Abstract
Plant B-type cyclin genes are expressed specifically in late G2- and M-phases during the cell cycle. Their promoters contain a common cis-acting element, called the MSA (M-specific activator) element, that is necessary and sufficient for periodic promoter activation. This motif also is present in the tobacco kinesin-like protein gene NACK1, which is expressed with timing similar to that of B-type cyclin genes. In this study, we show that G2/M-phase–specific activation of the NACK1 promoter also is regulated by the MSA element, suggesting that a defined set of G2/M-phase–specific genes are coregulated by an MSA-mediated mechanism. In a search for MSA binding factors by yeast one-hybrid screening, we identified three different Myb-like proteins that interact specifically with the MSA sequence. Unlike the majority of plant Myb-like proteins, these Myb proteins, NtmybA1, NtmybA2, and NtmybB, have three imperfect repeats in the DNA binding domain, as in animal c-Myb proteins. During the cell cycle, the level of NtmybB mRNA did not change significantly, whereas the levels of NtmybA1 and A2 mRNAs fluctuated and peaked at M-phase, when B-type cyclin genes were maximally induced. In transient expression assays, NtmybA1 and A2 activated the MSA-containing promoters, whereas NtmybB repressed them. Furthermore, expression of NtmybB repressed the transcriptional activation mediated by NtmybA2. Our data show that a group of plant Myb proteins that are structurally similar to animal c-Myb proteins have unexpected roles in G2/M-phase by modulating the expression of B-type cyclin genes and may regulate a suite of coexpressed genes.
INTRODUCTION
From yeast to humans, progression through the cell cycle is associated with the phase-specific transcription of defined sets of genes (McKinney and Heintz, 1991). Phase-specific transcription may contribute to an orderly progression through the cell cycle by ensuring that key proteins are produced in a strict temporal sequence. In most cases, cell cycle–dependent changes in transcript levels are regulated by promoter activity (Koch and Nasmyth, 1994; Müller, 1995; Ito, 1998).
During the G1-to-S transition, a set of specific genes are induced, including G1 cyclin genes and several genes involved in DNA synthesis in yeast (Koch and Nasmyth, 1994), animal (Müller, 1995), and plant cells (Ito, 1998). G1/S-phase–specific transcription is regulated by mechanisms involving the E2F/DP heterodimeric transcription factors in animal cells (Black and Azizkhan-Clifford, 1999). The recent identification of the plant homologs of E2F (Ramirez-Parra et al., 1999; Sekine et al., 1999; Albani et al., 2000) and DP (Magyar et al., 2000; Ramirez-Parra and Gutierrez, 2000) supports the idea that plants have evolved a mechanism for G1/S-phase–specific transcription that relies on genes analogous to those acting in animal cells.
Later in the cell cycle, during G2- and M-phases, another set of genes is expressed, among which B-type cyclin genes are the best characterized. During G2-phase, B-type cyclins interact with and activate the cyclin-dependent kinase, which is critical for entry into mitosis (Pines and Hunter, 1990). Mitotic B-type cyclin genes, human cyclin B1 and B2 (Piaggio et al., 1995; Brandeis and Hunt, 1996) and yeast CLB1 and CLB2 (Richardson et al., 1992), are induced in late S-phase and reach peak expression during G2- and M-phases. Similarly, plant B-type cyclin genes, B1 and B2 classes, are expressed specifically in late G2- and M-phases (Mironov et al., 1999; Ito, 2000). Unscheduled expression or overexpression of B-type cyclin genes often results in the formation of human cancers (Gong et al., 1994), increased growth of plant organs (Doerner et al., 1996), or lethal phenotypes (Lew et al., 1991). In yeast, G2/M-phase–specific transcription of CLB1 and CLB2 genes is controlled by a MADS box transcription factor, MCM1, in cooperation with a factor called SFF (Swi5 factor) (Althoefer et al., 1995; Maher et al., 1995; Breeden, 2000). Transcription of many other genes involved in mitosis is coregulated by a similar mechanism, probably mediated by MCM1/SFF (Spellman et al., 1998). The mechanisms that control G2/M-phase–specific promoter activation of human cyclin B1 are much debated (Cogswell et al., 1995; Katula et al., 1997; Hwang et al., 1998). It seems likely that the human cyclin B1 promoter is regulated by a complex mechanism involving multiple cis elements, none of which is sufficient for G2/M-phase–specific promoter activation (Hwang et al., 1998).
