Abstract
Starch is the major storage carbohydrate in higher plants and of considerable importance for the human diet and for numerous technical applications. In addition, starch can be accumulated transiently in chloroplasts as a temporary deposit of carbohydrates during ongoing photosynthesis. This transitory starch has to be mobilized during the subsequent dark period. Mutants defective in starch mobilization are characterized by high starch contents in leaves after prolonged periods of darkness and therefore are termed starch excess (sex) mutants. Here we describe the molecular characterization of the Arabidopsis sex1 mutant that has been proposed to be defective in the export of glucose resulting from hydrolytic starch breakdown. The mutated gene in sex1 was cloned using a map-based cloning approach. By complementation of the mutant, immunological analysis, and analysis of starch phosphorylation, we show that sex1 is defective in the Arabidopsis homolog of the R1 protein and not in the hexose transporter. We propose that the SEX1 protein (R1) functions as an overall regulator of starch mobilization by controlling the phosphate content of starch.
INTRODUCTION
During photosynthesis, a proportion of the photoassimilates are exported from the chloroplast stroma in the form of triose phosphates that serve as precursors for sucrose biosynthesis in the cytosol. A proportion also is retained within the chloroplast and stored transiently as starch. The accumulation of transitory starch in Arabidopsis is relatively constant throughout the light period and occurs concurrently with sucrose synthesis (Zeeman et al., 1998a). In the subsequent dark period, starch is mobilized and thereby provides a steady supply of carbon for export to sink organs and for energy metabolism (Zeeman and ap Rees, 1999). The alteration of net biosynthesis and net degradation of starch is reflected by the diurnal variation of leaf starch contents. Both starch synthesis and degradation are tightly controlled to adapt plant metabolism to changing environmental conditions.
Starch biosynthesis is controlled by the 3-phosphoglycerate (3-PGA)/Pi ratio, which increases when sucrose biosynthesis becomes limited. Because of the allosteric activation of ADP-glucose pyrophosphorylase (AGPase) by 3-PGA and its inhibition by Pi, starch biosynthesis approaches maximum rates when stromal 3-PGA/Pi ratios are high (Smith et al., 1997). This view has been reinforced by analysis of mutants defective in AGPase (Neuhaus and Stitt, 1990). On the contrary, our understanding of the mechanisms regulating starch breakdown is still fragmentary. Mutants that are unable to degrade starch provide a valuable tool for the study of regulatory mechanisms and the reaction pathways of starch breakdown.
Several Arabidopsis starch excess (sex) mutants (e.g., sex1, sex4, and dpe1) have been isolated by screening for plants that retain a high starch content even after prolonged periods of darkness (Caspar et al., 1991; Trethewey and ap Rees, 1994a; Eimert et al., 1995; Zeeman et al., 1998b; Critchley et al., 2001). sex4 is deficient in a distinct endoamylase that is involved in starch degradation (Zeeman et al., 1998a), and sex1 was proposed to be defective in the chloroplast envelope hexose transporter (Trethewey and ap Rees, 1994b). It was reasoned that a deficiency in hexose export from the chloroplast could cause an accumulation of the products of starch breakdown (e.g., glucose) within this organelle and thereby prevent efficient starch breakdown by an unknown mechanism. Recently, the putative chloroplast hexose transporter (pGlcT) was isolated and characterized at the molecular and biochemical levels (Weber et al., 2000). Attempts to complement the sex1 mutant phenotype by introducing a wild-type copy of the pGlcT gene failed, casting some doubt on the proposed cause of the observed mutant phenotype.
Here we report on the map-based cloning of the mutated gene in sex1 and demonstrate that sex1 is not defective in the plastidic hexose transporter but in a gene that encodes a protein with significant homology with the starch granule–bound protein R1 from potato (Lorberth et al., 1998). To investigate the role of the SEX1 (R1) protein further, we isolated additional Arabidopsis starch excess lines mutated at the SEX1 locus and studied the effect of these mutations on the amount and nature of the starch accumulated in the leaves.
RESULTS
Map-Based Cloning of SEX1
To study the role of SEX1, we isolated five new mutant alleles from different mutant populations and generated a segregating mapping population from a cross of sex1-1 (accession Columbia [Col-0]) to the Landsberg erecta wild type. By analysis of 713 F2 and F3 progeny showing the mutant phenotype with restriction fragment length polymorphism markers, the mutation could be mapped between markers g5957 and m241 on chromosome 1. These markers flank the bacterial artificial chromosome (BAC) clones F20B24 and T16B5. Inspection of the DNA sequence covered by these BAC clones for candidate genes revealed significant homology of a DNA region on BAC T16B5 with the starch granule–bound protein R1 from potato (Lorberth et al., 1998). A single sequence length polymorphism marker was generated for this region, and the mapping population was tested for the segregation pattern of the marker. No recombination events could be detected at this locus (see Methods).
Complementation of sex1 with the Wild-Type Gene
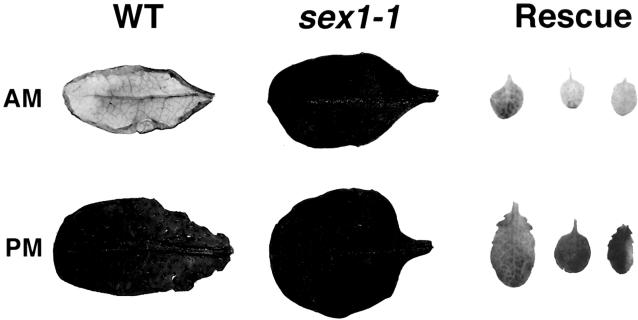
Using the single sequence length polymorphism marker as a probe, we screened an Arabidopsis genomic DNA phage library. We isolated a genomic DNA clone comprising the region homologous with the potato R1 gene. A SalI–XhoI fragment of this genomic DNA clone, including the SEX1 coding region flanked by 831 bp of 5′ and 1411 bp of 3′ noncoding region of the putative Arabidopsis SEX1 gene, was excised from the cloning vector and inserted into the binary vector pGREEN (Hellens et al., 2000). The construct was introduced into agrobacteria and then transformed into the mutant sex1-1. We obtained three independent kanamycin-resistant transgenic lines. The transformants were tested for their ability to degrade starch during a 12-hr dark period. Figure 1 shows that all transformants displayed a reversion of the starch excess phenotype, thus demonstrating that a defect in the SEX1 gene causes the starch excess phenotype of the Arabidopsis mutant sex1. The reversion of the starch excess phenotype was observed in plants of the same age and the same developmental stage compared with wild-type plants. The delay in development observed in sex1 compared with the wild type was not observed in the mutants complemented with the wild-type gene.
