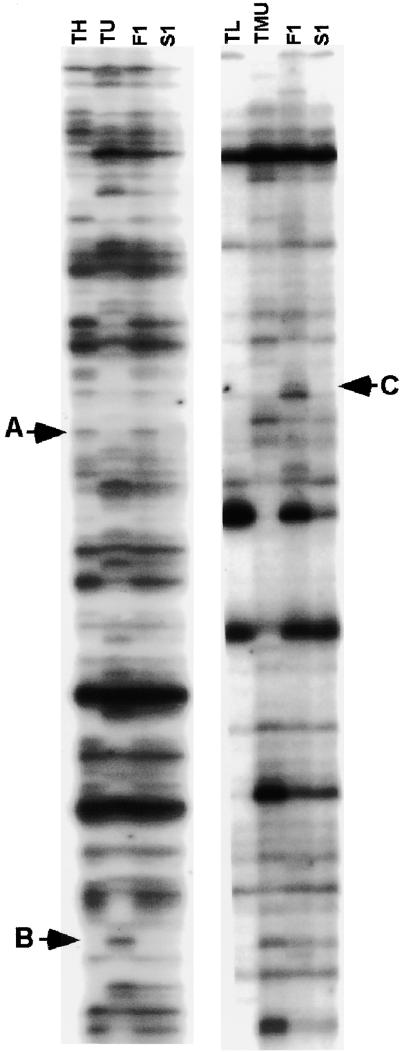
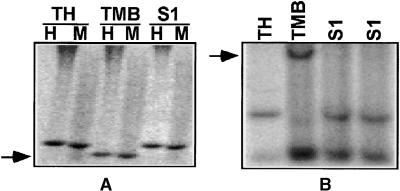
Figure 1.
AFLP Fingerprints of Genomic DNAs in Diploid and Allopolyploid Wheat.
The panel at left (EcoRI+AAC/MseI+CTA) included Ae. sharonensis (TH), Ae. umbellulata (TU), the F1 interspecific hybrid between TH and TU (F1), and the descendant allotetraploid (S1). The panel at right (EcoRI+ACC/MseI+CAG) included Ae. longissima (TL), T. urartu (TMU), the F1 hybrid, and the descendant allotetraploid (S1). Arrows indicate three different situations: disappearance of a band in only the allotetraploid plant (A); disappearance of a band in the F1 plant that was maintained in the allotetraploid plant (B); and a rare event in which a new band appeared in the F1 plant (C).
In addition to band disappearance, which was the most common event observed, we found five cases, among the 1832 AFLP bands analyzed in cross B, of new bands that were not seen in the parents that appeared in F1 and disappeared again in the allopolyploid (Figure 1, band C). Similarly, we found seven cases of bands that were present in the parents, disappeared in F1, and reappeared in the allotetraploid (data not shown). These deviations from additivity were probably caused by changes in methylation of the EcoRI site. Alterations in AFLP patterns caused by methylation changes are described in the MSAP analysis. Here, we have focused on the analysis of bands that disappeared in F1 and/or the allopolyploid.
DNA Gel Blot Validation of the AFLP-Detected Genetic Alterations
The AFLP-observed deviations from additivity could result from the heterozygosity of the parents, from artifacts of the polymerase chain reaction (PCR)-based AFLP analysis, or from methylation alteration, or they could be the result of random or reproducible genetic alterations induced by allopolyploidy. To distinguish between these various possibilities, we performed DNA gel blot analysis in independently synthesized allotetraploids. This analysis should reveal potential heterozygosity of the parents and indicate the reproducibility of the observed alterations.
Nine bands that disappeared, either in F1 or in the allopolyploid (all from cross B), were isolated from acrylamide AFLP gels (Figure 1) and used as probes for DNA gel blot analysis. The results of this analysis are shown in Table 2 and are described below. Eight of the nine bands originated from low copy DNA, and one was from high-copy DNA. Sequence analysis did not reveal any significant similarity for any of the nine sequences. In two cases, that of AFLP-isolated fragments 4 and 5 (AIF4 and AIF5), validation could not be determined, either because there was no polymorphism between the parents (AIF4) or because the band originated from high-copy DNA (AIF5) and discrete bands could not be analyzed. In all seven cases in which validation could be tested, the band elimination observed by AFLP was confirmed by DNA gel blot analysis (Figure 2). Note that, for all of the probes tested, the pattern of elimination was reproducible in three different S1 plants of each allotetraploid. This finding, as well as the reproducibility of migration patterns in the parents (data not shown), indicates that the deviation from additivity was not caused by the heterozygosity of the parents. Moreover, some of the enzymes used in the DNA gel blot analysis (DraI and EcoRV) are not particularly sensitive to methylation and band disappearance was not associated with the appearance of new bands, suggesting that for the cases studied here, the band disappearance in AFLP was not caused by cytosine methylation. In summary, AFLP is a robust and high throughput means to assess the induction of genomic rearrangements. Elimination was the most frequent genomic rearrangement; it was reproducible and could not be attributed to heterozygosity or methylation.
Table 2.
