Abstract
Peroxisomes are organelles that perform a wide range of metabolic functions in eukaryotic cells. However, their role in fungal pathogenesis is poorly understood. Here we report that ClaPEX6, an ortholog of PEX6, is required for the fungus Colletotrichum lagenarium to infect host plants. ClaPEX6 was identified in random insertional mutagenesis experiments aimed at elucidating genes involved in pathogenesis. Functional analysis, using a green fluorescent protein cassette containing the peroxisomal targeting signal1 (PTS1), revealed that import of PTS1-containing proteins is impaired in clapex6 mutants generated by targeted gene disruption. Failure of growth on fatty acids shows an inability of fatty acid β-oxidation in these mutants. These results indicate that disruption of ClaPEX6 impairs peroxisomal metabolism, even though clapex6 mutants show normal growth and conidiation in nutrient-rich conditions. The clapex6 mutants formed small appressoria with severely reduced melanization that failed to form infectious hyphae. These data indicate that peroxisomes are necessary for appressorium-mediated penetration of host plants. The addition of glucose increased the pathogenicity of clapex6 mutants, suggesting that the glucose metabolic pathway can compensate partially for peroxisomes in phytopathogenicity.
INTRODUCTION
Peroxisomes are single-membrane-bound organelles possessing multiple metabolic functions, including β-oxidation of fatty acids, glyoxylate metabolism, and metabolism of reactive oxygen species. They also are required for specific functions such as methanol assimilation in some yeast and penicillin biosynthesis in the filamentous fungus Penicillium chrysogenum (Müller et al., 1991; van den Bosch et al., 1992; Subramani, 1993). In plants, peroxisomes are known to differentiate into at least three different classes: glyoxysomes, leaf peroxisomes, and unspecialized peroxisomes (Beevers, 1979), and it has been shown that leaf peroxisomes contain some of the enzymes involved in photorespiration (Tolbert, 1982). Most peroxisomal matrix enzymes have been shown to contain one of two peroxisome targeting signals (PTSs) (Subramani, 1993). Extensive analysis of yeast mutants has identified >20 PEX genes whose products, peroxins, are required for peroxisome biogenesis (van der Klei and Veenhuis, 1997; Subramani, 1998). In humans, peroxisome biogenesis disorders, such as Zellweger syndrome, are a group of lethal inherited diseases (Lazarow and Moser, 1989). Human orthologs of the yeast PEX genes have been characterized as causative genes for peroxisome biogenesis disorders (Gould and Valle, 2000). Compared with studies of PEX genes in mammals and yeast, functional analysis of PEX genes in other organisms is limited to a few reports (Berteaux-Lecellier et al., 1995; Lin et al., 1999; Hayashi et al., 2000). There are no reports regarding the roles of PEX genes for fungal pathogens. Genetic and physiological studies have provided an increasing understanding of fungal infection mechanisms; however, it remains unclear whether peroxisomal functions are prerequisite for fungal phytopathogenicity.
Plant pathogenic fungi produce asexual spores called conidia for their reproduction. Conidia formed in lesions of infected plants are dispersed by wind or water splash. Once conidia land on aerial parts of the host plants, they start morphological development and secretion of the low-molecular-weight or enzymatic compounds required for infection. On the surface of host plants, conidia are exposed to limited nutrient conditions and require the use of storage compounds, such as lipids and carbohydrates, to infect their host plant. Many phytopathogens, including Colletotrichum species, which cause destructive anthracnose diseases on numerous crops and ornamental plants worldwide (Agrios, 1988), form a specific infection structure called an appressorium, which is pigmented by melanin. In Colletotrichum lagenarium, the causal agent of the cucumber anthracnose disease, melanization of appressoria was demonstrated to be essential for appressorium function (Kubo and Furusawa, 1991), and three melanin biosynthetic genes, PKS1, SCD1, and THR1, have been isolated and characterized (Takano et al., 1995, 1997b; Kubo et al., 1996; Perpetua et al., 1996).
In fungal phytopathogens, genes involved in pathogenicity have been identified by differential screening, by targeted deletion of candidate genes encoding a known physiological function, and by isolation of pathogenicity mutants. In Magnaporthe grisea, the causal agent of rice blast disease, darkly melanized appressoria similar to those of Colletotrichum species are formed. Analysis of signaling pathways and pathogenesis-related processes is being performed by many laboratories worldwide. Signaling pathways, cAMP, and mitogen-activated protein kinase (MAPK) pathways have been shown to play pivotal roles for fungal pathogenesis (Dean, 1997; Hamer et al., 1997; Adachi and Hamer, 1998; Thines et al., 2000). In C. lagenarium, the CMK1 MAPK gene has been shown to be required for germination, appressorium formation, and invasive growth in plant tissue (Takano et al., 2000). A recent report also demonstrated that disruption of the MAPK kinase gene affects germination and appressorium formation in a related fungus, C. gloeosporioides (Kim et al., 2000). Random insertional mutagenesis, including a restriction enzyme–mediated integration (REMI) method (Schiestl and Petes, 1991), has been used extensively for the isolation of pathogenicity genes in many phytopathogenic fungi (Lu et al., 1994; Dufresne et al., 1998; Sweigard et al., 1998; Tanaka et al., 1999; Urban et al., 1999). We have initiated insertional mutagenesis analysis of fungal pathogenicity in C. lagenarium.
In this study, we demonstrate that peroxisomal function is a prerequisite for fungal pathogenicity. We identify a novel insertional mutant of C. lagenarium generated by REMI, which is nonpathogenic to the host plant. Analysis of this mutant revealed that the loss of pathogenicity was caused by the disruption of a gene that exhibits significant homology with PEX6. Expression studies of green fluorescent protein (GFP)–containing PTS1 (for peroxisomal targeting signal1) showed that import of peroxisomal matrix proteins was impaired in pex6-deleted mutants of C. lagenarium. Also, the finding that the mutants grew poorly on fatty acid suggested a defect in fatty acid β-oxidation in peroxisomes. Further phenotypic analysis of the pex6 mutants demonstrated that peroxisomal metabolic function is essential for appressorium-mediated plant infection processes.
RESULTS
Isolation of ClaPEX6
REMI insertional mutagenesis was performed to isolate nonpathogenic mutants of C. lagenarium wild-type strain 104-T. Plasmid pCB1004 containing the hygromycin resistance gene (hph) with no homology with C. lagenarium DNA was used in REMI experiments. Isolated REMI transformants were tested for pathogenicity to host cucumber leaves. One mutant, X86, was identified that induced no lesions on cucumber leaves. DNA gel blot analysis revealed that a single copy plasmid was integrated into the genome of X86 (data not shown). A 6.8-kb HindIII fragment including both pCB1004 and the flanking genomic region was recovered from X86 by plasmid rescue. DNA gel blot analysis of the rescued genome fragment identified a restriction fragment length polymorphism between the wild-type and X86 genomic DNA digested with EcoRI (Figure 1, lanes 1 and 2). The wild-type 104-T contained a 7.0-kb EcoRI fragment, whereas X86 contained a 5.0-kb fragment. The rescued plasmid was introduced into the wild type to reproduce an insertional mutation of X86. Among 80 hygromycin-resistant transformants, six were nonpathogenic on cucumber (data not shown). DNA gel blot analysis demonstrated that the nonpathogenic transformants possessed the recombination event with the genomic region contained in the rescued plasmid. Recombination in the nonpathogenic transformants resulted in the disappearance of the 7.0-kb EcoRI fragment observed in the wild type (Figure 1, lanes 3 and 4), whereas transformants retaining pathogenicity maintained the 7.0-kb EcoRI fragment of the wild type (Figure 1, lanes 5 and 6). These results demonstrated that the pathogenicity defect in X86 was attributable to the insertional mutation to the rescued genomic region.