Figure 1.
Phenotype of the Mutant sex1-1 and Complementation of the Mutant with the Wild-Type (WT) Gene.
Leaves were harvested at the beginning (am) and the end (pm) of a 12-hr light period. After chlorophyll extraction with hot ethanol (80% [v/v]), starch was stained with iodine solution. The last column (complemented sex1-1) shows the rescue of the phenotype by transformation of sex1-1 with the wild-type gene.
Analysis of the Mutated Gene
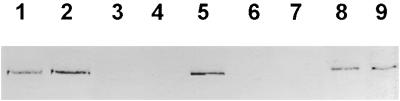
To further confirm the successful cloning of the SEX1 gene that codes for a protein of 1399 amino acid residues with a calculated molecular mass of 156.5 kD, we analyzed the site of mutation in three different sex1 mutant alleles. In sex1-1, a transition at position 3803 from G to A directs a substitution of a Gly residue to a Glu residue at amino acid position 1268. The deletion of two bases at positions 1824 and 1825 in the fast neutron–generated allele sex1-5 results in a truncated protein of 616 amino acid residues. Transition of G to A at position 3618 in the ethyl methanesulfonate–generated allele sex1-6 causes a premature stop resulting in a truncated protein of 1205 amino acid residues. The presence of the SEX1 protein in the mutant alleles sex1-1 to sex1-6 was analyzed using a polyclonal antiserum raised against the R1 protein from potato (Lorberth et al., 1998; Ritte et al., 2000a). As shown in Figure 2, SEX1 was not detectable in protein extracts from leaves of the mutant alleles sex1-2, sex1-3, sex1-5, and sex1-6. However, the amount of SEX1 protein was unaltered or reduced only slightly in sex1-1 and sex1-4. We were not able to detect significant amounts of the truncated proteins in the mutants sex1-5 and sex1-6, as predicted from the sequence analysis, suggesting that the truncated versions of the SEX1 protein are either less stable in vivo or more sensitive to proteolytic degradation during the isolation process.
Figure 2.
The SEX1 Protein Is Not Detectable in Leaf Extracts of sex1-2, sex1-3, sex1-5, and sex1-6.
Protein extracts of mature leaves from wild-type plants (Col-0, lane 1; RLD, lane 8), the different sex1 alleles (sex1-1, lane 2; sex1-2, lane 3; sex1-3, lane 4; sex1-4, lane 5; sex1-5, lane 6; sex1-6, lane 7), and potatoes (lane 9) were separated by SDS-PAGE and transferred to a membrane. The presence of SEX1 in the extracts was detected by an antiserum directed against R1 from potato.
Arabidopsis SEX1 shows 66.3% amino acid identity with the R1 protein from potato. A putative plastid-targeting sequence (amino acid residues 1 to 75) was detected by the program ChloroP (Emanuelsson et al., 1999), and the presence of processed SEX1 inside chloroplasts isolated from Arabidopsis leaves was demonstrated by protein gel blot analysis of total leaf and chloroplast proteins (data not shown).
We identified two other DNA regions in the Arabidopsis genome that displayed significant homology with the SEX1 gene. The first is located on chromosome 4 of Arabidopsis (hypothetical protein T22A6.280; accession number AL078637) and codes for a hypothetical protein of 1288 amino acid residues with a calculated molecular mass of 146.3 kD. It shows ∼50% amino acid identity with SEX1 but lacks a plastid-targeting signal (or any other detectable targeting signals) in the deduced amino acid sequence, suggesting that this gene encodes a cytosolic homolog of SEX1. One corresponding expressed sequence tag (EST) was isolated from a green silique library (accession number AV564246), suggesting a possible role of the gene in this particular tissue. The second homolog, located on chromosome 5 (hypothetical protein F9D12_19; accession number AF077407), encodes a hypothetical protein of 1186 amino acid residues with a calculated molecular mass of 130 kD. The deduced protein sequence shows ∼30% amino acid identity with SEX1. Analysis of the protein sequence with ChloroP revealed the presence of a putative plastid-targeting sequence. Thirteen corresponding ESTs were found in libraries from different tissues and developmental stages.
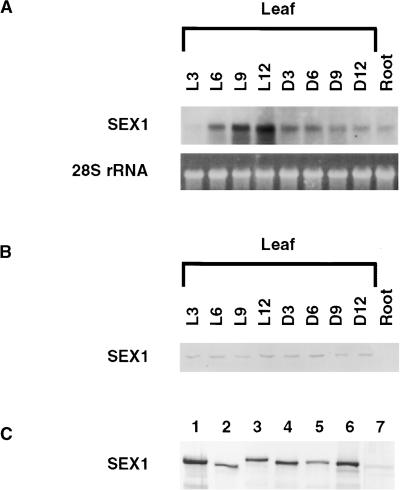
Figure 3A shows that the transcript level of the SEX1 mRNA displays a clear peak at the end of the light period. This finding is in agreement with those of Hammer et al. (2000), who observed that the SEX1 transcript was influenced by the circadian rhythms of the plant. To determine whether this fluctuation in transcript level translated to a change in the level of the SEX1 protein, we performed protein gel blotting on proteins extracted from leaves of Arabidopsis at intervals throughout a diurnal cycle. Figure 3B shows no difference in the amount of SEX1 protein observed, indicating that the diurnal variation of the SEX1 steady state transcript level is not represented at the protein level.
Figure 3.
SEX1 Can Be Detected in a Variety of Plant Species, and SEX1 Transcripts Show a Diurnal Cycle That Is Not Detected at the Protein Level.
(A) SEX1 transcript levels show a diurnal cycle with a peak at the end of the light period. Low expression of SEX1 in root extracts was detected. As a loading control, the amount of 28S rRNA is shown. L3 to L12 indicate hours in the light; D3 to D12 indicate hours in the dark.
(B) The amount of SEX1 in Arabidopsis leaves is constant throughout a diurnal cycle. No SEX1 protein was detected in root extracts.
(C) Protein gel blot analysis of extracts from different plant species: lane 1, potato; lane 2, Arabidopsis; lane 3, spinach; lane 4, fava bean; lane 5, barley; lane 6, maize; lane 7, S. aulata. Lanes 1 to 6 were loaded with 50 μg of total leaf protein, and lane 7 was loaded with 200 μg of a total protein extract of the unicellular green alga.