DNA Gel Blot Analysis of AIF Probes That Showed AFLP Band Disappearance in the F1 Hybrid and/or in the Allotetraploid Plants of Cross B (SlSl × AA)
| Probe | Primer Pairs | Fragment Size (bp) | Origin of Fragment | Eliminationa | Copy Number | Sequence Similarityb |
|---|---|---|---|---|---|---|
| AIF1 | E+AAC/M+CAG | 420 | urartu | + | Low | None |
| AIF2 | E+AAC/M+CAG | 202 | urartu | + | Low | None |
| AIF3 | E+AAC/M+CAG | 150 | urartu | + | Low | None |
| AIF4 | E+AAC/M+CAG | 80 | urartu | ND | Low | None |
| AIF5 | E+AGG/M+CTT | 530 | longissima | ND | High | None |
| AIF6 | E+AGG/M+CTT | 510 | urartu | + | Low | None |
| AIF7 | E+AGG/M+CTT | 450 | urartu | + | Low | None |
| AIF8 | E+AAC/M+CTT | 170 | longissima | + | Low | None |
| AIF9 | E+AAC/M+CTT | 165 | longissima | + | Low | None |
Elimination in F1 and/or the allotetraploid was validated (+) or could not be determined (ND). AIF1 to AIF7 were eliminated in the allotetraploid only, and AIF8 and AIF9 were eliminated in both F1 and the allotetraploid.
Similarity was considered significant for e values ≤ 0.005.
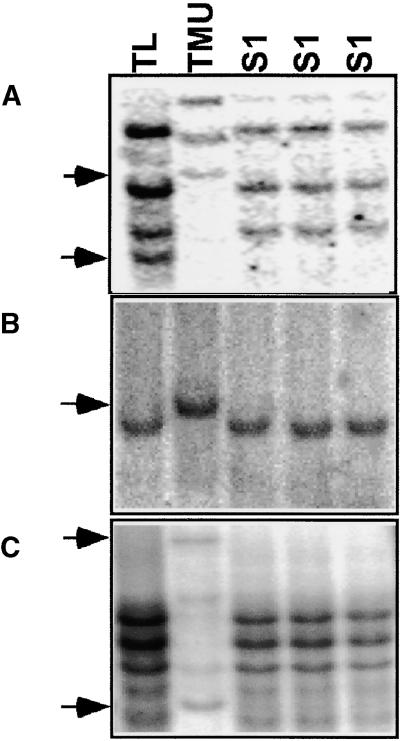
Figure 2.
DNA Gel Blot Hybridization with Three Different AIF Probes That Showed Band Disappearance with the AFLP Method.
Genomic DNA of Ae. longissima (TL; genome SlSl), T. urartu (TMU; genome AA), and three independently made newly synthesized allotetraploids (S1 generation; genome SlSlAA) was digested and blotted on a nylon membrane.
(A) AIF1 probe (Table 2) was isolated from T. urartu and hybridized to EcoRV-digested genomic DNA. Sequence elimination was found in both Ae. longissima (bottom arrow) and T. urartu (top arrow).
(B) AIF2 probe (Table 2) was isolated from T. urartu and hybridized to BamHI-digested genomic DNA. This was the only probe that showed a single-copy fragment in both parents. The T. urartu band, shown by the arrow, disappeared in the allotetraploid.
(C) AIF6 (Table 2) was isolated from T. urartu and hybridized to HindIII-digested genomic DNA. Two bands from T. urartu, shown by arrows, disappeared in the allotetraploid. This pattern of elimination also was shown with enzymes that are not sensitive to methylation, such as DraI, with all of the probes (data not shown).
Frequency of Allopolyploidy-Associated Epigenetic Alterations as Estimated by MSAP
The enzyme HpaII is sensitive to methylation of either cytosine residue at the recognition site (5′-CCGG), whereas its isoschizomer MspI is sensitive only to methylation of the external cytosine. Therefore, methylation of the internal cytosine would lead to a differential cleavage by the two isoschizomers and thus to the appearance of different MSAP fragments in the sequencing gel loaded with the amplification products from EcoRI+MspI versus EcoRI+ HpaII digests (Figure 3). Another, albeit less frequent, cause of the differential banding pattern, namely the presence of fragments in the EcoRI+HpaII digests and their absence in the EcoRI+MspI digests, has been observed (Figure 3) and has been attributed to hemimethylation of the external C, resulting in blocking of MspI digestion (McClelland et al., 1994).
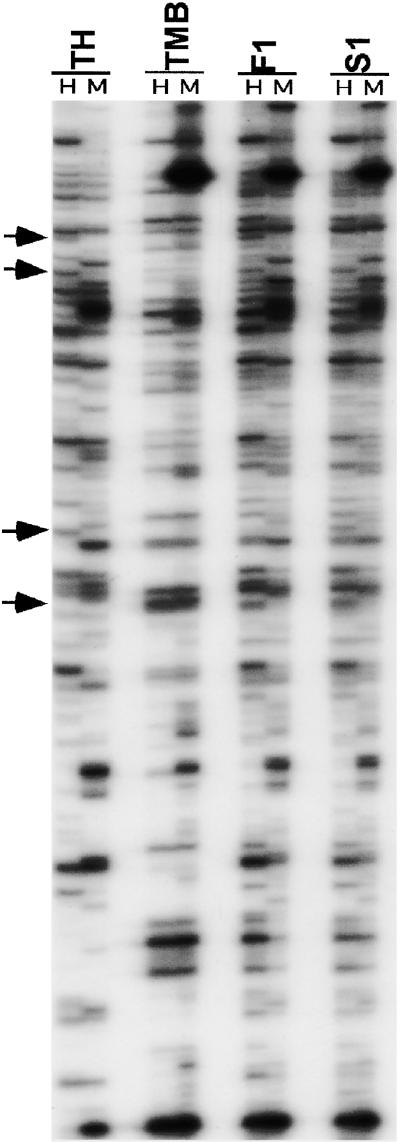
Figure 3.
MSAP Patterns Detected in the Two Diploid Parents, the F1 Hybrid, and the Allotetraploid.
The primer combination used was HM+TCAA/E+ACG. Lines 1 to 4 are Ae. sharonensis (TH), T. monococcum ssp aegilopoides (TMB), the F1 hybrid (F1), and the allotetraploid (S1), respectively. H and M refer to digestion with EcoRI+HpaII and EcoRI+MspI, respectively. Bands that showed alteration in methylation pattern in the F1 hybrid and/or the allotetraploid are shown by arrows.