Figure 1.
DNA Gel Blot Analysis of the Nonpathogenic Mutant X86 and Strains Transformed with the Rescued Plasmid of X86.
Genomic DNA from the wild-type strain 104-T (lane 1), X86 (lane 2), and four transformants of the wild type with the rescued plasmid (lanes 3 to 6) was digested with EcoRI, separated on a 1% agarose gel, and blotted. The blot was probed with a 1075-bp HindIII-ApaI genome fragment in the rescued plasmid. A single 7.0-kb EcoRI fragment was detected in the wild-type strain (arrow). In strain X86, the 7.0-kb fragment detected in the wild type disappeared but a 5.0-kb fragment was observed. Among the transformants with the rescued plasmid, two transformants (lanes 3 and 4) that lost pathogenicity did not contain the 7.0-kb fragment but contained the 5.0-kb fragment, which was common with X86. On the other hand, the other two transformants (lanes 5 and 6) that showed pathogenicity maintained the 7.0-kb fragment.
The rescued genomic region was used to isolate genome clones from the cosmid library of C. lagenarium. Cosmid inserts (ApaI, 9 kb; HindIII, 5 kb; and PstI, 2.7 kb) were subcloned into pBluescript KS-II. Sequence analysis of the rescued fragment and subclones revealed the existence of a large open reading frame spanning the whole region of the rescued fragment. The open reading frame consists of three exons (181, 3367, and 619 bp) and two introns (55 and 60 bp). The introns were confirmed by sequencing of cDNAs amplified by reverse transcription–polymerase chain reaction (PCR) (data not shown). The deduced amino acid sequence revealed high similarity with the sequences of members of Pex6 involved in peroxisome biogenesis (Figure 2). We designated this gene ClaPEX6. The deduced amino acid sequence of ClaPEX6 has high homology with Pex6 proteins from P. chrysogenum (50% identity), Pichia pastoris (36%), Yarrowia lipolytica (36%), Hansenula polymorpha (34%), Saccharomyces cerevisiae (34%), and human (31%). Also, the putative Pex6 ortholog of Arabidopsis (accession number AAD25809) showed 27% identity with ClaPex6. ClaPex6 has two AAA cassettes, although the first AAA cassette is not highly conserved. Each AAA cassette consists of putative ATP binding motifs, as described by Walker et al. (1982): a Walker A motif (THRNIGKA and GPPGTGKT) and a Walker B motif (KHVE and DELD) (Prosite accession number PS00017). ClaPex6 also contains the AAA-protein family signature (VFVIGATNRPDLLDPALLR) (accession number PS00674). ClaPex6 showed high similarity to other Pex6 proteins in the C-terminal region, including conserved motifs, although homology with P. chrysogenum Pex6 was found through the entire sequence of ClaPex6 (Figure 2). The sequence of the rescued fragment revealed that the plasmid insertion into X86 occurred in a position of the ClaPEX6 gene corresponding to the C-terminal region (at amino acid 1178) (Figure 2).
Figure 2.
Predicted Amino Acid Sequence of the C. lagenarium ClaPEX6 Gene.
The deduced amino acid sequence of C. lagenarium ClaPEX6 (C.l.) was aligned with Pex6 proteins from P. chrysogenum (P.c.), and S. cerevisiae (S.c.) by using the CLUSTAL W program (Thompson et al., 1994). Similar residues are shown on gray backgrounds. Gaps introduced for alignment are indicated by dashes. The ClaPex6 sequence contains two Walker A and Walker B motifs and one AAA-protein family signature motif. The plasmid insertion point in ClaPEX6 of strain X86 is indicated by the arrowhead. The sequence of ClaPEX6 has been deposited in GenBank (accession number AF343063).
Disruption of ClaPEX6
Strain X86 was not pathogenic on cucumber, indicating that the ClaPEX6 gene is involved in fungal pathogenicity. However, the insertional event in X86 occurred in a region near the C terminus of ClaPex6. This implies the possibility that ClaPex6 would not lose its function completely in strain X86. Therefore, we generated a ClaPEX6 deletion mutant (clapex6Δ) by one-step gene replacement. A gene replacement vector, pGDPEX6, containing the hph gene and both the 5′ and 3′ flanking regions of ClaPEX6, was constructed (Figure 3A). The region deleted by homologous recombination contained 2.8 kb of the coding region of ClaPEX6, including the highly conserved domains mentioned above. pGDPEX6 was introduced into the wild-type strain 104-T, and transformants were selected on hygromycin-containing medium.
Figure 3.
Gene Disruption of ClaPEX6.
(A) ClaPEX6 locus and gene replacement vector. By homologous recombination through double crossing over, the 2.8-kb region of ClaPEX6 was replaced by a hph cassette. A, ApaI; B, BamHI; P, PstI.
(B) DNA gel blot analysis of ClaPEX6 gene replacement mutants. Genomic DNA was isolated from the wild-type strain 104-T (lane 1), gene replacement transformants DPE1, DPE8, and DPE9 (lanes 2 to 4), and ectopic transformants DPE4 and DPE7 (lanes 5 and 6). Genomic DNA was digested with BamHI. The blot was hybridized with a 1.8-kb PstI fragment of the ClaPEX6 locus, indicated by the gray bar in (A).
To screen for clapex6Δ strains, we first investigated their pathogenicity to host cucumber leaves. Conidial suspensions from transformants were spotted on detached cucumber leaves and incubated for 7 days. Of 30 transformants tested, 21 did not induce lesions (Figure 4). These nonpathogenic transformants formed very few lesions even after 2 weeks of incubation (data not shown). DNA gel blot analysis was performed with three nonpathogenic transformants (DPE1, DPE8, and DPE9) and two pathogenic transformants (DPE4 and DPE7) (Figure 3B). The wild-type isolate 104-T and the transformants DPE4 and DPE7 maintained a 6.9-kb BamHI fragment (Figure 3B, lanes 1, 5, and 6). DPE4 and DPE7 contained one or two additional bands, indicating ectopic integration. In contrast, the nonpathogenic transformants DPE1, DPE8, and DPE9 did not contain the 6.9-kb BamHI wild-type fragment, but they did contain a common 5.5-kb BamHI fragment, consistent with the length expected from a gene replacement event (Figure 3B, lanes 2 to 4). These results demonstrated that the ClaPEX6 gene was disrupted in the nonpathogenic strains DPE1, DPE8, and DPE9. We concluded that the ClaPEX6 gene was essential for the pathogenicity of C. lagenarium. The clapex6Δ strains grew efficiently on nutrient-rich medium, although the growth rate was slightly lower than that of the wild type. Strain DPE1 grew at average 2.87 ± 0.03 cm (diameter) on potato dextrose agar (PDA) plates for 7 days, whereas the wild type grew at 3.32 ± 0.33 cm. Mycelial colonies formed by the mutants on PDA were darkly melanized similar to the wild type (data not shown). The amount of conidia produced by the clapex6 mutants was similar to that produced by the wild type (data not shown). These results indicate that the ClaPEX6 gene is dispensable for both growth and conidiation on nutrient-rich medium.
Figure 4.
Pathogenicity Assay of clapex6Δ Mutants.
Conidial suspensions of tested strains were spotted on the right half of detached cucumber leaves. On the left half of the leaves, the wild-type strain was inoculated as a positive control.
(A) clapex6 deletion mutant DPE1.
(B) Ectopic transformant DPE4.