We also tested the presence of the SEX1 protein in variety of higher plants and in a unicellular green alga. Using the polyclonal antiserum raised against R1 from potato, we demonstrated the presence of SEX1, in addition to potato and Arabidopsis, in leaf extracts from spinach, broad bean, barley, and maize as well as in extracts from the unicellular green alga Sphaerellopsis aulata (Figure 3C).
Starch Content and the Degree of Starch Phosphorylation in sex1 and the Wild Type
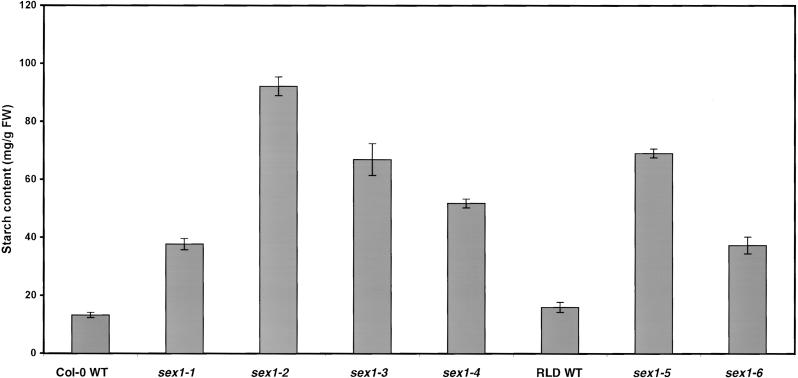
We measured the starch content of each of the mutant alleles of sex1 and the respective wild-type lines. The average content of starch in the different alleles of sex1 was three to seven times higher than in the wild type (Figure 4). The growth rate of the mutant plants was inversely correlated with the starch content. The highest rate of growth was observed in the wild-type lines, whereas the slowest growth was observed in sex1-2, sex1-3, and sex1-5 (data not shown).
Figure 4.
Starch Content of the Leaves of the Mutant Lines sex1-1 to sex1-6 and the Respective Wild-Type (WT) Lines.
All of the leaves from individual 6-week-old plants were harvested and boiled in ethanol. Starch in the ethanol-insoluble fraction was measured after enzymic digestion to glucose. Each value is the mean of four replicate samples ±se. FW, fresh weight.
In potato, the R1 protein, which is equivalent to SEX1, has been shown to influence the degree of starch phosphorylation (Lorberth et al., 1998). Antisense repression of the R1 transcript in potato led to a strong reduction of the amount of phosphate that was covalently bound to a small portion of the glucose monomers within amylopectin. It was demonstrated previously that most of the phosphate (60 to 70%) of potato tuber starch is bound to the C-6 position of glucose monomers within amylopectin, with the remainder bound to the C-3 position (Tabata and Hizukuri, 1971). Therefore, we analyzed the glucose 6-phosphate (Glc6P) content of starch isolated from the wild type and the mutant alleles sex1-1 to sex1-6 (Table 1). In sex1-2, sex1-3, and sex1-5, the Glc6P content was at or below the detection limit, whereas in sex1-1 and sex1-4, the Glc6P content of starch was approximately one-third of the wild-type level. In sex1-6, the amount of Glc6P was very close to the detection limit; therefore, we cannot definitely exclude a low residual Glc6P content of starch isolated from sex1-6.
Table 1.
Comparison of Glc6P Contents of Starch Isolated from Wild-Type Arabidopsis plants and from the Mutant Alleles sex1-1 to sex1-6
| Line | Phosphate Content (nmol Glc6P/μmol glucose) |
|---|---|
| Col-0 | 0.595 ± 0.200 |
| RLD | 0.471 ± 0.034 |
| sex1-1 | 0.162 ± 0.054 |
| sex1-2 | 0.032 ± 0.038 |
| sex1-3 | 0.027 ± 0.030 |
| sex1-4 | 0.155 ± 0.057 |
| sex1-5 | 0.003 ± 0.003 |
| sex1-6 | 0.044 ± 0.022 |
| Rescue | 0.481 ± 0.061 |
Starch was extracted from leaves that had been harvested at the end of the light period. Starch was hydrolyzed with acid, and Glc6P was determined by a coupled optical-enzymatic test. Numbers indicate ±se.
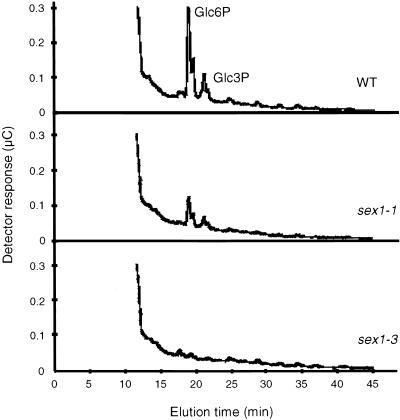
Glucose 3-phosphate (Glc3P) is not detectable enzymatically, although its amount (and that of Glc6P as well) can be estimated using high performance anion exchange chromatography (HPAEC) with pulsed amperometric detection (Blennow et al., 1998; Ritte et al., 2000b). HPAEC analysis of the hydrolyzed starches revealed reduction of the Glc6P content in sex1-1 to approximately one-fourth of the wild-type level (corresponding to the results obtained enzymatically; see Table 1), and the level of Glc3P was decreased to approximately the same extent. In starch from sex1-3 (a null allele lacking the SEX1 protein), neither Glc6P nor Glc3P was detectable (Figure 5). It seems likely that SEX1 is essential for the phosphorylation of both the C-6 and C-3 positions of the glucose moieties in starch and that the mutated versions of the SEX1 protein in the alleles sex1-1 and sex1-4 still have residual starch phosphorylating activity. However, the reduction of starch phosphate content by 70% caused by reduced SEX1 activity in these alleles apparently is sufficient to produce a starch excess phenotype. As shown in Table 1, in addition to rescuing the starch excess phenotype, transformation of sex1-1 with the wild-type SEX1 gene, under the control of its own promoter, restored the degree of starch phosphorylation to the level observed in the wild type.
Figure 5.
HPAEC Analysis with Pulsed Amperometric Detection of Hydrolyzed Starches Prepared from Leaves of Wild-Type (WT) Arabidopsis Plants and the Mutant Lines sex1-1 and sex1-3.
The analyzed samples are equivalent to 6 mg of starch. Glc6P and Glc3P were identified using the authentic sugar phosphates as standards (data not shown).