Seven pairs of selective primers produced 501 clear bands in the two parental lines Ae. sharonensis (genome SlSl) and T. monococcum ssp aegilopoides (genome AmAm) (Table 3). Each of the bands represented a recognition site cleaved by one or both of the isoschizomers. Of the 501 resolved bands, 159 were methylated in one or both parental lines (Table 3). The two parents showed essentially the same degree of methylation, namely 30.2% for Ae. sharonensis and 33.2% for T. monococcum ssp aegilopoides (data not shown). Therefore, on average, 31.7% (159 of 501) of the 5′-CCGG sites in the parental line genome were cytosine methylated in the leaves (Table 3).
Table 3.
Number of Bands Amplified Using Seven MSAP Selective Primer Combinations in the Parental Lines Ae. sharonensis (SlSl) and T. monococcum ssp aegilopoides (AmAm), the F1 Hybrid (SlAm), and the Allotetraploid (SlSlAmAm)
| Primer (HM+4/E+3)a |
Total Number of Bands in the Two Diploid Parentsb |
Polymorphic Sitesc |
Total Number of Methylated Sitesd |
Methylation Alteration in the F1 Hybrid |
Methylation Alteration in the Amphiploid |
|---|---|---|---|---|---|
| E+AAG | 76 | 2 | 31 | 2 | 3 |
| E+AGC | 65 | 4 | 28 | 3 | 0 |
| E+AGG | 64 | 3 | 18 | 8 | 8 |
| E+ACG | 72 | 7 | 20 | 0 | 0 |
| E+ACT | 78 | 4 | 22 | 0 | 2 |
| E+ACA | 70 | 9 | 15 | 1 | 2 |
| E+ACC | 76 | 8 | 25 | 0 | 3 |
| Total | 501 | 37 | 159 | 14 | 18 |
The selective primer HM+4 (5′-CATGAGTCCTGCTCGGTCAA [HM+TCCA]) was used in combination with each of the seven E+3 primers listed. The core sequence of EcoRI is identical to that used in the AFLP protocol by Vos et al. (1995).
Bands from four lanes were analyzed in each selective primer combination, two lanes for each parent (digested with either HpaII or MspI). Bands that were monomorphic (identical in both parents) were counted only once.
Sites showing conventional AFLP polymorphism, not related to methylation.
Bands were considered methylated if they showed polymorphism between the two isoschizomers. Bands with the same methylation pattern in both parents were scored only once.
The two parental lines, the F1 hybrid, and the allotetraploid were compared for their methylation patterns (Table 4, Figure 3). As mentioned for the AFLP analysis of genetic alterations, we expected that F1 and the allotetraploid would have the combined methylation pattern of both parents. Any deviation from additivity was considered an alteration in methylation pattern related to the F1 and/or the allotetraploid situation. Five major classes were identified among the methylated fragments (Table 4): (1) 21 bands (13.2% of all 159 methylated sites) were monomorphic, that is, they had the same pattern of methylation among parental lines, the F1 hybrid, and the allotetraploid; (2) 117 bands (73.5% of all methylated sites) showed differential cytosine methylation between the parents but no deviation from additivity in F1 or the allotetraploid; (3) three bands (1.8% of all methylated sites) showed differential cytosine methylation patterns between parents and F1 hybrids, but methylation was restored to the parental type in the allotetraploid; (4) 11 bands (6.9% of all methylated sites) showed alteration in cytosine methylation starting in the F1 hybrid that was maintained in the allotetraploid (interestingly, of those, 10 were from aegilopoides and one was from sharonensis); and (5) seven bands (4.4% of all loci) showed methylation alteration in the allotetraploid but not in the F1 hybrid.
Table 4.
Patterns of Cytosine Methylation in the Parental Lines, the F1 Hybrid, and the Allotetraploida
|
sharonensis (SlSl) |
aegilopoides (AmAm) |
F1 Hybrid (SlAm) |
Allotetraploid (SlSlAmAm) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Methylation Patterns | HpaII | MspI | HpaII | MspI | HpaII | MspI | HpaII | MspI | Number of Sites | Total (%) |
| Monomorphic | − | + | − | + | − | + | − | + | 21 | 21 (13.2) |
| Polymorphic between parents, no change in F1 and amphiploid |
+ | − | − | − | + | − | + | − | 47 | |
| − | + | − | − | − | + | − | + | 35 | ||
| − | − | − | + | − | + | − | + | 24 | 117 (73.5) | |
| − | + | + | + | + | + | + | + | 6 | ||
| − | − | + | − | + | − | + | − | 5 | ||
| Methylation alteration in F1 only | + | − | − | − | − | − | + | − | 2 | 3 (1.8) |
| + | + | − | + | − | − | − | + | 1 | ||
| Methylation alteration in F1 and amphiploid | − | − | − | + | − | − | − | − | 4 | 11 (6.9) |
| + | − | − | − | − | − | − | − | 1 | ||
| − | − | + | + | + | − | + | − | 6 | ||
| Methylation alteration in amphiploid only |
− | + | − | − | − | + | − | − | 3 | 7 (4.4) |
| − | − | − | + | − | + | − | − | 3 | ||
| + | − | − | − | + | − | − | − | 1 | ||
a +, band present; −, band absent. Each row represents a different MSAP banding pattern that was observed in the parents-F1-allotetraploid combination.