In plants, periodic expression of B-type cyclin genes also is regulated, at least in part, by a periodic change in the activity of their promoters in the cell cycle (Shaul et al., 1996; Ito et al., 1997; Colón-Carmona et al., 1999; Tréhin et al., 1999). Promoter analysis of the Nicotiana sylvestris cyclin B1 gene, Nicsy;CycB1;1, showed the presence of a 23-bp element that acts as a cell cycle phase–independent transcriptional activator (Tréhin et al., 1999). This element may be important for “quantitative” control, which determines the level of transcription, but is not associated with “qualitative” control, which determines the timing of transcription. We have analyzed the promoter of the cyclin B1 gene, Catro; CycB1;1 (CYM), from Catharanthus roseus and showed that the timing of promoter activation during the cell cycle is determined by a single type of cis element called MSA (M-specific activator), which is necessary and sufficient for periodic promoter activation (Ito et al., 1998a). MSA-like motifs are found in B-type cyclin promoters from various plant species. The tobacco kinesin-like protein genes NACK1 and NACK2, which are expressed with timing similar to that of B-type cyclin genes in the cell cycle, also contain MSA-like motifs in their promoters. This finding suggests that a defined set of G2/M-phase–specific genes might be coregulated by a common MSA-mediated mechanism in plants (Ito et al., 1998a).
In this report, we demonstrate that the MSA-like motifs present in the NACK1 promoter are functional cis elements that direct G2/M-phase–specific gene expression and describe the cloning and characterization of the Myb-related transcription factors NtmybA1, NtmybA2, and NtmybB, which bind specifically to MSA elements. These proteins, like animal c-Myb proteins, contain three imperfect repeat sequences in their N-terminal DNA binding domains, in contrast to the majority of plant Myb proteins, which contain only two such sequences (Jin and Martin, 1999). The data presented in this article indicate that three repeat plant Myb proteins are involved in MSA-mediated G2/M-phase–specific transcription by binding to the MSA element and modulating its activity.
RESULTS
Functional Analysis of MSA-Like Sequences in the Promoter of a G2/M-Phase–Specific Gene
In our previous work, the MSA element was identified by analyzing the promoter of a cyclin B1 gene (Catro;CycB1;1) from C. roseus using synchronized cell cultures of tobacco BY2 cells (Ito et al., 1998a). The Catro;CycB1;1 promoter contains four copies of the MSA motif (designated RT1, RT2, RT3, and RT4), and similar motifs were found in the promoters of other B-type cyclin genes: Glyma;CycB1;3 (cyc4Gm) from soybean, Arath;CycB1;2 (cyc1bAt) and Arath; CycB2;1 (cyc2aAt) from Arabidopsis, and Nicta;CycB1;3 (NtCYM) from tobacco. This motif also was present in the promoters of tobacco NACK1 and NACK2 genes, which encode kinesin-like proteins (Ito et al., 1998a). Accumulation of NACK1 and NACK2 transcripts occurs at essentially the same times in the cell cycle as that of the tobacco cyclin B1 gene Nicta;CycB1;3 (data not shown).