Starch Composition and Structure in sex1 and the Wild Type
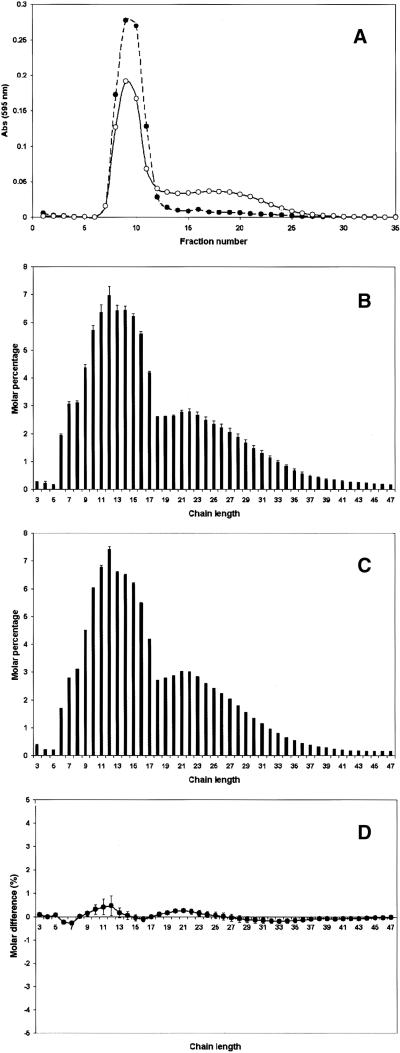
We analyzed starch isolated from wild-type and sex1-1 leaves to determine whether there were any changes in the composition (amylose-to-amylopectin ratio) and structure (distribution of chain lengths in amylopectin) of the starch. Using size exclusion chromatography, we found that sex1-1 starch had an appreciably higher amylose content compared with the wild-type starch (Figure 6A). We used fluorophore-assisted PAGE to analyze the distribution of chain lengths in the amylopectin fraction of the starch. The chain length distributions in the wild type and mutant were very similar (Figures 6B and 6C). A difference plot revealed a trend toward a slight increase in the number of short chains in sex1-1 starch (degree of polymerization 10 to 24) and a decrease in longer chains (degree of polymerization 31 to 46) compared with the wild type (Figure 6D); however, these changes were not statistically significant.
Figure 6.
Analysis of the Composition and Structure of Starch from sex1-1 and the Wild Type.
(A) Separation of amylose and amylopectin from wild-type (closed circles) and sex1-1 (open circles) starch by Sepharose CL2B chromatography. Starch (0.5 mg) was dissolved in 0.5 M NaOH, applied to a 5-mL column, and eluted with 0.1 M NaOH. The absorbance (Abs) of the fractions after the addition of an iodine solution was measured at 595 nm.
(B) Analysis of the chain length distribution of amylopectin from wild type using fluorophore-assisted PAGE. Starch samples were debranched with isoamylase, derivatized with the fluorophore 8-amino-1,3,6-pyrenetrisulphonic acid, and subjected to gel electrophoresis in an Applied Biosystems DNA sequencer. Data were analyzed using GeneScan software. Peak areas of chains between 3 and 47 glucose residues in length were summed, and the areas of individual peaks are expressed as percentages of the total. The values are means ±se of three independent replicate samples.
(C) Analysis of the chain length distribution of amylopectin from sex1-1, as described in (B).
(D) Percentage molar difference plot derived by subtracting the molar percentages of wild-type amylopectin chain lengths from those of sex1-1 (B). The se values were added together.
SEX1 Contains a Sequence Motif That Is Homologous with the Nucleotide Binding Site of Dikinases
To determine the putative functions of the SEX1 protein, a BLASTP search (Altschul et al., 1997) was performed, and the amino acid sequence was screened for known motifs. In a previous report, homology of the C-terminal region of the R1 protein with a bacterial phosphoenolpyruvate (PEP) synthetase (PPS; pyruvate:water dikinase; EC 2.7.9.2) was found (Lorberth et al., 1998), suggesting that R1 might have enzymatic activity. An extended search for motifs revealed significant homology (28% identity, 46% similarity) of the C-terminal 300 amino acid residues of SEX1 with the N-terminal region (residues 4 to 350) of PPS. In addition, a search of the SEX1 protein sequence against the PROSITE (Hofmann et al., 1999) and Pfam (Bateman et al., 1999) protein motif databases detected significant homology (expected value of 0.0026) of the SEX1 C-terminal region (residues 1079 to 1398) with the N-terminal Pfam-A domain PPDK_N_term of pyruvate:phosphate dikinase (PPDK; EC 2.7.9.1), which converts pyruvate to PEP. This motif also can be found in N-terminal regions of PPS proteins.
The N-terminal 400 amino acid residues of both PPDK and PPS contain the nucleotide binding domains of these enzymes (Carroll et al., 1994; Herzberg et al., 1996). An alignment of the SEX1 C-terminal region with the N-terminal regions of PPDKs from higher plants and bacteria and PPSs from bacteria identified several amino acid residues that are well conserved in all members of these protein families (Figure 7). Interestingly, these residues have been localized to the active site of the nucleotide binding region of PPDK from Clostridium symbiosum (Herzberg et al., 1996; McGuire et al., 1998; see Discussion for details), the only member of these protein families for which crystal structure data are available (Herzberg et al., 1996). The PEP/pyruvate binding regions of both PPDK and PPS, which are located on the C-terminal domains of these enzymes, however, did not show any homology with the SEX1 protein.
Figure 7.
Amino Acid Alignment of Phosphoenol Pyruvate Synthase (Pyruvate: Water Dikinase), Pyruvate: Phosphate Dikinase, and SEX1.
Identical amino acid residues are boxed, and similar residues are shaded. The alignment shows the N-terminal nucleotide binding domains of PPDK and PPS and the C-terminal putative nucleotide binding domain of SEX1. PPSA SYNY3, Phosphoenol Pyruvate Synthase A, Synechocystis spec. PCC 6803; PPSA PYRFU, Phosphoenol Pyruvate Synthase A, Pyrococcus furiosus; PPSA ECOLI, Phosphoenol Pyruvate Synthase A, Escherichia coli; PODK Ath, Pyruvate: Phosphate Dikinase, A. Thaliana.