Appressoria Formed by clapex6 Mutants Are Defective in Host Penetration
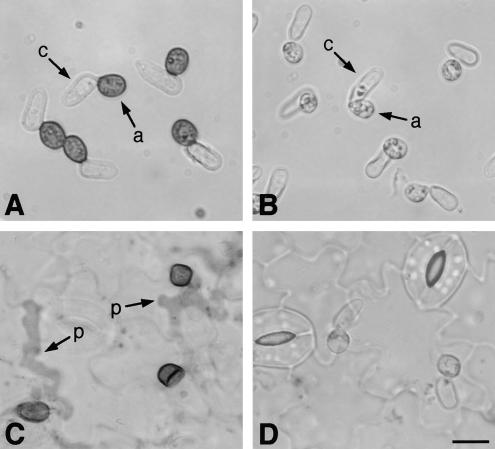
The clapex6Δ strains germinated effectively on a glass surface, and their germ tubes differentiated into swollen appressoria, indicating that the clapex6 mutants retain the ability to undergo the early steps for appressorium formation. However, appressoria formed by the clapex6 mutants were not identical to those of the wild type. Appressoria of the clapex6 mutants were smaller than those of the wild-type strain (Figures 5A and 5B, Table 1). Furthermore, melanization of appressoria in the clapex6 mutants was severely reduced (Figures 5A and 5B), although their mycelium was as darkly melanized as that of the wild type (data not shown). We investigated whether appressoria formed by clapex6 mutants are able to penetrate the host plant. Conidia of the wild-type 104-T formed darkly melanized appressoria on the host plant. Three to 4 days after inoculation, the appressoria formed penetration hyphae into the epidermal cells (Figure 5C). The clapex6 mutants germinated and formed appressoria effectively on the plant surface, but their appressoria were slightly smaller and showed a decrease in melanization, which was similar to the phenotype on a glass surface (Figures 5B and 5D). These phenotypes also were observed in the original mutant X86 produced by REMI mutagenesis (data not shown).
Figure 5.
Appressoria Formed by clapex6 Mutants and Their Function.
(A) and (B) Conidial suspensions of the wild type and clapex6 mutant, respectively, were spotted on glass slides and incubated for 12 hr. Appressoria of the clapex6 mutant are smaller and show severe reduction of melanization.
(C) and (D) Conidia were inoculated on cucumber cotyledons, incubated for 4 days, and stained with lactophenol aniline blue.
(A) and (C) show the wild-type strain 104-T; (B) and (D) show the clapex6 mutant DPE1. a, appressorium; c, conidium; p, penetration hyphae. Bar in (D) = 10 μm for (A) to (D).
Table 1.
Effect of Glucose and Scytalone on the C. lagenarium clapex6 Mutant
| Appressorium
|
Lesion Formationd (%) |
|||
|---|---|---|---|---|
| Strain | Compounda | Melanizationb | Sizec (μm) | |
| 104-T | Water | +++ | 6.91 ± 0.39 | 99.62 |
| DPE1 | Water | −/+ | 6.08 ± 0.56 | 1.01 |
| DPE1 | Glucose | ++ | 6.75 ± 0.38 | 21.20 |
| DPE1 | Scytalone | ++ | 6.13 ± 0.41 | 1.41 |
Conidia were incubated in water, 1 mM glucose solution, or 1 mM scytalone solution.
Appressoria of the clapex6 mutant were faintly melanized (−/+). Addition of glucose or scytalone restored melanization of the clapex6 mutant (++), although the degree of melanization did not reach the wild-type level (+++).
Diameters of appressoria were measured with a ×100 objective. Each value represents the average of >100 appressoria.
Conidial suspension was spotted on detached cucumber leaves and incubated for 7 days, and the number of lesions was counted. The percentage of lesion formation was based on the total number (>200) of spotted conidial suspensions.
Appressoria formed by the clapex6 mutants had the ability to penetrate an artificial substrate, cellulose membrane, suggesting that the mutants retain basic appressorium-mediated penetration ability (data not shown). However, appressoria of the clapex6 mutants did not form any penetration hyphae into the host plant, indicating that the mutants have a defect in appressorial penetration of plants (Figure 5D). To assess the ability of the clapex6 mutants to grow invasively inside the host plant cells independent of penetration, the mutants were inoculated through wounded sites of the plants. As a result, the clapex6 mutants formed lesions on the wounded leaves effectively (data not shown). This indicated that the clapex6 mutants retained the ability for invasive growth in the host plant. From these results, we concluded that the pathogenicity defect in clapex6 mutants was caused by the failure of penetration into the host plant, which resulted from formation of nonfunctional appressoria.
A Defect in the Transport of PTS1-Containing Proteins in clapex6 Mutants
Because the identified gene showed high similarity to PEX6 genes in other organisms, we focused on peroxisome biogenesis in C. lagenarium. First, electron microscopic analysis was performed to investigate the cell structure in conidia of the wild type (Figure 6). Conidia of the wild-type strain contained numerous lipid bodies that were electron translucent. Conidia had a nucleus, mitochondria, and vacuoles with heterogenous contents. Electron microscopic analysis also indicated that conidia of C. lagenarium contained peroxisomes that were distinguished from other organelles. The organelles regarded as peroxisomes were observed frequently at the peripheral region of conidia rather than at the central region (Figure 6). Peroxisomal proteins are synthesized in the cytosol and are imported post-translationally into peroxisomes. Peroxisome matrix proteins usually contain either PTS1 or PTS2 (Subramani, 1993). The PTS1 signal, which consists of the C-terminal tripeptide SKL or a derivative thereof, is present on the vast majority of these matrix proteins. The PTS2 signal is located near the N terminus. We assessed the import of peroxisome matrix proteins in clapex6 mutants by determining the intracellular localization of GFP (eGFP) fused to the PTS1 sequence (GFP-PTS1). Vector pBAGFPPTS1 containing GFP-PTS1 with the bialaphos-resistant (bar) gene was transformed into the wild-type 104-T and the clapex6 mutant DPE1. Bialaphos-resistant transformants were selected, and the subcellular sites of GFP-PTS1 fluorescence in transformants were investigated by fluorescence microscopy. We also generated both wild-type and mutant strains, with intact GFP as a control.
Figure 6.
Ultrastructure of Conidia of C. lagenarium.
Conidia of the wild-type strain of C. lagenarium were fixed with 5% KMnO4 and processed for electron microscopy. The arrowhead indicates the peroxisome. L, lipid body; M, mitochondrion; N, nucleus; V, vacuole. Bar = 1 μm.