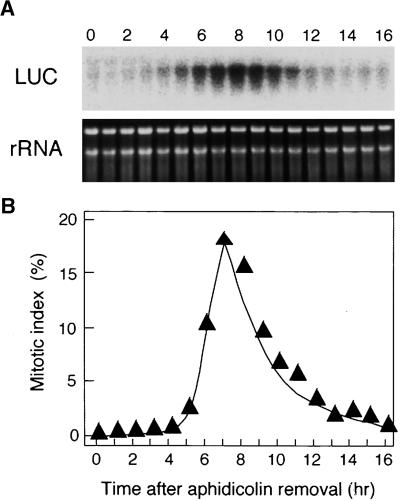
To investigate whether the MSA-like sequences in the NACK1 promoter function as MSA, the activities of wild-type and mutant NACK1 promoters were examined in tobacco BY2 cells. The wild-type NACK1 promoter contains two MSA-like sequences. A fusion between the wild-type NACK1 promoter and the firefly luciferase reporter gene (LUC) was introduced into BY2 cells by Agrobacterium tumefaciens–mediated transformation, and the resulting stable transformants were synchronized by aphidicolin treatment. Changes in NACK1 promoter activity were monitored by determining LUC mRNA levels during the cell cycle by RNA gel blot analysis (Figure 1). The LUC mRNA was undetectable during S-phase, just after release from the aphidicolin block, but levels then increased in parallel with the increase in the mitotic index. A peak in LUC mRNA level was observed at 7 to 8 hr, after which the level decreased rapidly (Figure 1). This G2/M-phase–specific accumulation of LUC mRNA indicated that NACK1 promoter activity is G2/M-phase specific.
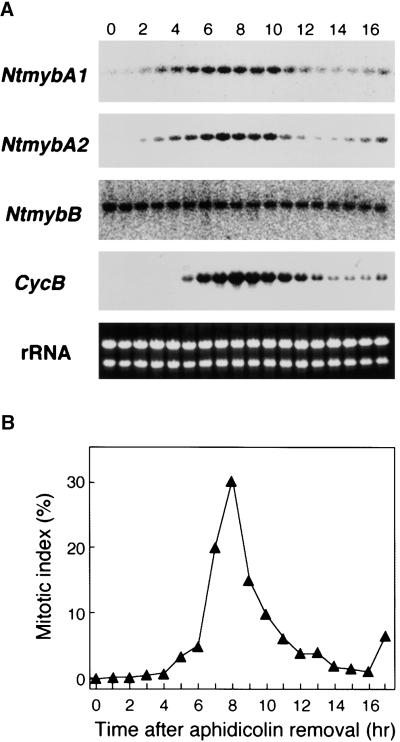
Figure 1.
G2/M-Phase–Specific Activation of the NACK1 Promoter.
(A) G2/M-phase–specific transcription of the NACK1 promoter–LUC transgene in BY2 cells. Tobacco BY2 cells transformed with the NACK1 promoter (−496 to +68)–LUC construct were arrested by aphidicolin treatment. Aphidicolin was removed (0 hr), and cells were harvested at 1-hr intervals. Total RNA was isolated and hybridized with the LUC coding region as a probe. Ethidium bromide staining of the rRNA is shown as well.
(B) Change in mitotic index during synchronous cultures. The efficiency of cell synchronization was checked by measuring the mitotic index at 1-hr intervals.
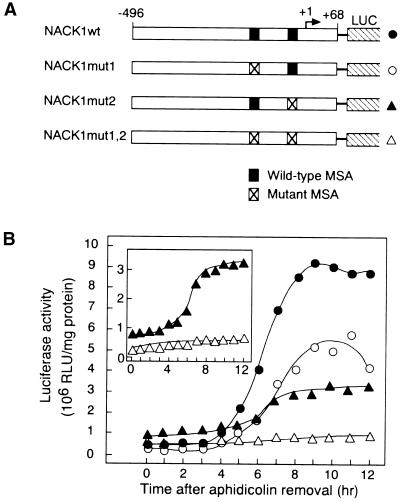
To examine the role of MSA-like sequences in the NACK1 promoter, the motifs were mutated by changing two bases (AC to TT) in the central core pentamer, AACGG (Figure 2A). The mutant promoters were fused to the LUC reporter and transformed into BY2 cells. The resulting transgenic BY2 cell lines were synchronized, and LUC enzymatic activity was assayed (Figure 2B). Confirming the results from the mRNA analysis, the wild-type promoter directed a G2/M-phase–specific increase of LUC activity. The enzyme activity did not decline after M-phase, possibly because of the low turnover rate of LUC protein in BY2 cells. A similar pattern of change in LUC activity was observed when the Catro; CycB1;1 promoter–LUC construct was analyzed in our previous study (Ito et al., 1998a). When either one of the two MSA-like sequences in the NACK1 promoter was mutated, there was a significant reduction in maximal LUC activity, although the G2/M-phase–specific increase in LUC activity was still significant (Figure 2B, inset). Only when both of the MSA-like sequences were mutated was the G2/M-phase–specific increase of reporter activity abolished (Figure 2B). Therefore, we concluded that both of the MSA-like sequences in the NACK1 promoter function as MSA elements and are required for G2/M-phase–specific promoter activity in BY2 cells.