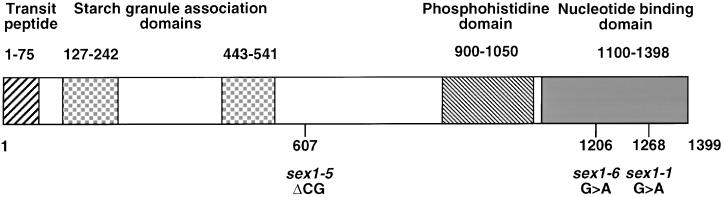
In the N-terminal domain of the mature SEX1 protein, we detected two regions of homology (23% identity, 41% similarity; amino acid residues 127 to 242 and 443 to 541) with the N-terminal region (amino acid residues 196 to 370) of a putative plastidic α-amylase (T17F3.14, chromosome 1, accession number AC010675) from Arabidopsis (T.-S. Yu and J. Chen, unpublished data) that might represent the starch binding domain and/or the phosphorylation domain of SEX1 (Figure 8). This domain of the putative plastidic α-amylase, however, is not present in other amylases from bacteria, animals, fungi, or plants.
Figure 8.
Protein Domains of the SEX1 Protein.
SEX1 carries a leader peptide that directs the precursor protein to the plastid stroma. The positions of the putative starch binding domains and of the starch phosphorylation domain are indicated. The mutations in different mutant alleles of SEX1 are shown at bottom.
DISCUSSION
Using a positional cloning approach, we identified the mutated gene in the Arabidopsis starch excess mutant sex1. The starch excess phenotype of sex1 could be complemented by transformation of the mutant with a wild-type copy of the SEX1 gene. In four alleles of sex1, the absence of the SEX1 protein was demonstrated by protein gel blot analysis. The deduced SEX1 amino acid sequence shows significant homology with that of the R1 protein from potato that was shown recently to influence the degree of starch phosphorylation in leaves and tubers of potato and, when expressed in Escherichia coli, in bacterial glycogen as well (Lorberth et al., 1998). As proposed earlier for potato, our results clearly demonstrate the crucial role of SEX1 (R1) in determining the phosphate content of starch, which appears to be tightly linked to the degradability of starch. Semicrystalline starch is hydrophobic, and it may be difficult for hydrolytic enzymes to act on intact starch granules. It can be assumed that phosphorylation changes the surface charge distribution and increases the hydrophilicity and hydration of starch and that only when phosphorylated might starch (except cereal endosperm starch) be accessible to degrading enzymes in vivo.
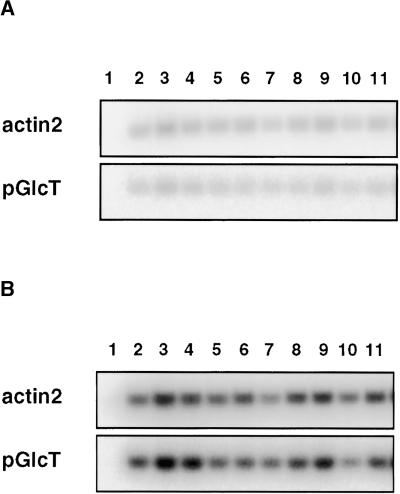
In contrast to an earlier report (Trethewey and ap Rees, 1994b), our results demonstrate that the primary lesion in sex1 is not in the plastidic hexose transporter but in the SEX1 (R1) protein, which was described only recently (Lorberth et al., 1998). Using quantitative reverse transcriptase–mediated polymerase chain reaction, we found no significant differences in the transcript levels of the putative plastidic glucose translocator (Weber et al., 2000) between wild-type plants and the different alleles of sex1 (Figure 9). Accordingly, the putative plastidic glucose transporter is not deregulated transcriptionally in these mutants. However, other possible pleiotropic effects of sex1 on the activity of this transporter cannot be excluded. It is conceivable that the transporter is regulated post-transcriptionally by R1 or by downstream factors of the starch degradation pathway. If so, this would have wide-reaching implications for signal transduction pathways between the chloroplast and the cytosol.
Figure 9.
Quantitative RT-PCR Analysis of the Expression of the Plastidic Hexose Transporter in Different sex1 Alleles and in Wild-Type Plants.
(A) DNA gel blot of RT-PCR products obtained after 16 cycles of PCR amplification.
(B) DNA gel blot of RT-PCR products obtained after 21 cycles of PCR amplification.
Total RNA was isolated at two different times (end of a 12-hr-light period [pm] and end of a 12-hr-dark period [am]) from the accessions Col-0 and RLD and from the mutants alleles sex1-1, sex1-5, and sex1-6. The RNA was reverse transcribed into cDNA, and the cDNAs encoding the putative plastidic hexose transporter (pGlcT) and actin2 (control) were amplified by specific oligonucleotide primers. After size separation on agarose gels, the PCR products were blotted onto nylon membranes and the membranes were hybridized to specific probes for pGlcT and actin2. Lane 1, water control; lane 2, Col-0, am; lane 3, sex1-1, am; lane 4, sex1-6, am; lane 5, RLD, am; lane 6, sex1-5, am; lane 7, Col-0, pm; lane 8, sex1-1, pm; lane 9, sex1-6, pm; lane 10, RLD pm; lane 11, sex1-5, pm.
The absence of detectable phosphate in the mutant lines lacking SEX1 suggests strongly that this protein plays a crucial role in determining the phosphate content of starch during starch synthesis, as proposed previously by Lorberth et al. (1998). In the mutant lines in which the SEX1 protein was detectable, the phosphate content of the starch was reduced by ∼70%. The good inverse correlation between the degree of phosphorylation of the starch and the severity of the starch excess phenotype indicates that the amount of phosphate bound to starch may be a vital factor in the regulation of starch degradation.
Our analysis of the starch accumulated in sex1 leaves revealed that there was no significant alteration of the distribution chain lengths of amylopectin. However, the amylose content was increased significantly. This may be attributable to the increase in the level of GBSS1 (the isoform of starch synthase required for amylose synthesis) in sex1 leaves relative to the soluble starch synthases, which make amylopectin (S.C. Zeeman, unpublished data). Therefore, it seems unlikely that a change in the nature of the starch, other than the phosphate content, can account for the reduction in starch degradation and the consequent starch excess phenotype.