DNA Gel Blot Validation of the MSAP-Detected Epigenetic Alterations
DNA gel blot analysis was performed to confirm the methylation patterns detected by MSAP using 12 MSAP-isolated fragments (MIFs) as probes. These included five fragments (of 11 total) that showed methylation alteration in both the F1 hybrid and the allotetraploid and all seven fragments that showed methylation alteration in the allotetraploid only (Table 4). Total DNA from leaves was digested with either HpaII or MspI. All 12 fragments were sequenced and analyzed for similarity to known sequences in the database. Three of the five fragments that showed methylation alteration in the F1 hybrid (MIFs 1 to 5) showed high similarity to repetitive DNA sequences and highly repeated retrotransposons, and two fragments showed no significant similarity to known sequences (Table 5).
Table 5.
DNA Gel Blot Analysis of the Fragments That Showed Methylation Alteration in the F1 Hybrid of the Cross Ae. sharonensis (SlSl) and T. monococcum ssp aegilopoides (AmAm) and in the Derived Allotetraploid
| Methylation Alterationb
|
|||||||
|---|---|---|---|---|---|---|---|
| Probe | Primer (HM+4/E+3)a | Fragment Size (bp) | Origin of the Fragment | F1 | Amphiploid | Copy Number | Sequence Similarityc |
| MIF1 | E+AAG | 210 | sharonensis | + | + | High | Barley retrotransposon |
| MIF2 | E+AAG | 260 | aegilopoides | + | + | Low | None |
| MIF3 | E+AGG | 100 | sharonensis | + | + | High | Barley retrotransposon |
| MIF4 | E+AGG | 150 | aegilopoides | + | + | High |
Ceratitis capitata unknown repetitive DNA fragment |
| MIF5 | E+ACA | 134 | aegilopoides | + | + | High | None |
| MIF6 | E+AAG | 350 | aegilopoides | − | + | Low | None |
| MIF7 | E+ACT | 110 | aegilopoides | − | + | High | None |
| MIF8 | E+ACT | 240 | sharonensis | − | + | Low | None |
| MIF9 | E+ACA | 270 | aegilopoides | − | + | High | None |
| MIF10 | E+ACC | 120 | aegilopoides | − | + | Low | None |
| MIF11 | E+ACC | 405 | aegilopoides | − | + | Low | None |
| MIF12 | E+ACC | 275 | aegilopoides | − | + | High | None |
The selective primer HM+4 (5′-CATGAGTCCTGCTCGGTCAA [HM+TCCA]) was used in combination with each of the four E+3 primers listed. The core sequence of EcoRI is identical to that used in the AFLP protocol by Vos et al. (1995).
+, methylation alteration; −, no alteration.
Similarity was considered significant for e values ≤ 0.005.
Sequence analysis of the seven fragments that showed methylation alteration in the allotetraploid (MIFs 6 to 12) showed no significant similarity to known sequences (Table 5). DNA gel blot analysis of these fragments showed that three of these seven fragments were high-copy number and the rest were low-copy number (Table 5). Methylation alteration in the allotetraploid was validated for all four low-copy fragments using DNA gel blot analysis. An example is shown in Figure 4A for MIF8 as a probe (Table 5). Furthermore, the four low-copy fragments were hybridized to membranes containing the same DNA samples digested with methylation-insensitive restriction enzymes. In these cases, there was no alteration in the DNA gel blot hybridization pattern. An example is shown in Figure 4B using MIF8 as a probe. Similarly, altered methylation in F1 and/or the allopolyploid was validated for all five low-copy probes tested. These results show perfect agreement between DNA gel blot analysis and methylation alteration in the allopolyploid detected by MSAP. The high-copy probes led to a smear on the autoradiogram, leaving us unable to assess methylation alteration (data not shown).
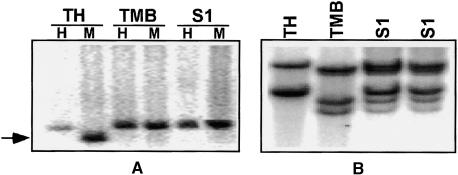
Figure 4.
DNA Gel Blot Analysis Using MIF8 Probe That Showed an Alteration in MSAP Pattern in the Allotetraploid (S1) of the Cross between Ae. sharonensis (TH) and T. monococcum ssp aegilopoides (TMB).
(A) Each DNA sample was digested with EcoRI+HpaII (H) and EcoRI+MspI (M), and the two digests were loaded on the gel side by side (HpaII digest on the left and MspI digest on the right). The arrow points to the fragment whose methylation is altered in the allotetraploid.
(B) DNA samples were digested with DraI (not sensitive to cytosine methylation) using the same hybridization probe. MIF8 was obtained using primer combination HM+TCAA/E+ACT (Table 5).
Note that using MSAP, we also recovered fragments that were not differentially methylated but that showed band disappearance in F1 and/or the allopolyploid, as was found using AFLP (Figures 1 and 2). DNA gel blot analysis of two such bands showed elimination in the allopolyploid using both membranes that included DNA samples digested by isoschizomers (Figure 5A) and membranes that included the same DNA samples digested with nonsensitive restriction enzymes (Figure 5B). These two bands were both low copy and showed no significant similarity to known sequences.
Figure 5.
DNA Gel Blot Analysis of a Band Isolated from MSAP That Did Not Show Alteration in Methylation but Rather Showed Band Disappearance.
The bands obtained by both isoschizomers in parent T. monococcum ssp aegilopoides (TMB) were eliminated in the first generation of the allotetraploid (S1). TH, Ae. sharonensis.
(A) Each DNA sample was digested with EcoRI+HpaII (H) and EcoRI+MspI (M), and the two digests were loaded on the gel side by side (HpaII digest on the left and MspI digest on the right).
(B) DNA samples were digested with EcoRV using the same hybridization probe. The probe was obtained using primer combination HM+TCAA/E+AAG. Arrows indicate the band of TMB that disappeared in the allotetraploid.