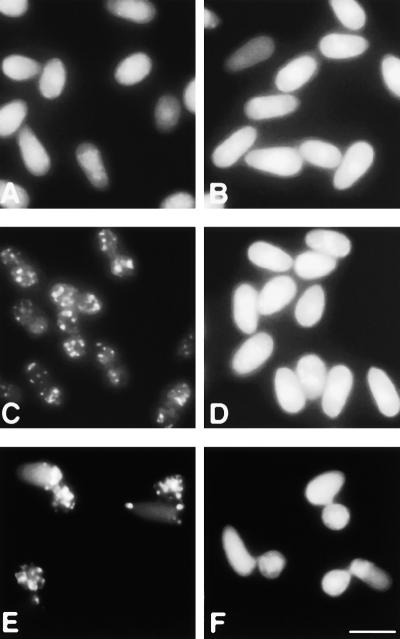
The transformants expressing GFP-PTS1 or GFP exhibited identical phenotypes in growth, conidiation, appressorium formation, and pathogenicity to those of corresponding parental strains (data not shown). Localization of GFP fluorescence was investigated in conidia of each strain by fluorescence microscopy (Figure 7). When GFP-PTS1 was expressed in the wild type, abundant punctate fluorescence was observed clearly, with no fluorescence being observed in the cytosol (Figure 7C). These fluorescent dots of GFP-PTS1 were observed at the peripheral region of conidia more frequently than at the central region, which appeared to be consistent with the results of electron microscopic analysis (Figures 6 and 7C). Conversely, wild-type strains expressing GFP showed diffuse fluorescence without a punctate pattern (Figure 7A). Occasionally, several spots were found in conidia showing diffuse fluorescence when GFP was highly expressed. These results suggest that GFP-PTS1 is recognized correctly and transported into the peroxisomes in the wild type of C. lagenarium. In contrast, when GFP-PTS1 was expressed in the clapex6 mutant, punctate fluorescence was not observed. The clapex6 mutant expressing GFP-PTS1 showed diffuse fluorescence, which was similar to the case of GFP expression in the mutant (Figures 7B and 7D). Similar results were obtained from observations of hyphal structure in each strain (data not shown). These results indicated that organelles visualized by GFP-PTS1 represent peroxisomes and that clapex6 mutants are defective in the import of peroxisome matrix proteins containing the PTS1 signal. We investigated the subcellular distribution of peroxisomes labeled by GFP-PTS1 during appressorium formation. First, conidia formed germ tubes, and germ tubes differentiated into swollen appressoria accompanied by the formation of vacuoles inside conidia. Growth and fusion of vacuoles resulted in the movement of cytoplasm from conidia to appressorial cells. At this stage, the majority of peroxisomes were localized in appressoria, not in conidia (Figure 7E). On the other hand, the clapex6 mutant showed diffuse fluorescence in both conidia and appressoria (Figure 7F).
Figure 7.
Intracellular Localization of the GFP-PTS1 Fusion Protein in the clapex6 Mutant.
(A) and (B) Green fluorescence of the intact GFP gene expressed in preincubated conidia of the wild type and the clapex6 mutant.
(C) and (D) Subcellular localization of GFP-PTS1 expressed in preincubated conidia.
(E) and (F) Subcellular localization of GFP-PTS1 in appressoria.
(A), (C), and (E) show the wild-type strain; (B), (D), and (F) show the clapex6 mutant. Bar in (F) = 10 μm for (A) to (F).
Roles of Peroxisomes for Appressorium-Mediated Infection
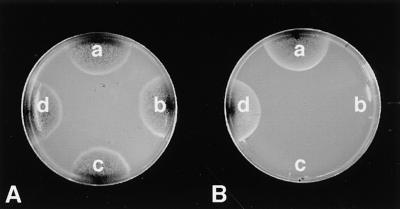
Peroxisome organelles contribute in various ways to cellular metabolism. The β-oxidation pathway for degradation of fatty acids to acetyl-CoA is one of the main metabolic pathways in peroxisomes. To investigate peroxisomal metabolic functions in clapex6 mutants, we assessed the β-oxidation pathway in the mutants by using a lipid-containing medium (Figure 8). It has been reported in several yeast species that most peroxisome-defective mutants are unable to grow on fatty acids (such as oleate) as sole carbon sources because of their defects in β-oxidation of fatty acids (Spong and Subramani, 1993; Nuttley et al., 1994). We first investigated the growth ability of the wild type on fatty acid medium containing Tween 80, of which oleic acid is the major component. The wild type was able to grow on this fatty acid medium. The growth rate of the wild type on the fatty acid medium was similar to that on glucose medium as a sole carbon source (Figure 8). This result indicates that the wild-type strain of C. lagenarium is able to use fatty acids by the β-oxidation pathway of fatty acids. In contrast to the wild type, the clapex6 mutants were completely defective in growth on medium containing Tween 80 (Figure 8B), whereas they showed growth similar to the wild type on glucose medium (Figure 8A). They did not form any mycelia on the Tween 80 medium even after extended incubation (20 days; data not shown). These results strongly suggested that clapex6 mutants have a defect in the fatty acid β-oxidation pathway in peroxisomes.
Figure 8.
Growth of clapex6 Mutants on Fatty Acids.
Growth ability of the wild type (104-T), the clapex6 mutants (DPE1 and DPE8), and the ectopic transformant (DPE4) was investigated on glucose medium and fatty acid medium as a sole carbon source. Fatty acid medium contains 0.5% Tween 80. The clapex6 mutants failed to grow on the fatty acid medium. Plate A contained glucose medium; plate B contained fatty acid medium. a, wild-type strain; b, DPE1; c, DPE8; d, DPE4.
Acetyl-CoA produced by β-oxidation is transported to the mitochondrion, and it replenishes intermediates of the citric acid cycle in S. cerevisiae (van Roermund et al., 1995, 1999). Because clapex6 mutants were defective in the β-oxidation pathway, we hypothesized that the pathogenicity defect in clapex6 mutants would be the result of the inability to supply citric acid cycle intermediates by β-oxidation in peroxisomes. The glycolytic pathway, which metabolizes glucose, supplies intermediates of the citric acid cycle independent of β-oxidation in peroxisomes. Therefore, we investigated appressorium formation of the clapex6 mutants in the presence of glucose. The wild-type strain formed normal appressoria in 1 mM glucose solution (data not shown). When conidia of the clapex6 mutant DPE1 were incubated in the glucose solution, they formed appressoria whose size and melanization increased compared with appressoria formed without glucose (Figures 9A and 9B, Table 1). On the other hand, scytalone, an intermediate product of the melanin biosynthesis pathway, restored appressorial melanization but not the size of appressoria in the clapex6 mutant (Figure 9C, Table 1).
Figure 9.
Partial Restoration of the Appressorium Phenotype in clapex6 Mutants by the Addition of Glucose and an Intermediate Product of Melanin.
Conidia of the clapex6 mutant were incubated for 12 hr at 24°C in water, glucose solution, or melanin intermediate (scytalone) solution.
(A) Water.
(B) 1 mM glucose.
(C) 1 mM scytalone.
Bar in (C) = 10 μm for (A) to (C).
The finding that glucose restored the phenotype of appressoria in the clapex6 mutant suggested that β-oxidation in peroxisomes of the wild-type strain supports the citric acid cycle during appressorium formation and contributes to the appressorium maturation steps. However, even in the glucose solution, the pigment intensity of melanin in the mutant appressoria still was lower than that of the wild type, suggesting that restoration by glucose was incomplete (Figures 5 and 9). To assess whether the addition of glucose can restore the pathogenicity of the clapex6 mutants, conidia of the wild type and the clapex6 mutant DPE1 suspended in water, 1 mM scytalone, or 1 mM glucose solution were tested for the ability to infect cucumber leaves (Table 1). The wild-type strain induced lesions efficiently when conidia were suspended in water, scytalone solution, or glucose solution (Table 1 and data not shown). DPE1 formed lesions only in 1% of inoculated spots with water or scytalone solution. In contrast, inoculated with glucose, DPE1 formed lesions in ∼20% of inoculated spots, indicating that glucose metabolism partially compensated for the roles of peroxisomes in appressorium-mediated plant infection (Table 1).
DISCUSSION
We identified the PEX6 ortholog gene (ClaPEX6) of the fungal pathogen C. lagenarium by molecular analysis of the nonpathogenic REMI mutant. The human PEX6 gene has been identified as a gene responsible for peroxisome biogenesis disorder of complementation group C (group 4 in the United States) (Tsukamoto et al., 1995). The C. lagenarium clapex6Δ strains generated by gene replacement showed growth and conidiation similar to those of the wild type on nutrient-rich medium. This demonstrates that C. lagenarium PEX6 is dispensable for growth and conidiation in nutrient-rich conditions. On the other hand, Kiel et al. (2000) suggested the possibility that PEX6 is necessary for cell viability in the filamentous fungus P. chrysogenum. In the original mutant X86, the plasmid was inserted into a position corresponding to the C-terminal region of ClaPex6. Sequencing of the rescued plasmid revealed that the inserted vector sequence produced an in-frame stop codon at 77 to 79 bp downstream from the insertion point in the ClaPEX6 gene of X86 (data not shown). This suggests that strain X86 would produce a truncated ClaPex6 protein that contains conserved domains, including two AAA cassettes and an AAA-protein signature. The finding that the phenotype of X86 is similar to that of clapex6 deletion mutants suggests an essential role of the C-terminal region of the Pex6 protein, although this region has no characterized motifs.