In contrast to transitory starch and storage starch found in potatoes and many other tubers, starch phosphate monoesters (Glc6P residues) are not detectable enzymatically in storage starch of the cereal endosperm (Blennow et al., 2000; Ritte et al., 2000b). It has been shown that most of the phosphate found in cereal endosperm starch is contained in phospholipids that are complexed with the amylose fraction of the starch (Kossmann and Lloyd, 2000). However, nuclear magnetic resonance studies indicate that some phosphate esters also may be present in endosperm starch (Lim et al., 1994), and the occurrence of SEX1 (R1) in dry seed of barley and maize has been demonstrated (Ritte et al., 2000b). These findings suggest a yet unknown function of SEX1 in starch mobilization in tissues surrounding the cereal endosperm. Amyloplasts of the cereal endosperm disintegrate during drying of the endosperm, making endosperm starch accessible to α-amylases that are secreted by the aleurone layer of the seed coat. Recently, it was demonstrated that starch synthesis in cereal endosperm can be driven by the supply of ADP-glucose derived from a cytosolic isoform of the AGPase that in many instances is less sensitive to the 3-PGA/Pi ratio (Denyer et al., 1996; Thorbjørnsen et al., 1996a, 1996b). Metabolism of transitory starch, on the other hand, occurs in intact plastids, and the biosynthesis of transitory starch is controlled tightly by the 3-PGA/Pi ratio. Obviously, the different mode of starch synthesis and breakdown in cereal endosperm adds another level to the control of starch metabolism that does not involve the action of SEX1.
The R1 protein of potato was identified initially by its ability to bind to starch granules. Recently, it was shown that the binding of SEX1 (R1) to transitory starch granules in the dark (during net starch degradation) and its dissociation from the granules in the light (during net starch synthesis) coincides with diurnal fluctuations in the size of the leaf starch pool (Ritte et al., 2000a) and therefore might have a regulatory function. However, in ex1-1 leaves harvested after several hours in the dark, the binding of SEX1 to starch granules was not impaired (J. Chen, unpublished observation), and none of the mutations could be localized to the putative N-terminal starch binding domain (Figure 8).
In all three sequenced alleles, the putative C-terminal nucleotide binding domain (amino acid residues 1110 to 1399) was either deleted or mutated. This domain shows significant homology with the nucleotide binding domains of PPDK and PPS. The PPDK from C. symbiosum has been crystallized and its tertiary structure determined (Herzberg et al., 1996). Interestingly, those residues that are suggested to participate in the binding of the γ-phosphate of ATP are conserved in SEX1, PPDK, and PPS (Figure 7). The C-terminal residues D321, E323, and R337 in the C. symbiosum PPDK correspond to the N-terminal residues D1380, E1382, and R1396 of Arabidopsis SEX1. Residues K22 and R92 in the bacterial PPDK are related to residues K1095 and R1159 of SEX1. It can be assumed that the phosphate donor for starch phosphorylation by SEX1 is a nucleoside triphosphate (e.g., ATP or GTP), although, at present, there is no direct experimental evidence for a starch phosphorylating activity of SEX1 and the metabolite that acts as a donor of the phosphate group has not been identified.
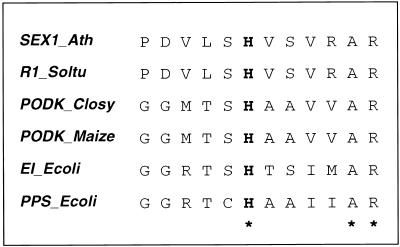
In addition to the homology of the C-terminal domain of SEX1 with the N terminus of bacterial and plant PPDK and bacterial PPS, we identified a small stretch of amino acids that might represent a phosphohistidine domain of SEX1 (Figure 10). This domain is located ∼100 amino acid residues upstream of the putative C-terminal nucleotide binding domain (amino acid residues 998 to 1009). A similar phosphohistidine domain is found in PPDK at approximately the same distance from the nucleotide binding domain. PPDK (and probably also PPS and the protein E1 of the bacterial phosphotransferase system) transfers a phosphate residue from ATP that is bound to the N-terminal nucleotide binding domain toward a substrate bound to the C-terminal PEP/pyruvate binding site via a phosphohistidine intermediate. The catalytic histidine that mediates the phosphoryl group transfer from ATP to Pi and pyruvate (in PPDK) or from ATP to water and pyruvate (in PPS) resides on an 18-kD domain that is located between the substrate binding domain and the nucleotide binding domain.
Figure 10.
Putative Phosphohistidine Domains of SEX1 from Arabidopsis and R1 from Potato.
The regions surrounding the putative phosphohistidines of SEX1 from Arabidopsis (SEX1_Ath) and potato (R1_Soltu) are aligned with the phosphohistidine sites of the pyruvate:phosphate dikinases from C. symbiosum (PODK_Closy) and maize (PODK_Maize), the protein EI of the E. coli phosphotransferase system (EI_Ecoli), and the pyruvate:water dikinase from E. coli (PPS_Ecoli). The putative phosphohistidine is printed in boldface, and residues conserved in all protein classes are marked with asterisks.
We suggest that SEX1 follows a similar mechanism by transferring a phosphate residue from a nucleotide bound to the C-terminal nucleotide binding domain to its substrate starch via a phosphohistidine intermediate. Analogous to the phosphotransfer in PPDK, alternatively, the histidine could interact with the putative C-terminal nucleotide binding site and the putative N-terminal starch binding site. Interestingly, it has been shown that the nucleotide binding domain and the PEP/pyruvate binding domain of PPDK are functionally independent, thus showing that there are separate active sites for the partial reactions of ATP hydrolysis and substrate phosphorylation (Xu et al., 1995). It is tempting to speculate that the SEX1 protein has evolved from the fusion of a nucleotide binding domain and a phosphohistidine domain to a starch binding domain, thus leading to a protein with starch phosphorylating activity. Thus, SEX1 might act as a starch:water dikinase, rendering starch accessible to degradation. However, the possibility remains that SEX1 affects another enzyme that phosphorylates starch directly.
Obviously, the putative C-terminal nucleotide binding domain is vital for the physiological function of SEX1, but (reversible) binding of SEX1 to starch granules also may be required. Although we did not analyze the interaction of SEX1 with other proteins that are bound to the starch granules, it can be assumed that the degree of starch phosphorylation mediated by SEX1 controls the activities of other yet uncharacterized proteins that require the presence of phosphate esters before they can attack an intact starch granule. This initial attack might be required before degrading enzymes (e.g., amylases) (Kakefuda and Preiss, 1997; Zeeman et al., 1998a) are able to proceed with starch mobilization.
Starch synthesis and degradation are regulated tightly, and both processes have to be coordinated with the carbohydrate metabolism in the cytosol. This coordination requires signals that transmit the carbohydrate status of the cytosol to the plastid metabolism. It remains an open challenge to identify signals and components of the signal chain that initiate the degradation of transitory starch via the action of SEX1.