DISCUSSION
In previous works, we have shown that a limited set of loci could undergo rapid elimination in newly synthesized allopolyploids from the wheat group (Feldman et al., 1997; Liu et al., 1998b; Ozkan et al., 2001). These findings were based on the analysis of fragments that had the unexpected property of not being present in all genomes of polyploid wheat (chromosome-specific or genome-specific sequences). In this study, we have addressed the frequency of genetic and epigenetic changes in interspecific and intergeneric F1 hybrids and in newly synthesized allopolyploids and have sequenced some of the loci involved. This was done by analyzing a large and unbiased sample of wheat loci using the AFLP method (Vos et al., 1995) or the derived MSAP method (Xiong et al., 1999). The banding patterns obtained by both methods were found to be reproducible; moreover, AFLP or MSAP bands showing unexpected inheritance in the F1 hybrid or the allopolyploid were isolated, sequenced, and used as probes for DNA gel blot analysis. In all cases, the DNA gel blot data corroborated the AFLP or MSAP data, indicating that AFLP and MSAP provide reliable and high throughput methods to test for genetic and epigenetic changes, respectively.
The Observed Genetic and Epigenetic Alterations Are Not the Result of Heterozygosity
The unexpected inheritance of AFLP and MSAP bands in the F1 hybrids and the allopolyploids was not attributable to the heterozygosity of the parents or to PCR artifacts, as supported by the following evidence. The diploid parents of the allopolyploid were collected as single plants and selfed, and a single plant was used for each cross. The accessions used to produce the hybrids were checked by DNA gel blot for heterozygosity and were found to be homozygous at 50 loci (H. Ozkan, unpublished data). The independently made F1 and allopolyploids had similar AFLP and MSAP banding patterns. Bands that were present in F1 and disappeared in the allopolyploid could not disappear as a result of genetic segregation because chromosome doubling from dihaploid parents should produce homozygous lines. Moreover, band disappearance was not caused by chromosome loss because plants were checked cytologically and found to be euploid, as expected. Finally, the DNA gel blot validation of band elimination (Figures 2 and 5) did not provide evidence for heterozygosity. Therefore, the observed reproducibility in band elimination or methylation suggests that some sequences in the genome have unknown features that make them the target for sequence elimination or for methylation or demethylation in interspecific F1 hybrids and/or allopolyploids. These data are consistent with the findings of Ozkan et al. (2001), who analyzed a limited number of loci in 35 different interspecific and intergeneric hybrids and their derived amphiploids.
Frequency and Timing of Genetic and Epigenetic Alterations
The results shown in Table 1 indicate that a high fraction of the genome can be eliminated in newly synthesized allopolyploids and that the parental genomes are not affected equally. Note that DNA loss frequencies can be determined only for polymorphic bands, whereas the appearance of new bands, which was very rare, could be determined irrespective of the polymorphism of the parents. In cross A, 14% of the polymorphic bands (24 of 171) from Ae. sharonensis disappeared compared with only 0.5% (one of 202) for Ae. umbellulata. In cross B, there was less difference between the two parents, but overall the elimination rates were similar. The finding that one genome can eliminate up to 14% of its loci in a single generation (assuming that loss frequency is similar for monomorphic and polymorphic bands) shows that allopolyploidy leads not only to the establishment of new species in one step but also to the rapid evolution of the individual genomes in the allopolyploid background. Such rapid evolution could explain the elusive nature of the B genome of wheat (Talbert et al., 1995). These findings also suggest that genomes do not have a one-way ticket to obesity (Bennetzen and Kellogg, 1997) but that mechanisms exist for DNA elimination as discussed below. The timing of elimination was dependent on the genomic combination: most of the elimination occurred in the F1 hybrid of cross A, whereas in cross B, most of the elimination occurred after chromosome doubling. These data show that large-scale genomic rearrangements can occur extremely rapidly, perhaps as early as in the zygote of interspecific and intergeneric F1 plants. Indeed, when deletion occurred in F1 (Ozkan et al., 2001; this work), it was usually an all-or-nothing event, and there was no weak hybridization signal that could suggest the chimeric nature of the F1 tissues from which the genomic DNA was extracted. This suggests that elimination was a very early event. Note that there may be a difference between sequences that disappear in the F1 hybrid and those that disappear after chromosome doubling, as found by Ozkan et al. (2001) for chromosome-specific versus genome-specific sequences. Our work extends previous works on wide hybrids in cereals and other species showing that genomic rearrangements could occur, usually in repetitive DNA, and that new RFLP fragments could be observed (Lin et al., 1985; Lapitan et al., 1988; Svitashev et al., 1995).
Alterations in cytosine methylation also were associated with the hybrid and allopolyploid condition (Table 4, Figure 3). Few events (1.8%) were found whereby the methylation pattern was altered in F1 but was restored to the parental type in the allopolyploid. Most events of methylation pattern alteration had occurred in the F1 hybrid (6.9%) and were maintained in the allopolyploid or occurred only in the allopolyploid (4.4%). These results are higher than reported for methylation alterations in F1 of intraspecific crosses; in rice, Xiong et al. (1999) reported, using MSAP, that methylation patterns were altered for 4.1% of the sites analyzed in the F1 hybrids of crosses between cultivars. An interesting observation, which needs to be substantiated by a larger sample, is that 10 of 11 bands that showed methylation alteration in the F1 hybrid and the allopolyploid were from the genome of one parent (aegilopoides; Table 4), suggesting that there may be a difference between genomes in their ability to be modified (demethylated or hypermethylated) in the polyploid background. Methylation alterations in F1 involved mostly repetitive DNA (Table 5), with some examples from retrotransposons. Although we did not find examples of known genes, the methylation alterations found here might be related to the allopolyploidy-associated changes in methylation of coding sequences (Liu et al., 1998a) and to the gene silencing recently described for Arabidopsis using cDNA-AFLP (Comai, 2000). The demethylation of retrotransposons shown in this work (Table 5) might be consistent with transposon activation observed as the spread of dispersed repetitive DNA, including retroelements, to new genomes since polyploid formation in cotton (Zhao et al., 1998). Similarly, demethylation may affect DNA transposon activity, as was shown for several maize elements, such as Mutator, Activator, and Suppressor-mutator (Chandler and Walbot, 1986; Dennis et al., 1986; Banks et al., 1988). On the other hand, we did not find new AFLP bands corresponding to retroelements or DNA transposons (Table 2), suggesting that there was no large-scale occurrence of new transposon insertion in the F1 hybrid or the allopolyploid.