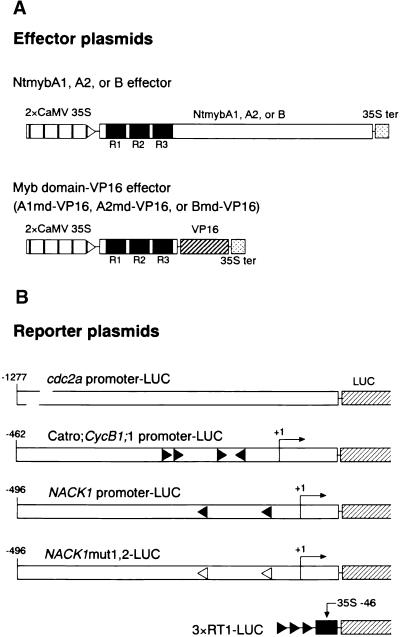
Figure 2.
G2/M-Phase–Specific Activation of the NACK1 Promoter Is Dependent on the MSA Elements.
(A) Scheme of NACK1 promoter–LUC derivatives used for stable transformation of BY2 cells. Numbers indicate nucleotide positions relative to the transcriptional start site (+1). Wild-type and mutant MSA sequences are shown by closed boxes and crossed boxes, respectively.
(B) Changes in LUC enzymatic activity during the cell cycle. The transgenic BY2 cells were synchronized by aphidicolin treatment and harvested at 1-hr intervals for LUC assays. Promoter constructs are designated with the symbols shown at right in (A). The inset shows LUC activity of NACK1mut2 and NACK1mut1,2 constructs with an expanded scale. RLU, relative light unit.
Isolation of cDNAs Encoding MSA Binding Proteins
To isolate cDNAs encoding DNA binding proteins that interact with the MSA motif, we used the yeast one-hybrid screening system. We constructed two yeast strains carrying the yeast HIS3 reporter gene with either a six-copy tandem repeat of MSA elements from the Catro;CycB1;1 promoter (CYM-HIS3) or a three-copy tandem repeat of MSA elements from the NACK1 promoter (NACK1-HIS3). The yeast cells carrying the CYM-HIS3 reporter were transformed with an expression library of cDNA fragments of mRNAs prepared from actively dividing BY2 cells. The cDNA fragments were fused to the transcriptional activation domain of yeast GAL4. Approximately 8 × 106 yeast transformants were screened, and 26 HIS+ colonies were obtained. Of these, 15 plasmids conferred a HIS+ phenotype to yeast cells with the NACK1-HIS3 reporter. DNA sequence analysis of the cDNA inserts led to the classification of these 15 cDNA clones into five distinct cDNA groups. Among them, cDNAs encoding three different Myb-related proteins were obtained, which were designated NtmybA1, NtmybA2, and NtmybB.