Analysis by GFP-PTS1 expression indicated that the import of PTS1-containing peroxisome matrix proteins into peroxisomes was impaired in clapex6 mutants. It has been demonstrated in several organisms that peroxisomal membrane proteins do not use the sorting machinery of peroxisome matrix proteins to reach their target organelle but follow alternative pathways (Baerends et al., 2000). In humans and several yeast species such as P. pastoris, H. polymorpha, S. cerevisiae, and Y. lipolytica, cells lacking Pex6p contain peroxisomal vesicle structures, although, in several other organisms, the size and number of peroxisomes in the mutants are different from those in the wild type (Spong and Subramani, 1993; Nuttley et al., 1994; Purdue and Lazarow, 1995; Yahraus et al., 1996; Titorenko and Rachubinski, 1998; Kiel et al., 1999). This indicates that pex6 mutants in these organisms retain the ability to import peroxisomal membrane proteins. In contrast, these mutants show severe reduction in the import of peroxisome matrix proteins, which is consistent with the phenotype of the C. lagenarium pex6 mutants. These findings suggest the possibility that the pex6 mutants of C. lagenarium contain peroxisomal vesicle structures like the pex6 mutants in other organisms.
Studies in human and P. pastoris have shown that PEX6 is required for the stability of the PTS1 receptor, indicating a direct role of PEX6 for matrix protein import (Dodt and Gould, 1996; Yahraus et al., 1996). In Y. lipolytica, of which pex6 mutants showed severe reduction of peroxisomes in both size and number, it has been reported that PEX6 is involved in peroxisome membrane biogenesis (Titorenko and Rachubinski, 1998; Titorenko et al., 2000). A detailed function of ClaPEX6 for peroxisome biogenesis in C. lagenarium remains to be elucidated. Fatty acid degradation by the β-oxidation pathway has been shown to be confined to peroxisomes in yeast and plants, whereas this metabolic pathway is present in both peroxisome and mitochondrion in mammalian cells (Beevers, 1982; Schulz, 1991; Kunau et al., 1995). Both yeast and plants are able to degrade fatty acids completely within their peroxisomes, whereas mammals require the mitochondrial β-oxidation system to oxidize long-chain fatty acids because the peroxisomal β-oxidation system seems to function only as a chain-shorting system of very-long-chain fatty acids. In addition, enzymatic studies in the filamentous fungus Aspergillus nidulans have suggested that β-oxidation can progress completely in peroxisomes and does not occur in mitochondria (Valenciano et al., 1996). We showed that the C. lagenarium pex6 mutants were able to grow on glucose but not on fatty acid (oleic acid is not a very-long-chain fatty acid), indicating that the mutants lacked the ability for fatty acid β-oxidation. This result strongly suggests that β-oxidation of fatty acid in C. lagenarium takes place exclusively in peroxisomes and not in mitochondria.
Peroxisomal Metabolism for Appressorium-Mediated Plant Infection
Appressoria formed by clapex6 mutants were smaller compared with the wild type and showed severe reduction in melanization. Melanization of appressoria is essential for appressorial penetration in C. lagenarium. In C. lagenarium, melanin is synthesized from the pentaketide pathway in which 1,8-dihydroxynaphthalene is the intermediate precursor of the polymer (Kubo and Furusawa, 1991). It was demonstrated recently that a 1,8-dihydroxynaphthalene–melanin intermediate can be synthesized in vitro from malonyl-CoA that is produced mainly from acetyl-CoA (Fujii et al., 2000). We believe that the reduction of appressorial melanization in clapex6 mutants is attributable to the lack of acetyl-CoA used for synthesis of malonyl-CoA; this belief is supported by the finding that the addition of acetyl-CoA partially complemented appressorial melanization in the clapex6 mutants (data not shown). This suggests that melanin biosynthesis in appressoria depends mainly on acetyl-CoA produced by fatty acid β-oxidation in peroxisomes. In contrast, clapex6 mutants formed darkly melanized colonies on nutrient-rich medium. We believe that metabolic functions of peroxisomes make no significant contribution to mycelial melanization on nutrient medium.
From studies in other organisms, including yeast, acetyl-CoA produced by fatty acid β-oxidation is believed to be transported from peroxisomes to mitochondria to replenish citric acid cycle intermediates in C. lagenarium. Glucose that can supply citric acid cycle intermediates via the glycolytic pathway restored the phenotype of appressoria in the clapex6 mutants. This suggests that part of the acetyl-CoA produced in peroxisomes during appressorium formation is transported to the mitochondrion for the citric acid cycle and that this contribution of peroxisomes is essential for the formation of functional appressoria under poor nutrient conditions, such as on the host plant surface. In the presence of the melanin intermediate scytalone, restoration of the phenotype was restricted to appressorial melanization. The melanized appressoria of the mutants with glucose showed partial restoration of pathogenicity, whereas those with scytalone showed no pathogenicity. We believe that appressorium-mediated penetration into plants requires expansion of the appressoria to a certain size as well as appressorial melanization. The pathogenicity of the clapex6 mutants did not attain wild-type levels even when inoculated with glucose, indicating that glucose cannot completely restore peroxisome functions for pathogenicity in clapex6 mutants. The incomplete restoration of pathogenicity in clapex6 mutants might be the result of a negative effect of fatty acid accumulation or of a defect in other peroxisomal metabolic functions, such as degradation of H2O2, which glucose would not be able to complement.
It is known that acetyl-CoA from β-oxidation of fatty acids can be used for the synthesis of glycerol through the gluconeogenesis pathway. In Magnaporthe and Colletotrichum species, melanin-pigmented appressoria generate enormous turgor, which is an important factor for penetration by appressoria (Howard et al., 1991; Bechinger et al., 1999). In M. grisea, turgor in appressoria was estimated to be generated by the accumulation of intracellular glycerol (de Jong et al., 1997). Intracellular glycerol concentration increases after the development of appressoria, indicating that glycerol synthesis for turgor production occurs inside appressoria of M. grisea (de Jong et al., 1997). Our GFP-PTS1 assay of C. lagenarium revealed that fully developed appressoria contained abundant peroxisomes. This finding suggests the possibility that β-oxidation in peroxisomes inside appressoria contributes to glycerol synthesis, although quantitative analysis of glycerol is necessary in the wild type and clapex6 mutants.
When conidia of fungal pathogens attach on host plant surfaces, they must be subjected to poor nutrient conditions. Under this environment, they have to precede morphological developments such as appressorium formation for infection. It is plausible, therefore, that conidia of fungal pathogens contain storage compounds (e.g., lipid bodies and glycogen) that would be used for infection-related morphological developments (Hawker and Madelin, 1976; Thines et al., 2000). In this report, ultrastructural analysis indicated that dormant conidia of C. lagenarium have abundant lipid bodies. Also, in M. grisea, the activity of triacylglycerol lipase, which generates glycerol and fatty acids from triacylglycerol, has been shown to be induced during appressorium formation and maturation (Thines et al., 2000), which suggests that abundant fatty acids are produced during this process. Our results present direct evidence that metabolic functions of peroxisomes, especially β-oxidation of fatty acids, are essential for appressorium-mediated infection in fungal pathogens. The specific role of PEX6 in the infection process also suggests that peroxisome metabolic pathways will present novel targets for fungal disease control.