Underlying Mechanism of Rapid and Reproducible Allopolyploidy-Associated Sequence Elimination
Our finding that a significant fraction of the genome can undergo elimination in F1 and in newly synthesized allotetraploids suggests that both interspecific and intergeneric hybridity and allopolyploidy are shocks that trigger a rapid and massive genomic response in wheat. Surprisingly, this response was reproducible; that is, the same loci were always involved in the same patterns of sequence elimination (Table 2, Figure 5; see also Ozkan et al., 2001). We did not find evidence for a random mutator effect, for example, through point mutations, recombination, and transposable element activities. Such rearrangements would have caused new RFLPs or AFLP fragments (compared with the parents) to appear in the F1 hybrid and the allopolyploids. Elimination occurred rapidly and in a reproducible manner. A number of mechanisms could explain the type of elimination observed here: (1) gene conversion between the homoalleles (with rather long conversion tracts); (2) excision of the eliminated sequence and subsequent loss, via mechanisms such as transposon excision, V(D)J recombination, or site-specific recombination, as was shown in Tetrahymena (Godiska and Yao, 1990); and (3) crossing over between direct repeats that flank the eliminated sequence and loss of the excised circle. Gene conversion is the least likely mechanism because it would require very long conversion tracts and recombination between homologous chromosomes. Moreover, mapping of some eliminated sequences using nullisomic-tetrasomic lines (e.g., WPG90 used by Feldman et al., 1997) showed that these sequences were present in only one set of chromosomes and were absent in the homologs.
At this time, we have no support for or against the other mechanisms, and experiments are under way in our laboratory to determine the deletion or recombination breakpoints and thus gain more insight into the underlying elimination mechanism. In any case, this work shows that sequence elimination is the major and immediate response of the wheat genome to wide hybridization and allopolyploidy, that it can affect a large fraction of the genome (up to 14% of the polymorphic fragments), and that it is reproducible. In this respect, allopolyploidy somehow fits McClintock's definition of a genomic shock that “initiates a highly programmed sequence of events within the cell that serves to cushion the effects of the shock” (McClintock, 1984). Future studies may reveal whether the well-orchestrated and reproducible sequence elimination reported here is an example of preprogrammed adaptation of the genome to amphiploidy or a fortuitous response with no adaptive significance.
METHODS
Plant Material
The plant material for this study consists of three different combinations of newly synthesized allotetraploids, F1 hybrids, and their homozygous diploid parents. The first cross involved Aegilops sharonensis (genome SlSl) × Ae. umbellulata (genome UU), the second cross involved Ae. longissima (genome SlSl) × Triticum urartu (genome AA), and the third cross involved Ae. sharonensis (genome SlSl) × T. monococcum ssp aegilopoides (genome AmAm). The synthetic allotetraploids were obtained after colchicine treatment of the F1 plants. Chromosome number was determined in root tip mitosis of the F1 hybrids and the allotetraploids. The parental lines and the synthetic allopolyploids are maintained in the plant collection of our laboratory, and their production is described by Ozkan et al. (2001).
Amplified Fragment Length Polymorphism (AFLP) Analysis
The AFLP procedure was performed according to the protocol of Gibco BRL according to Vos et al. (1995), with minor modifications. Genomic DNA was extracted from young fresh leaves by the cetyl-trimethyl-ammonium bromide procedure (Kidwell and Osborn, 1992). One-half microgram of genomic DNA was digested at 37°C for 2 hr. The restriction-ligation reaction contained 12 units of MseI, 10 units of EcoRI, 1 unit of T4-DNA ligase, 5 pmol of EcoRI adaptor, 50 pmol of MseI adaptor, 0.5 M NaCl, 5 μg of BSA, and 10 × DNA ligase buffer (1 × DNA ligase buffer is 50 mM Tris-HCl, pH 7.5, 10 mM MgCl, 10 mM dithiothreitol, 1 mM ATP, 25 mg/ml bovine serum albumin) in a final volume of 10 μL. The digested-ligated DNA was diluted 1:10 with 90 μL of double-distilled water.
The adaptor sequences were as follows: MseI adaptors, 5′-TACTCAGGACTCAT-3′ and 5′-GACGATGAGTCCTGAG-3′; EcoRI adaptors, 5′-CTCGTAGACTGCGTACC-3′ and 5′-AATTGGTACGCAGTC- TAC-3′. Preselective amplification was performed with primers complementary to the core of the adaptor sequences; the EcoRI preselective primer was 5′-GACTGCGTACCAATTCA-3′, and the MseI preselective primer was 5′-GATGAGTCCTGAGTAAC-3′. The polymerase chain reaction (PCR) contained 50 ng of restricted-ligated DNA, 50 ng of EcoRI preselective primer, 50 ng of MseI preselective primer, 1 unit of Taq DNA polymerase, 2 μL of 10 × Taq DNA polymerase buffer (1 × Taq DNA polymerase buffer is 50 mM Tris HCl, pH 8.0, 100 mM NaCl, 0.1 mM EDTA, 1 mM DDT, 50% glycerol), 2 μL of 25 mM MgCl2, and 2.5 mM deoxynucleotide triphosphates in a final volume of 20 μL. The PCR conditions were 20 cycles of 30 sec at 94°C, 60 sec at 56°C, and 60 sec at 72°C. After preamplification, the PCR products were diluted 1:20 with 190 μL of double-distilled water.