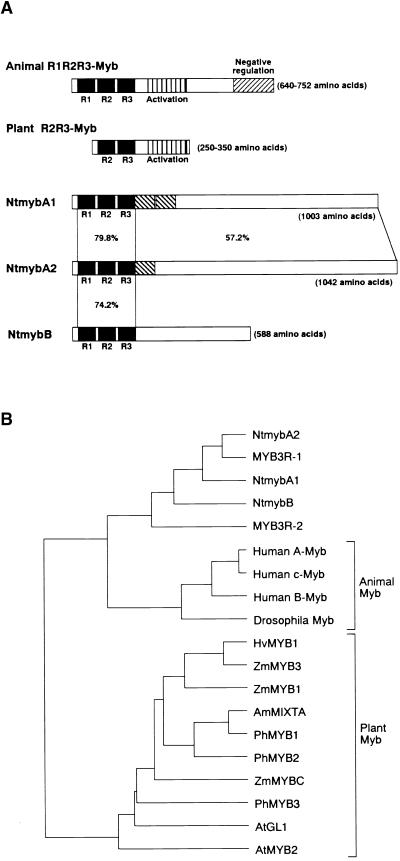
Some of the cDNAs contained complete open reading frames of NtmybA2 and NtmybB, whereas all NtmybA1 cDNAs were partial clones truncated at their 3′ ends. The 3′ region of NtmybA1 cDNA was obtained by rapid amplification of 3′ cDNA ends (RACE) and sequenced. NtmybA1, A2, and B potentially encode proteins of 1003, 1042, and 588 amino acids, respectively, all of which contain a conserved domain structurally related to the “Myb domain” (Figure 3A). The Myb domain is composed of three imperfect repeat sequences (R1, R2, and R3) of 50 to 53 amino acids in the mammalian Myb protein c-Myb and the related proteins A-Myb and B-Myb (R1R2R3-Myb), whereas most of the plant Myb proteins identified thus far contain only two repeats (R2R3-Myb). Interestingly, the proteins encoded by the tobacco cDNAs, like animal c-Myb proteins, were R1R2R3 type (Figure 3A). Other members of this group of Myb genes (MYB3R-1/pc-Myb1 and MYB3R-2/pc-Myb2 from Arabidopsis) have been reported recently after systematic sequencing of the genome (Braun and Grotewold, 1999; Kranz et al., 2000), but their functions are unknown.
Figure 3.
Structures of NtmybA1, A2, and B Proteins.
(A) Scheme showing structures of Myb proteins. Structures of NtmybA1, A2, and B are shown together with functional domains of animal R1R2R3-Myb and typical plant R2R3-Myb proteins. R1, R2, and R3 motifs in DNA binding Myb domains are shown as closed boxes. Repeat sequences found in NtmybA1 are shown as hatched boxes. Amino acid identities within and outside of the Myb domains are shown for NtmybA1, A2, and B.
(B) Phylogenetic tree of Myb proteins. The tree was constructed with the UPGMA tree program (Genetyx Software Development Co. Ltd., Tokyo, Japan) based on amino acid sequences of the R2 and R3 motifs in the Myb domains of Ntmyb proteins and other members of the Myb family isolated from plants and animals: c-Myb, A-Myb, and B-Myb from human; Drosophila melanogaster Myb; MYB3R-1, MYB3R-2, AtMYB2, and AtGL1 from Arabidopsis; ZmMYB1, ZmMYB3, and ZmMYBC from maize; PhMYB1, PhMYB2, and PhMYB3 from petunia; AmMIXTA from Antirrhinum majus; and HvMYB1 from barley.
In the Myb domain, NtmybA1, A2, and B showed highest similarity to each other and to Arabidopsis MYB3R-1 and MYB3R-2, with identity scores of 71 to 90%. A phylogenetic tree was constructed based on the amino acid sequences of the R2 and R3 Myb motifs of animal and plant Myb proteins (Figure 3B). The results indicate that the tobacco Ntmyb proteins and the Arabidopsis MYB3R proteins are far more similar to animal R1R2R3-Myb proteins than to plant R2R3-Myb proteins. When the amino acid sequences of the Myb domains were compared, the Ntmyb proteins showed 62 to 64% identity with human c-Myb but only 35 to 45% identity with plant R2R3-Myb proteins. However, this phylogenetic analysis also revealed that plant R1R2R3-type Myb proteins did not group with any animal R1R2R3-type Myb proteins (A-Myb, B-Myb, and c-Myb) but formed a separate branch. The tree shows that NtmybA1 and A2 are more closely related to each other than to NtmybB and that Arabidopsis MYB3R-1 is more similar to tobacco NtmybA1 and A2 than to Arabidopsis MYB3R-2.