METHODS
Plant Material
M2 seed of ethyl methanesulfonate– and fast neutron–mutagenized Arabidopsis thaliana were obtained from Lehle Seeds (Round Rock, TX). Plants were grown on soil at 23°C in a 12-hr light/dark cycle or as indicated. Screening of mutants was performed as described (Caspar et al., 1991). Restriction fragment length polymorphism markers, expressed sequence tag (EST) clones, and seed of sex1-1 were obtained from the Arabidopsis Biological Resource Center (Columbus, OH).
Protein Analysis
SDS-PAGE and protein gel blotting were performed as described (Ritte et al., 2000a).
Analysis of Starch
Isolation of starch granules was performed as described (Ritte et al., 2000a). Starch-bound glucose 6-phosphate (Glc6P) and glucose 3-phosphate (Glc3P) were analyzed by high performance anion exchange chromatography with pulsed amperometric detection as described by Ritte et al. (2000b); however, a CarboPac PA-100 column (Dionex, Sunnyvale, CA) was used. Alternatively, Glc6P was determined enzymatically (Nielsen et al., 1994). Starch content was determined by enzymatic hydrolysis of the starch to glucose as described by Zeeman et al. (1998a). Analysis of starch composition by Sepharose CL2B chromatography was performed as described by Denyer et al. (1995). Analysis of the chain length distribution of amylopectin by fluorophore-assisted PAGE was conducted as described by Edwards et al. (1999). The starch was prepared from a batch of ∼100 plants. Three aliquots of this starch were boiled to disperse the starch and treated with isoamylase to debranch the amylopectin, and the reducing ends were labeled with fluorophore. Each sample was run three times on an Applied Biosystems (Foster City, CA) sequencer and analyzed with GeneScan software (Applied Biosystems, Foster City, CA) to obtain the peak areas. The mean peak area was taken as the value of that sample.
Molecular Techniques
Standard DNA and RNA techniques were used as described (Sambrook et al., 1989). Plant transformation was achieved by in planta vacuum infiltration (Bechtold et al., 1993). The cDNA sequence of SEX1 was determined by sequencing 10 overlapping reverse transcription–mediated polymerase chain reaction (RT-PCR) products amplified with Pfu DNA polymerase from cDNA that was prepared by RT of leaf RNA. The same procedure was used to sequence the mutant alleles. Single sequence length polymorphism markers indicative of the SEX1 loci of the Arabidopsis accessions Columbia (Col-0) and Landsberg erecta were generated by testing randomly different primer combinations deduced from the sequence of bacterial artificial chromosome (BAC) T16B5. The primers R1-2f-w (5′-ATTAGATAAACTACGATTGCC-3′) and R1-2-rev (5′-TAAGTCGATTTTGGA-CGAACC-3′) amplified a DNA fragment of 1362 bp in Col-0 and a DNA fragment of ∼1700 bp in Landsberg erecta.
RT-PCR was performed as described by Kofler et al. (2000) with the exception that nonsaturating PCR conditions were used. The actin2 gene-specific primers actin2-Sfor (5′-TGTACGCCAGTGGTCGTACAACC-3′) and actin2-Brev (5′-GAAGCAAGAATGGAA-CCACCG-3′) amplified a cDNA fragment of 600 bp, whereas the pGlcT gene-specific primers A12T7-2fw (5′-CAACAGCTCAGAGTG-TGCAG-3′) and A12T3-3fw (5′-CCATCCTGCCTCCGGCTCAG-3′) amplified a cDNA fragment of 428 bp. The PCR program was 94°C for 2 min followed by 16 or 21 cycles of 94°C for 30 sec, 54°C for 30 sec, and 72°C for 48 sec each. To visualize RT-PCR products that were not detectable by ethidium bromide staining, the amplification products were blotted onto nylon membranes and detected by hybridization to specific probes.
GenBank Accession Numbers
The sequences reported in this article have been deposited in the GenBank database (accession numbers AC007354 and AF312027).
Acknowledgments
We gratefully acknowledge the donation of EST G3G2T7 and the markers pARMS CD3-179 and RFLP CD3-315 by the Arabidopsis Biological Resource Center and the donation of seed by the Nottingham Arabidopsis Stock Centre. The mutant alleles sex1-2, sex1-3, and sex1-4 were isolated originally in laboratory of the late Prof. Tom ap Rees. We thank Dr. Nora Eckermann for performing the HPAEC analysis and Katrin Fischer for help with the analysis of starch phosphorylation. We gratefully acknowledge Dr. Antje Rottmann (Universität Potsdam, Institut für Organische Chemie und Strukturanalytik) for providing Glc3P. This work was supported by a grant from the National Science Council, Taiwan, Republic of China (NSC 88-2311-B-001-034) and by the Academia Sinica, Taipei, Taiwan (to J.C.), by grants from the Biotechnology and Biological Sciences Research Council of the United Kingdom (to S.C.Z and A.M.S.) and the Gatsby Charitable Foundation (to S.C.Z), and by grants from the Deutsche Forschungsgemeinschaft (to U.-I.F. and A.W.) and the Fonds der Chemischen Industrie (to U.-I.F.).