Selective amplification was performed with the 32P-end-labeled EcoRI selective primers and the unlabeled MseI selective primers. The labeling reaction contained 50 ng of EcoRI selective primer, 100 μCi of γ-32P-ATP, 10 units of T4 polynucleotide kinase, and 10 μL of 5 × polynucleotide kinase buffer (1 × polynucleotide kinase buffer is 50 mM Tris HCl, 1 mM DDT, 0.1 mM EDTA, 50% glycerol [v/v], pH 7.5) in a final volume of 50 μL. Samples were incubated at 37°C for 1 hr and then heated to 70°C for 10 min. The sequence of the selective primers is similar to that of the preselective primers, with the addition of two variable nucleotides at the 3′ end. The AFLP primers are identified by the last three specific nucleotides of the selective primers, namely EcoRI+ANN primers and MseI+CNN primers.
The selective amplification reaction contained 50 ng of template DNA from the preselective amplification, 1 ng of labeled EcoRI selective primer, 5 ng of MseI selective primer, 1 unit of Taq DNA polymerase, 2 μL of Taq DNA polymerase buffer, 2 μL of 25 mM MgCl2, and 2.5 mM deoxynucleotide triphosphates in a final volume of 20 μL. The PCR cycles were one cycle of 2 min at 94°C, 30 sec at 65°C, and 2 min at 72°C, followed by 10 cycles each of annealing temperature of 1°C less than the former one, and 25 cycles of 1 sec at 94°C, 30 sec at 56°C, and 2 min at 72°C. The PCR products were mixed with 20 μL of dye (98% formamide, 10 mM EDTA, 0.1% bromphenol blue, and 0.1% xylene cyanol). The denatured PCR products were separated on 6% denaturing polyacrylamide (20:1 acrylamide:bisacrylamide, 7.5 M urea, and 1 × Tris-borate-EDTA buffer, pH 7.8) standard sequencing gel (43 cm in length) at 55 W for 1.5 hr. Gels were dried and exposed to x-ray film (Fuji Photo Film, Tokyo, Japan) for 6 to 12 hr at room temperature.
Methylation-Sensitive Amplification Polymorphism (MSAP) Analysis
The original protocol for detecting AFLP provided by Vos et al. (1995) was modified to incorporate the use of methylation-sensitive restriction enzymes. The modified protocol involved the use of the isoschizomers HpaII and MspI in combination with EcoRI. The adapter and the basic primer sequences for the EcoRI end were the same as those provided in the original protocol (Vos et al., 1995). A new double-strand fragment, referred to as the HpaII-MspI adaptor, was designed for each of the isoschizomer digestions using oligonucleotides 5′-GATCATGAGTCCTGCT-3′ and the complementary strand. The basic primer sequence for the HpaII-MspI digestions (HM+0) was designed accordingly as 5′-ATCATGAGTCCTGCTCGG-3′. Primers with three selective nucleotides for the EcoRI ends (E+3) and four selective nucleotides for the HpaII-MspI ends (HM+4) were designed as shown in Table 3. All of the adaptor and primer sequences were synthesized by the nucleic acid synthesis unit at the Weizmann Institute of Science.
Scoring of AFLP and MSAP bands
Each AFLP and MSAP gel was run twice. For the AFLP analysis, the replicate gel was run from the same DNA sample but from a different restriction-ligation-amplification reaction. For the MSAP analysis, the replicate gel was derived from an independent DNA extraction. Moreover, with both AFLP and MSAP gels, the upper part and the lower part of the gel, where resolution is not satisfactory, was not used for band scoring. The summary of the various types of bands shown in Tables 1, 3, and 4 is based only on the high-resolution, middle part of the gel, which was reproducible in all cases.
Cloning of Fragments Subjected to Genetic or Epigenetic Alterations and Validation by DNA Gel Blot Analysis
Fragments that showed evidence of polyploidy-associated alterations were excised from AFLP gels and reamplified using the following PCR conditions: 3 min at 94°C, 30 sec at 94°C, 1 min at 56°C, 1 min at 72°C, followed by 34 cycles. The AFLP-isolated fragments (AIFs) and the MSAP-isolated fragments (MIFs) were ligated into pGEM-T easy vector (Promega), transformed to Escherichia coli strain XL-1 Blue, and sequenced using universal T7 and/or T3 primers. The AIFs and the MIFs were labeled with phosphorus-32 and used as hybridization probes for DNA gel blot analysis. The procedure of DNA gel blot analysis followed essentially the method described by Liu et al. (1997), except that methylation analyses used either HpaII or MspI.
Sequence Analysis
Sequences obtained in this study were analyzed for similarity to known sequences in public databases using the BLAST package 2.0 on the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/BLAST/).
Acknowledgments
We thank an anonymous referee for constructive comments. This work was supported by a United States–Israel Binational Science Foundation fund grant. H.O. was supported by the Turkish Council of Higher Education and by a short-term European Molecular Biology Organization fellowship. K.K. and H.S. were supported by a fellowship from the Feinberg Graduate School for Ph.D. and M.Sc., respectively.