In addition to the Myb repeats, the NtmybA1 protein contains a 56–amino acid sequence that is repeated twice just C-terminal to the Myb domain, although the significance of this sequence is unknown (Figure 3A). Outside of the Myb domain, all three Ntmyb proteins showed no significant similarity to any other Myb proteins or to any proteins identified previously, except that NtmybA1 and A2 showed similarity to a limited region of MYB3R-1. A 126–amino acid sequence outside of the Myb domain of MYB3R-1 (amino acid positions 563 to 688) showed 61 to 63% identity to the C-terminal regions of NtmybA1 and A2. However, even outside of the conserved Myb domain, NtmybA1 and A2 share 57% amino acid identity, whereas NtmybB shows no significant similarity in this region to NtmybA1 or A2 (Figure 3A). The similarities in primary sequence may indicate that NtmybA1 and A2 share similar biological functions that could differ from that of NtmybB. Based on structural similarity, Arabidopsis MYB3R-1 also may have a function similar to that of NtmybA1 and A2.
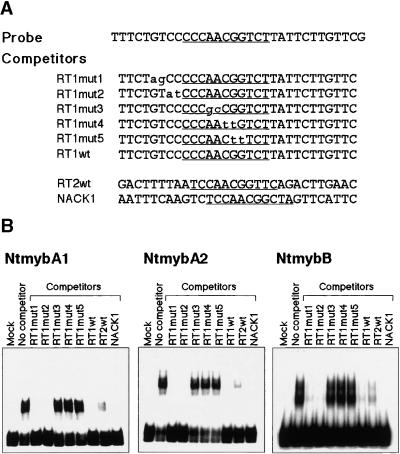
NtmybA1, A2, and B Bind Specifically to the MSA Motif
To test the specificity of DNA binding to NtmybA1, A2, and B, we conducted electrophoretic mobility shift assays with proteins translated in vitro. As shown in Figure 4, all three Myb proteins bound to the wild-type 30-bp Catro;CycB1;1 promoter sequence containing an MSA element (RT1). To determine the nucleotides important for binding to this sequence, we used a series of competitors with two base mutations (Figure 4A). The oligonucleotides with mutations outside of the MSA element competed strongly for binding with the wild-type probe. However, any mutation within the MSA element abolished the ability to compete for binding to the wild-type MSA element (Figure 4B). Binding of the Ntmyb proteins to the MSA element was not context specific: different regions of the Catro;CycB1;1 promoter that contain other MSA elements (RT2) or another MSA-containing promoter (NACK1) also competed for binding (Figure 4B). These results indicate that all three Ntmyb proteins can bind to functional MSA elements but do not bind to elements containing mutations that were shown to eliminate the function of MSA in vivo (Ito et al., 1998a). Specific binding to the MSA motif also was observed using truncated versions of Ntmyb proteins that contained only the Myb domain (data not shown), indicating that DNA binding ability and sequence specificity are determined by the Myb domains, like other Myb transcription factors in animals and plants.
Figure 4.
Characterization of the DNA Binding Affinity of the Ntmyb Proteins.
(A) Nucleotide sequences of oligonucleotides used as probe and competitors in electrophoretic mobility shift assays. Sequences of MSA elements are underlined.
(B) Electrophoretic mobility shift assays of Ntmyb proteins. Binding reaction mixtures were incubated with the probe and either mock-translated product (Mock) or in vitro synthesized NtmybA1, A2, and B in the presence or absence of a 40-fold molar excess of unlabeled oligonucleotide competitors.