METHODS
Fungal Strains, Media, and Fungal Transformation
Colletotrichum lagenarium (syn C. orbiculare) strain 104-T (stock culture of the laboratory of Plant Pathology, Kyoto University) was used as the wild-type strain. All C. lagenarium cultures were maintained on 3.9% (w/v) potato dextrose agar (PDA) medium (Difco, Detroit, MI) at 24°C in the dark. Fatty acid medium contained 1.6% yeast nitrogen base without amino acids (Difco), 1% NH4NO3, 0.5% (v/v) Tween 80, and 1.5% agar. pH was adjusted to 6.0 with Na2HPO4. Glucose medium contained 1.6% yeast nitrogen base without amino acids (Difco), 1% NH4NO3, 2% glucose, and 1.5% agar. Protoplast preparation and transformation of C. lagenarium were performed basically according to the method described previously (Kubo et al., 1991). In restriction enzyme–mediated integration (REMI) mutagenesis, vector pCB1004 containing the hph gene was transformed into fungal protoplasts with restriction enzymes. Strain X86 was identified from screening of transformants generated with 50 units of XhoI. Transformants with the hph gene or the bar gene were selected on regeneration medium containing 100 μg/mL hygromycin B (Wako Pure Chemicals, Osaka, Japan) and 25 μg/mL bialaphos (Meiji Seika Kaisha, Ltd., Tokyo, Japan), respectively.
Genomic DNA Gel Blot Analysis
Total genomic DNA of C. lagenarium strain 104-T was isolated from mycelia as described previously (Takano et al. 1997a). Gel electrophoresis and restriction enzyme digestion were performed using standard procedures (Sambrook et al., 1989). DNA probes were labeled with α-32P-dCTP (Amersham Pharmacia Biotech, Buckinghamshire, UK) with the BcaBEST labeling kit (Takara, Ohtsu, Japan) or labeled with digoxigenin-dUTP (Boehringer Mannheim) with the BcaBEST digoxigenin labeling kit (Takara). DNA gel blot analysis was performed as described previously (Takano et al., 1997a).
Isolation of ClaPEX6 by Plasmid Rescue and Sequence Analysis
Genomic DNA of strain X86 was isolated, digested with HindIII, and subjected to electrophoresis. On the basis of the result that DNA gel blot analysis using pCB1004 as a probe identified 6.8-kb HindIII fragment in X86 (data not shown), ∼6 to 8 kb of digested DNAs were isolated from loading gel, self-ligated, and transformed to Escherichia coli, which resulted in the generation of E. coli possessing a rescued plasmid. A 1.0-kb ApaI-HindIII genomic fragment from the rescued plasmid was used to isolate genomic clones containing ClaPEX6 from a C. lagenarium cosmid library. Three overlapping genomic clones containing ClaPEX6 were subcloned into pBSII(KS−). DNA sequence was determined using the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Warrington, UK) and an automated DNA sequencer (model 310; Applied Biosystems).
Plasmid Constructs
To construct the gene replacement vector pGDPEX6, the 1.8-kb PstI fragment containing the 5′ flanking region of ClaPEX6 was introduced into the PstI site of pCB1636 (Sweigard et al., 1997) containing the hph gene to produce plasmid pCB5PEX6. The 2.3-kb fragment containing the 3′ flanking region of ClaPEX6 was amplified by polymerase chain reaction (PCR) with primers PX6AAP (5′-CGCAAGGGGCCCATGGATTGACTGCCACT-3′) and PX6S3 (5′-AGCGACGCGATGCTCAA-3′). The primer PX6AAP contained a terminal ApaI site. PCR was performed with LA Taq polymerase with GC-rich buffer (Takara). The amplified fragment was digested with ApaI and introduced into the ApaI site of pCB5PEX6 to give the plasmid pGDPEX6. Plasmids pBAGFPPTS1 and pBAGFP were constructed using pCB1531 containing a bialaphos-resistant gene (bar) (Sweigard et al., 1997). Expression of both GFP-PTS1 and green fluorescent protein (GFP) genes was controlled by a 221-bp short promoter region of the melanin gene SCD1 because expression of GFP under the SCD1 short promoter resulted in constitutive GFP fluorescence at all fungal stages examined (Y. Takano, E. Oshiro, and T. Okuno, unpublished data). The SCD1 terminator was amplified by PCR with primers SD1THS (5′-GCCCAAGCTTAGCGGCGTGCT-3′) and SD1TCA (5′-CGGGATCGATCCGGGAGGCTGAATC-3′). SD1THS and SD1TCA contain a terminal HindIII site and ClaI site, respectively.
The amplified product was digested with HindIII and ClaI and introduced into pCB1531 to produce pBAT. The SCD1 promotor region was amplified with primers SD1PNS (5′-CAGGTTGCGGCCGCGTGTTTTGCGGCAGTCC-3′) and SD1PXBA (5′-CGGGTCTAGACTGATAGGTGGGATATT-3′). SD1PNS and SD1PXBA contain a ter-minal NotI site and XbaI site, respectively. The amplified product was digested with NotI and XbaI and introduced into pBAT to produce pBATP. The entire GFP (EGFP) open reading frame fragment with the PTS1 sequence (SKL) was amplified from pCB16EGFP (Y. Takano, E. Oshiro, and T. Okuno, unpublished data) with primers EGFPX (5′-GCCCTCTAGACAGACACAATGGTGAGCAAGGGCGAG-3′) and GFPPTS1B (5′-GGCGGATCCTTACAGCTTCGACTTGTACAGCTC-GTCCAT-3′) and introduced into the XbaI-BamHI site of pBATP, which resulted in pBAGFPPTS1. The intact GFP fragment was amplified with primers EGFPX and GFPSTOP (5′-GGCGGATCCTTACTT-GTACAGCTCGTCCAT-3′) and introduced into pBATP to produce pBAGFP. Each PCR amplification was performed with KOD-Plus DNA polymerase (Toyobo, Tsuruga, Japan) according to the manufacturer's instructions.
Pathogenicity Tests
Inoculation to cucumber leaves (Cucumis sativus) was performed as described previously (Takano et al., 1997a). In the case of inoculation through wounded sites, 20 μL of conidial suspension (5 × 105 conidia/mL) was spotted on the wounded sites produced by a 26 G1/2 needle. Inoculated leaves were placed in humid Petri dishes, and symptoms were observed after 7 days of incubation at 24°C.
Microscopy
For conidial gemination and appressorium formation, conidia were harvested from 7-day-old cultures on PDA medium. Ten microliters of conidial suspension (5 × 105 conidia/mL in 0.1% yeast extract solution) was placed on a glass slide (eight-well multitest slide; ICN Biomedicals, Aurora, OH) and incubated in a humid environment at 24°C in the dark. After 1 hr of incubation, 0.1% yeast extract solution was changed to distilled water followed by incubation for 11 hr (Y. Takano, E. Oshiro, and T. Okuno, unpublished data) and samples were observed on a microscope (Axioskop; Carl Zeiss, Jena, Germany). Images were captured with a chilled charge-coupled device camera (Argus 50; Hamamatsu Photonics, Hamamatsu, Japan). In the case of glucose and scytalone treatment, conidia were incubated with 1 mM glucose and 1 mM scytalone solution, respectively. For investigation of appressorium-mediated penetration, 30 μL of conidial suspension (5 × 105 conidia/mL) was spotted onto the lower epidermis of cucumber cotyledons. After 3 days of incubation in a humid environment, the epidermal layers were peeled off, stained with lactophenol aniline blue, and observed under light microscopy (Takano et al., 1997a). For observation of GFP fluorescence, cells were viewed on the microscope equipped with an epifluorescent optic. In the case of observation of dormant conidia, conidia collected from cultures were observed directly. For observation of appressoria, conidia were incubated on the glass slide for 6 hr as described above.