References
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Finn, R.D., and Sonnhammer, E.L.L. (1999). Pfam 3.1: 1313 multiple alignments match the majority of proteins. Nucleic Acids Res. 27, 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, G., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. 316, 1194–1199. [DOI] [PubMed] [Google Scholar]
- Blennow, A., Bay-Schmidt, M.A., Olsen, C.E., and Møller, B.L. (1998). Analysis of starch-bound glucose 3-phosphate and glucose 6-phosphate using controlled acid treatment combined with high-performance anion-exchange chromatography. J. Chromatogr. 829, 385–391. [Google Scholar]
- Blennow, A., Engelsen, S.B., Munck, L., and Møller, B.L. (2000). Starch molecular structure and phosphorylation investigated by a combined chromatographic and chemometric approach. Carbohydr. Polym. 41, 163–174. [Google Scholar]
- Carroll, L.J., Xu, Y., Thrall, S.H., Martin, B.M., and Dunaway-Mariano, D. (1994). Substrate binding domains in pyruvate phosphate dikinase. Biochemistry 33, 1134–1142. [DOI] [PubMed] [Google Scholar]
- Caspar, T., Lin, T.-P., Kakefuda, G., Benbow, L., Preiss, J., and Somerville, C. (1991). Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol. 95, 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley, J.H., Zeeman, S.C., Takaha, T., Smith, A.M., and Smith, S.M. (2001). A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 26, 89–100. [DOI] [PubMed] [Google Scholar]
- Denyer, K., Barber, L.M., Burton, R., Hedley, C.L., Hylton, C.M., Johnson, S., Jones, D.A., Marshall, J., Smith, A.M., Tatge, H., Tomlinson, K., and Wang, T.L. (1995). The isolation and characterization of novel low-amylose mutants of Pisum sativum L. Plant Cell Environ. 18, 1019–1026. [Google Scholar]
- Denyer, K., Dunlap, F., Thorbjørnsen, T., Keeling, P., and Smith, A.M. (1996). The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial. Plant Physiol. 112, 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, A., Fulton, D.C., Hylton, C.M., Jobling, S.A., Gidley, M., Rossner, U., Martin, C., and Smith, A.M. (1999). A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J. 17, 251–261. [Google Scholar]
- Eimert, K., Wang, S.M., Lue, W.L., and Chen, J.C. (1995). Monogenic recessive mutations causing both late floral initiation and excess starch accumulation in Arabidopsis. Plant Cell 7, 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer, S.L., Hogenesch, J.B., Straume, M., Chang, H.-S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Herzberg, O., Chen, C.H.C., Kapadia, G., McGuire, M., Carroll, L.J., Noh, S.J., and Dunaway-Mariano, D. (1996). Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites. Proc. Natl. Acad. Sci. USA 93, 2652–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, K., Bucher, P., Falquet, L., and Bairoch, A. (1999). The PROSITE database, its status in 1999. Nucleic Acids Res. 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakefuda, G., and Preiss, J. (1997). Partial purification and characterization of a diurnally fluctuating novel endoamylase from Arabidopsis thaliana leaves. Plant Physiol. Biochem. 35, 907–913. [Google Scholar]
- Kofler, H., Häusler, R.E., Schulz, B., Gröner, F., Flügge, U.I., and Weber, A. (2000). Molecular characterisation of a new mutant allele of the plastidic phosphoglucomutase in Arabidopsis, and complementation of the mutant with the wild-type cDNA. Mol. Gen. Genet. 263, 978–986. [DOI] [PubMed] [Google Scholar]
- Kossmann, J., and Lloyd, J. (2000). Understanding and influencing starch biochemistry. Crit. Rev. Plant Sci. 19, 171–226. [PubMed] [Google Scholar]
- Lim, S.T., Kasemsuwan, T., and Jane, J.L. (1994). Characterization of phosphorous in starch by P-31-nuclear magnetic-resonance spectroscopy. Cereal Chem. 71, 488–493. [Google Scholar]
- Lorberth, R., Ritte, G., Willmitzer, L., and Kossmann, J. (1998). Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat. Biotechnol. 16, 473–477. [DOI] [PubMed] [Google Scholar]
- McGuire, M., Huang, K., Kapadia, G., Herzberg, O., and Dunaway- Mariano, D. (1998). Location of the phosphate binding site within Clostridium symbiosum pyruvate:phosphate dikinase. Biochemistry 7, 13463–13474. [DOI] [PubMed] [Google Scholar]
- Neuhaus, H.E., and Stitt, M. (1990). Control analysis of photosynthate partitioning: Impact of reduced activity of ADP-glucose pyrophosphorylase or plastid phosphoglucomutase on the fluxes to starch and sucrose in Arabidopsis thaliana (L.) Heynh. Planta 182, 445–454. [DOI] [PubMed] [Google Scholar]
- Nielsen, T.H., Wischmann, B., Enevoldsen, K., and Møller, B.L. (1994). Starch phosphorylation in potato-tubers proceeds concurrently with de-novo biosynthesis of starch. Plant Physiol. 105, 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte, G., Lorberth, R., and Steup, M. (2000. a). Reversible binding of the starch-related R1 protein to the surface of transitory starch granules. Plant J. 21, 387–391. [DOI] [PubMed] [Google Scholar]
- Ritte, G., Eckermann, N., Haebel, S., Lorberth, R., and Steup, M. (2000. b). Compartmentation of the starch-related R1 protein in higher plants. Starch Staerke 52, 179–185. [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Smith, A.M., Denyer, K., and Martin, C. (1997). The synthesis of the starch granule. Annu. Rev. Plant Physiol. 48, 65–87. [DOI] [PubMed] [Google Scholar]
- Tabata, S., and Hizukuri, S. (1971). Studies on starch phosphate. II. Isolation of glucose 3-phosphate and maltose phosphate by acid hydrolysis of potato starch. Starch Staerke 23, 267–272. [Google Scholar]
- Thorbjørnsen, T., Villand, P., Kleczkowski, L.A., and Olsen, A.-O. (1996. a). A single gene encodes two different transcripts for the ADP-glucose pyrophosphorylase small subunit from barley (Hordeum vulgare). Biochem. J. 313, 149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorbjørnsen, T., Villand, P., Denyer, K., Olsen, A.-O., and Smith, A.M. (1996. b). Distinct isoforms of ADPglucose pyrophosphorylase occur inside and outside the amyloplasts in barley endosperm. Plant J. 10, 243–250. [Google Scholar]
- Trethewey, R.N., and ap Rees, T. (1994. a). A mutant of Arabidopsis thaliana lacking the ability to transport glucose across the chloroplast envelope. Biochem J. 301, 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trethewey, R.N., and ap Rees, T. (1994. b). The role of the hexose transporter in the chloroplasts of Arabidopsis thaliana L. Planta 195, 168–174. [Google Scholar]
- Weber, A., Servaites, J.C., Geiger, D.R., Kofler, H., Hille, D., Gröner, F., Hebbeker, U., and Flügge, U.I. (2000). Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell 12, 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., McGuire, M., Dunaway-Mariano, D., and Martin, B.M. (1995). Separate site catalysis by pyruvate phosphate dikinase as revealed by deletion mutants. Biochemistry 34, 2195–2202. [DOI] [PubMed] [Google Scholar]
- Zeeman, S.C., Northrop, F., Smith, A.M., and ap Rees, T. (1998. a). A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J. 15, 357–365. [DOI] [PubMed] [Google Scholar]
- Zeeman, S.C., Umemoto, T., Lue, W.L., Au-Yeung, P., Martin, C., Smith, A.M., and Chen, J. (1998. b). A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 10, 1699–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman, S.C., and ap Rees, T. (1999). Changes in carbohydrate metabolism and assimilate export in starch-excess mutants of Arabidopsis. Plant Cell Environ. 22, 1445–1453. [Google Scholar]