Transcript Accumulation of NtmybA1 and A2 Is Regulated by the Cell Cycle
Changes in the transcript levels of NtmybA1, A2, and B were examined in tobacco BY2 cells synchronized by aphidicolin treatment (Figure 5). As shown in Figure 5B, the removal of aphidicolin resulted in synchronous progression through the cell cycle, with a clear peak in the mitotic index after 8 hr. NtmybB mRNA was present throughout the cell cycle, and the levels were relatively constant (Figure 5A). The levels of NtmybA1 and A2 mRNAs changed in parallel during the cell cycle. The levels of NtmybA1 and A2 mRNAs were below detection just after aphidicolin release, started to increase at 3 to 4 hr, and reached a maximum at 8 hr. Subsequently, the levels decreased sharply at the completion of cell division (Figure 5A). To relate this transcript accumulation pattern to that of B-type cyclin genes, the blot was reprobed with Nicta;CycB1;3, a tobacco cyclin B1 cDNA (Ito et al., 1997) (Figure 5A, CycB). The pattern of accumulation of NtmybA1 and A2 transcripts in the cell cycle was very similar to that of Nicta;CycB1;3, but NtmybA1 and A2 mRNAs appeared 1 to 2 hr earlier than that of Nicta;CycB1;3 (Figure 5A). The G2/M-phase–specific transcript accumulation observed for NtmybA1 and A2 is consistent with the idea that these two Myb proteins may play a role in promoter activation of B-type cyclin genes.
Figure 5.
Change in the Levels of Transcripts for NtmybA1, A2, and B during the Cell Cycle.
(A) RNA gel blot analysis of NtmybA1, A2, and B. Tobacco BY2 cells were synchronized by aphidicolin treatment. Cells were collected at 1-hr intervals after release from aphidicolin block. Gene-specific probes for NtmybA1, A2, and B were used for hybridization. To detect Nicta;CycB1;3 (CycB) mRNA, full length cDNA was used as a probe. Ethidium bromide staining of the rRNA is shown as well.
(B) Change in mitotic index during synchronous cultures. The efficiency of cell synchronization was checked by measuring the mitotic index at 1-hr intervals.
In Situ Localization of Ntmyb mRNAs
To determine if Ntmyb mRNAs are expressed in the same cells at the same time as their presumptive target genes, we performed in situ hybridization on tobacco shoot apices. Longitudinal sections of shoot apices of 40-day-old tobacco plants were hybridized with digoxigenin-labeled antisense RNA probes of Nicta;CycB1;3 and NtmybA1, A2, and B cDNAs (Figure 6). NtmybA1 and A2 probes gave a scattered signal in isolated cells in the meristematic region, which is a characteristic feature of cell cycle–regulated transcripts (Fobert et al., 1994; Kouchi et al., 1995). The Nicta;CycB1;3 mRNA (Figure 6, CycB) also was localized in a similar manner. In contrast, NtmybB mRNA was detected uniformly in the meristematic region, indicating that the transcript level of NtmybB is cell cycle independent.
Figure 6.