Electron Microscopy
Conidia from 7-day-old cultures on PDA medium were collected and suspended in 0.1 M phosphate buffer, pH 7.2 (0.1 M KH2PO4 and 0.1 M Na2HPO4) and fixed with 5% KMnO4 solution for 3 hr at room temperature. After fixation, conidia were washed five times in 0.1 M phosphate buffer. Conidia were collected by centrifugation and dehydrated gradually through an ethanol series and 100% propylene oxide. Propylene oxide was replaced with LR White Resin (London Resin Company Ltd., Berkshire, UK) and allowed to harden at 55°C for 2 days. Ultrathin sections were prepared using a diamond knife and examined with an electron microscope (H-7100 FA; Hitachi, Tokyo, Japan).
GenBank Accession Numbers
Data accession numbers for P. chrysogenum and S. cerevisiae Pex6p are AF233277 and P33760, respectively.
Acknowledgments
We thank Kazuyuki Mise for valuable suggestions and support during the course of this work. We acknowledge Ralph A. Dean and Thomas K. Mitchell for critical reading of the manuscript. We are grateful to Nobuyuki Fuchigami for providing bialaphos and Yasuyuki Kubo for providing scytalone. This work was supported in part by Grant 10760031 from the Ministry of Education, Science, Sports, and Culture of Japan and a Grant-in-Aid (JSPS-RFTF96L00603) from the Research for the Future program of the Japan Society for the Promotion of Science.
References
- Adachi, K., and Hamer, J.E. (1998). Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea. Plant Cell 10, 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrios, G.N. (1988). Plant Pathology, 3rd ed. (San Diego, CA: Academic Press).
- Baerends, R.J.S., Faber, K.N., Kiel, J.A.K.W., van der Klei, J., Harder, W., and Veenhuis, M. (2000). Sorting and function of peroxisomal membrane proteins. FEMS Microbiol. Rev. 24, 291–301. [DOI] [PubMed] [Google Scholar]
- Bechinger, C., Giebel, K.-F., Schnell, M., Leiderer, P., Deising, H.B., and Bastmeyer, M. (1999). Optical measurements of invasive forces exerted by appressoria of a plant pathogenic fungus. Science 285, 1896–1899. [DOI] [PubMed] [Google Scholar]
- Beevers, H. (1979). Microbodies in higher plants. Annu. Rev. Plant Physiol. 30, 159–193. [Google Scholar]
- Beevers, H. (1982). Glyoxysomes in higher plants. Ann. NY Acad. Sci. 386, 243–251. [Google Scholar]
- Berteaux-Lecellier, V., Picard, M., Thompson-Coffe, C., Zicker, D., Panvier-Adoutte, A., and Simonet, J.-M. (1995). A nonmammalian homolog of the PAF1 gene (Zellweger syndrome) discovered as a gene involved in caryogamy in the fungus Podospora anserina. Cell 81, 1043–1051. [DOI] [PubMed] [Google Scholar]
- Dean, R.A. (1997). Signal pathways and appressorium morphogenesis. Annu. Rev. Phytopathol. 35, 211–234. [DOI] [PubMed] [Google Scholar]
- de Jong, J.C., McCormack, B.J., Smirnoff, N., and Talbot, N.J. (1997). Glycerol generates turgor in rice blast. Nature 389, 244–245. [Google Scholar]
- Dodt, G., and Gould, S.J. (1996). Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: Evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135, 1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne, M., Bailey, J.A., Dron, M., and Langin, T. (1998). clk1, a serine/threonine protein kinase-encoding gene, is involved in pathogenicity of Colletotrichum lindemuthianum on common bean. Mol. Plant-Microbe Interact. 11, 99–108. [DOI] [PubMed] [Google Scholar]
- Fujii, I., Mori, Y., Watanabe, A., Kubo, Y., Tsuji, G., and Ebizuka, Y. (2000). Enzymatic synthesis of 1,3,6,8-tetrahydroxynaphthalene solely from malonyl coenzyme A by a fungal iterative type I polyketide synthetase PKS1. Biochemistry 39, 8853–8858. [DOI] [PubMed] [Google Scholar]
- Gould, S.J., and Valle, D. (2000). Peroxisome biogenesis disorders: Genetics and cell biology. Trends Genet. 16, 340–345. [DOI] [PubMed] [Google Scholar]
- Hamer, J.E., Xu, J.-R., Urban, M., Adachi, K., Lau, G., Tenjo, F., and Bhargava, T. (1997). Signal transduction and gene expression during early stage of fungal pathogenesis in the rice blast fungus. In Molecular Genetics of Host-Specific Toxins in Plant Disease, K. Kohmoto and O.C. Yoder, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 225–244.
- Hawker, L.W., and Madelin, M.F. (1976). The dormant spore. In The Fungal Spore, D.J. Weber and W.M. Hess, eds (New York: John Wiley), pp. 1–72.
- Hayashi, M., Nito, K., Toriyama-Kato, K., Kondo, M., Yamaya, T., and Nishimura, M. (2000). AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 19, 5701–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, R.J., Frerrari, M.A., Roach, D.H., and Money, N.P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA 88, 11281–11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel, J.A.K.W., Hilbrands, R.E., van der Klei, I.J., Rasmussen, S.W., Salomons, F., van der Heide, M., Faber, K.N., Cregg, J.M., and Veenhuis, M. (1999). Hansenula polymorpha Pex1p and Pex6p are peroxisome-associated AAA proteins that functionally and physically interact. Yeast 15, 1059–1078. [DOI] [PubMed] [Google Scholar]
- Kiel, J.A.K.W., Hilbrands, R.E., Bovenberg, R.A.L., and Veenhuis, M. (2000). Isolation of Penicillium chrysogenum PEX1 and PEX6 encoding AAA proteins involved in peroxisome biogenesis. Appl. Microbiol. Biotechnol. 54, 238–242. [DOI] [PubMed] [Google Scholar]
- Kim, Y.-K., Kawano, T., Li, D., and Kolattukudy, P.E. (2000). A mitogen-activated protein kinase kinase required for induction of cytokinesis and appressorium formation by host signals in the conidia of Colletotrichum gloeosporioides. Plant Cell 12, 1331–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, Y., and Furusawa, I. (1991). Melanin biosynthesis: Prerequisite for successful invasion of the plant host by appressoria of Colletotrichum and Pyricularia. In The Fungal Spore and Disease Initiation in Plants and Animals, G.T. Cole and H.C. Hoch, eds (New York: Plenum Publishing), pp. 205–217.
- Kubo, Y., Nakamura, H., Kobayashi, K., Okuno, T., and Furusawa, I. (1991). Cloning of a melanin biosynthetic gene essential for appressorial penetration of Colletotrichum lagenarium. Mol. Plant-Microbe Interact. 4, 440–445. [DOI] [PubMed] [Google Scholar]
- Kubo, Y., Takano, Y., Endo, N., Yasuda, N., Tajima, S., and Furusawa, I. (1996). Cloning and structural analysis of the melanin biosynthesis gene SCD1 encoding scytalone dehydratase in Colletotrichum lagenarium. Appl. Environ. Microbiol. 62, 4340–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunau, W.H., Dommes, V., and Schulz, H. (1995). Beta-oxidation of fatty acids in mitochondria, peroxisomes and bacteria: A century of continued progress. Prog. Lipid Res. 34, 267–342. [DOI] [PubMed] [Google Scholar]
- Lazarow, P.B., and Moser, H.W. (1989). Disorders in peroxisome biogenesis. In The Metabolic Bases of Inherited Disease, 6th ed, C.R. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, eds (New York: McGraw Hill), pp. 1479–1509.