References
- Allard, R.W., Garcia, P., Saenzdemiera, L.E., and Delavega, M.P. (1993). Evolution of multilocus genetic-structure in Avena-Hirtula and Avena-Barbata. Genetics 135, 1125–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Banks, J.A., Masson, P., and Fedoroff, N. (1988). Molecular mechanisms in the developmental regulation of the maize Suppressor-mutator transposable element. Genes Dev. 2, 1364–1380. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., and Kellogg, E.A. (1997). Do plants have a one-way ticket to genomic obesity? Plant Cell 9, 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V.L., and Walbot, V. (1986). DNA modification of a maize transposable element correlates with loss of activity. Proc. Natl. Acad. Sci. USA 83, 1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai, L. (2000). Genetic and epigenetic interactions in allopolyploid plants. Plant Mol. Biol. 43, 387–399. [DOI] [PubMed] [Google Scholar]
- Dennis, E.S., Gerlach, W.L., and Peacock, W.J. (1986). Excision of the Ds controlling element from the Adh1 gene of maize. Maydica XXXI, 47–57.
- Feldman, M. (2001). The origin of cultivated wheat. In The Wheat Book, A. Bonjean and W. Angus, eds (Paris: Lavoisier Tech. & Doc), pp. 1–56.
- Feldman, M., Liu, B., Segal, G., Abbo, S., Levy, A.A., and Vega, J.M. (1997). Rapid elimination of low-copy DNA sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 147, 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiska, R., and Yao, M.C. (1990). A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell 61, 1237–1246. [DOI] [PubMed] [Google Scholar]
- Kidwell, K.K., and Osborn, T.C. (1992). Simple plant DNA isolation procedures. In Plant Genomes: Methods for Genetic and Physical Mapping, J.S. Beckmann and T.C. Osborn, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–13.
- Lapitan, N.L.V., Sears, R.G., and Gill, B.S. (1988). Amplification of repeated DNA sequences in wheat × rye hybrids regenerated from tissue culture. Theor. Appl. Genet. 56, 17–23. [DOI] [PubMed] [Google Scholar]
- Lin, S.S., Ho, T.D., and Harlan, J.R. (1985). Rapid amplification and fixation of new restriction sites in ribosomal DNA repeats in the derivatives of a cross between maize and Tripsacum dactyloides. Dev. Genet. 6, 101–112. [Google Scholar]
- Liu, B., Segal, G., Vega, J.M., Feldman, M., and Abbo, S. (1997). Isolation and characterization of chromosome-specific DNA sequences from a chromosome arm genomic library of common wheat. Plant J. 11, 959–965. [Google Scholar]
- Liu, B., Vega, J.M., Segal, G., Abbo, S., Rodova, H., and Feldman, M. (1998. a). Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. I. Changes in low-copy noncoding DNA sequences. Genome 41, 272–277. [DOI] [PubMed] [Google Scholar]
- Liu, B., Vega, J.M., and Feldman, M. (1998. b). Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41, 535–542. [DOI] [PubMed] [Google Scholar]
- Masterson, J. (1994). Stomatal size in fossil plants: Evidence for polyploidy in the majority of angiosperms. Science 264, 421–424. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., Scheid, O.M., and Matzke, A.J.M. (1999). Rapid structural and epigenetic changes in polyploid and aneuploid genomes. Bioessays 21, 761–767. [DOI] [PubMed] [Google Scholar]
- McClelland, M., Nelson, M., and Raschke, E. (1994). Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 22, 3640–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801. [DOI] [PubMed] [Google Scholar]
- Ozkan, H., Levy, A.A., and Feldman, M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops–Triticum) group. Plant Cell 13, 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakuma, T., Ogihara, Y., and Tsujimoto, H. (1995). Genome rearrangement of repetitive sequences in processes of hybridization and allopolyploidization in Triticinae. In 8th International Wheat Genetics Symposium, Z. Li and Z. Xin, eds (Beijing: China Agriculture Scientech Press), pp. 563–566.
- Scheid, O.M., Jakovleva, L., Afsar, K., Maluszynska, J., and Paszkowski, J. (1996). A change of ploidy can modify epigenetic silencing. Proc. Natl. Acad. Sci. USA 93, 7114–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, K.M., Lu, P., Tang, K.L., and Osborn, T.C. (1995). Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92, 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring, J. (1997). Vertebrate evolution by interspecific hybridization: Are we polyploid? FEBS Lett. 400, 2–8. [DOI] [PubMed] [Google Scholar]
- Stebbins, G.L. (1971). Chromosomal Evolution in Higher Plants. (New York: Addison-Wesley).
- Svitashev, S.K., Vershinin, A.V., Trunova, S.A., Pershina, L.A., and Shumny, V.K. (1995). Molecular analysis of the genomes of wide hybrids in cereals. Hereditas 122, 25–31. [Google Scholar]
- Talbert, L.E., Blake, N.K., Storlie, E.W., and Lavin, M. (1995). Variability in wheat based on low-copy DNA-sequence comparisons. Genome 38, 951–957. [DOI] [PubMed] [Google Scholar]
- Vos, P., Hogers, R., Bleeker, M., Reijans, M., Vandelee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M., and Zabeau, M. (1995). AFLP: A new technique for DNA-fingerprinting. Nucleic Acids Res. 23, 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J.F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42, 225–249. [PubMed] [Google Scholar]
- Wolfe, K.H., and Shields, D.C. (1997). Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713. [DOI] [PubMed] [Google Scholar]
- Xiong, L.Z., Xu, C.G., Maroof, M.A.S., and Zhang, Q.F. (1999). Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Gen. Genet. 261, 439–446. [DOI] [PubMed] [Google Scholar]
- Zhao, X.P., Si, Y., Hanson, R.E., Crane, C.F., Price, H.J., Stelly, D.M., Wendel, J.F., and Paterson, N.H. (1998). Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Res. 8, 479–492. [DOI] [PubMed] [Google Scholar]