- Lin, Y., Sun, L., Nguyen, L.V., Rachubinski, R.A., and Goodman, H.M. (1999). The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284, 328–330. [DOI] [PubMed] [Google Scholar]
- Lu, S., Lyngholm, L., Yang, G., Bronson, C., Yoder, O.C., and Turgeon, B.G. (1994). Tagged mutations at the Tox1 locus of Cochliobolus heterostrophus by restriction enzyme-mediated integration. Proc. Natl. Acad. Sci. USA 91, 12649–12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, W.H., Van der Krift, T.P., Krouwer, A.J.J., Wosten, H.A.B., Van der Voort, L.H.M., Smaal, E.B., and Verkleij, A.J. (1991). Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J. 10, 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttley, W.M., Brade, A.M., Eitzen, G.A., Veenhuis, M., Aitchison, J.D., Szilard, R.K., Glovert, J.R., and Rachubinski, R.A. (1994). PAY4, a gene required for peroxisome assembly in the yeast Yarrowia lipolytica, encodes a novel member of a family of putative ATPases. J. Biol. Chem. 269, 556–566. [PubMed] [Google Scholar]
- Perpetua, N.S., Kubo, Y., Yasuda, N., Takano, Y., and Furusawa, I. (1996). Cloning and characterization of a melanin biosynthetic THR1 reductase gene essential for appressorial penetration of Colletotrichum lagenarium. Mol. Plant-Microbe Interact. 9, 323–329. [DOI] [PubMed] [Google Scholar]
- Purdue, P.E., and Lazarow, P.B. (1995). Identification of peroxisomal membrane ghosts with an epitope-tagged integral membrane protein in yeast mutants lacking peroxisomes. Yeast 11, 1045–1060. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schiestl, R.H., and Petes, T.D. (1991). Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88, 7585–7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, H. (1991). Beta-oxidation of fatty acids. Biochim. Biophys. Acta 1081, 109–120. [DOI] [PubMed] [Google Scholar]
- Spong, A.P., and Subramani, S. (1993). Cloning and characterization of PAS5: A gene required for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J. Cell Biol. 123, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani, S. (1993). Protein import into peroxisomes and biogenesis of the organelle. Annu. Rev. Cell Biol. 9, 445–478. [DOI] [PubMed] [Google Scholar]
- Subramani, S. (1998). Components involved in peroxisome import, biogenesis, proliferation, turnover and movement. Physiol. Rev. 78, 171–188. [DOI] [PubMed] [Google Scholar]
- Sweigard, J., Chumly, F., Carrol, A., Farrall, L., and Valent, B. (1997). A series of vectors for fungal transformation. Fungal Genet. Newsl. 44, 52–55. [Google Scholar]
- Sweigard, J.A., Carroll, A.M., Farrall, L., Chumley, F.G., and Valent, B. (1998). Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol. Plant-Microbe Interact. 11, 402–412. [DOI] [PubMed] [Google Scholar]
- Takano, Y., Kubo, Y., Shimizu, K., Mise, K., Okuno, T., and Furusawa, I. (1995). Structural analysis of PKS1, a polyketide synthase gene involved in melanin biosynthesis in Colletotrichum lagenarium. Mol. Gen. Genet. 249, 162–167. [DOI] [PubMed] [Google Scholar]
- Takano, Y., Kubo, Y., Kawamura, C., Tsuge, T., and Furusawa, I. (1997. a). The Alternaria alternata melanin biosynthesis gene restores appressorial melanization and penetration of cellulose membranes in the melanin-deficient albino mutant of Colletotrichum lagenarium. Fungal Genet. Biol. 21, 131–140. [PubMed] [Google Scholar]
- Takano, Y., Kubo, Y., Kuroda, I., and Furusawa, I. (1997. b). Temporal transcriptional pattern of three melanin biosynthesis genes, PKS1, SCD1, and THR1, in appressorium-differentiating and nondifferentiating conidia of Colletotrichum lagenarium. Appl. Environ. Microbiol. 63, 351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, Y., Kikuchi, T., Kubo, Y., Hamer, J.E., Mise, K., and Furusawa, I. (2000). The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant-Microbe Interact. 13, 374–383. [DOI] [PubMed] [Google Scholar]
- Tanaka, A., Shiotani, H., Yamamoto, M., and Tsuge, T. (1999). Insertional mutagenesis and cloning of the genes required for biosynthesis of the host-specific AK-toxin in the Japanese pear pathotype of Alternaria alternata. Mol. Plant-Microbe Interact. 12, 691–702. [DOI] [PubMed] [Google Scholar]
- Thines, E., Weber, R.W.S., and Talbot, N.J. (2000). MAP kinase and protein kinase A–dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12, 1703–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko, V.I., and Rachubinski, R.A. (1998). Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol. Cell. Biol. 18, 2789–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko, V.I., Chan, H., and Rachubinski, R.A. (2000). Fusion of small peroxisomal vesicles in vitro reconstructs an early step in the in vivo multistep peroxisome assembly pathway of Yarrowia lipolytica. J. Cell Biol. 148, 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert, N.E. (1982). Leaf peroxisomes. Ann. NY Acad. Sci. 386, 254–268. [Google Scholar]
- Tsukamoto, T., Miura, S., Nakai, T., Yokota, S., Shimozawa, N., Suzuki, Y., Orii, T., Fujiki, Y., Sakai, F., Bogaki, A., Yasumo, H., and Osumi, T. (1995). Peroxisome assembly factor-2, a putative ATPase cloned by functional complementation on a peroxisome-deficient mammalian cell mutant. Nat. Genet. 11, 395–401. [DOI] [PubMed] [Google Scholar]
- Urban, M., Bhargava, T., and Hamer, J.E. (1999). An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 18, 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenciano, S., De Lucas, J.R., Pedregosa, A., Monistrol, I.F., and Laborda, F. (1996). Induction of β-oxidation enzymes and microbody proliferation in Aspergillus nidulans. Arch. Microbiol. 166, 336–341. [DOI] [PubMed] [Google Scholar]
- van den Bosch, H., Schutgens, R.B.H., Wanders, R.J.A., and Tager, J.M. (1992). Biochemistry of peroxisomes. Annu. Rev. Biochem. 61, 157–197. [DOI] [PubMed] [Google Scholar]
- van der Klei, I.J., and Veenhuis, M. (1997). Yeast peroxisomes: Function and biogenesis of a versatile cell organelle. Trends Microbiol. 5, 502–509. [DOI] [PubMed] [Google Scholar]
- van Roermund, C.W.T., Elgersma, Y., Singh, N., Wanders, R.J.A., and Tabak, H.F. (1995). The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J. 14, 3480–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund, C.W.T., Hettema, E.H., van den Berg, M., Tabak, H.F., and Wanders, R.J.A. (1999). Molecular characterization of carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2p. EMBO J. 18, 5843–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.E., Saraste, M., Runswick, M.J., and Gay, N.J. (1982). Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahraus, T., Braverman, N., Dodt, G., Kalish, J.E., Morrell, J.C., Moser, H.W., Valle, D., and Gould, S.J. (1996). The peroxisome biogenesis disorder group 4 gene, PXAAA1, encodes a cytoplasmic ATPase required for stability of the PTS1 receptor. EMBO J. 15, 2914–2923. [PMC free article] [PubMed] [Google Scholar]