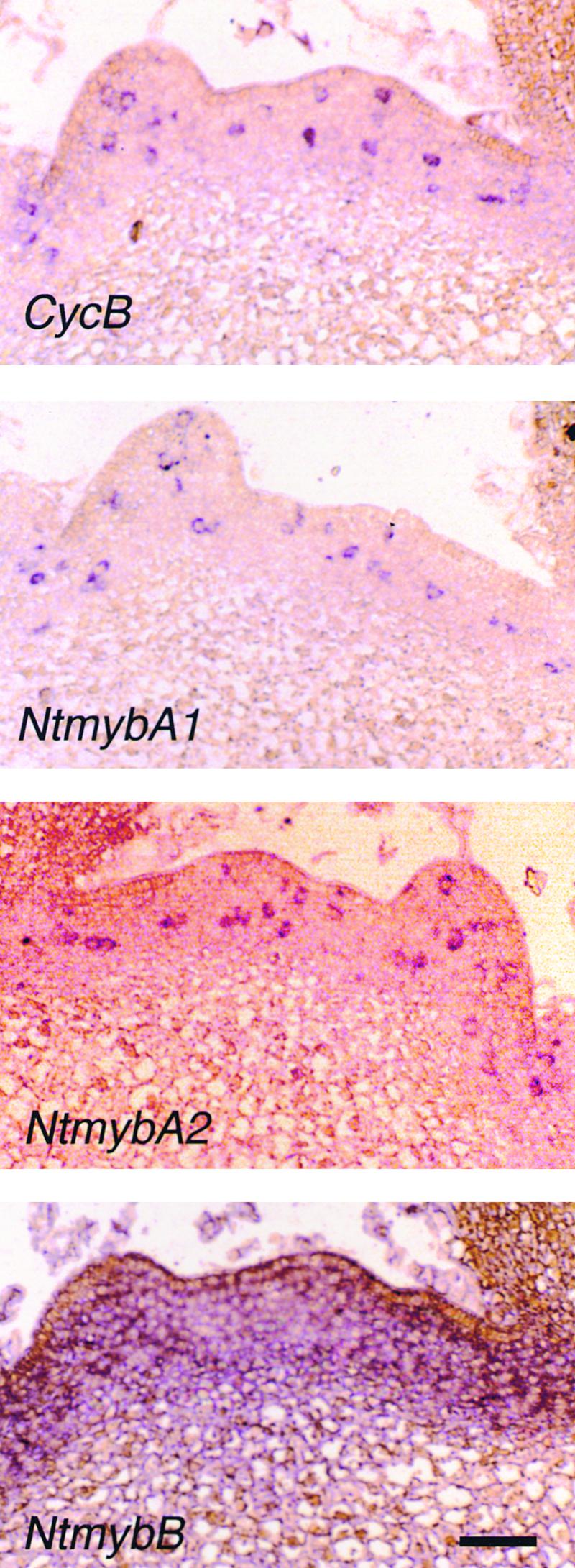
Localization of the Transcripts of NtmybA1, A2, and B in Tobacco Shoot Apices.
Longitudinal sections of shoot apices were hybridized with digoxigenin-labeled antisense RNA probes of Nicta;CycB1;3 (CycB) and NtmybA1, A2, and B and viewed under a bright field. Hybridization signals are visible as purple color development. Sense RNA probes did not produce any hybridization signals (data not shown). Bar = 50 μm.
To relate the period of transcript accumulation to the cell cycle phase, tissues were counterstained with a DNA-specific dye, 4′,6-diamidino-2-phenylindole (DAPI). Figure 7 shows mRNA localization of NtmybA1 and A2 and Nicta; CycB1;3 together with DAPI-stained nuclei from the same sections. The mitotic index in the overall meristematic region containing cells in which Nicta;CycB1;3 transcript was observed was no more than 15%. However, much higher mitotic indices were observed in the cell populations showing signals for either Nicta;CycB1;3 or NtmybA1 or A2 mRNA. The mitotic index of cells labeled with the Nicta; CycB1;3 probe was 78%, consisting of 43% prophase, 23% metaphase, 6% anaphase, and 5% telophase (Table 1). Similar but slightly lower mitotic indices were observed among cells showing signals for NtmybA1 (67%: 60% prophase and 7% metaphase) and NtmybA2 (70%: 66% prophase and 4% metaphase). Neither anaphase nor telophase nuclei were observed among cells labeled with NtmybA1 or A2 probe (Table 1). These results indicate that NtmybA1 and A2 mRNAs are accumulated at a similar time, mainly in M-phase, and that this timing is slightly earlier than that of Nicta; CycB1;3 mRNA. These patterns of cell cycle–regulated transcript accumulation in shoot apices are consistent with the results of RNA gel bot analysis (Figure 5). In the meristematic regions, nearly 100% of prophase cells were labeled with Nicta;CycB1;3, NtmybA1, and A2 probes. This result is consistent with the idea that transcription factors (NtmybA1 and A2) and their target activity are present in the same type of cells.
Figure 7.
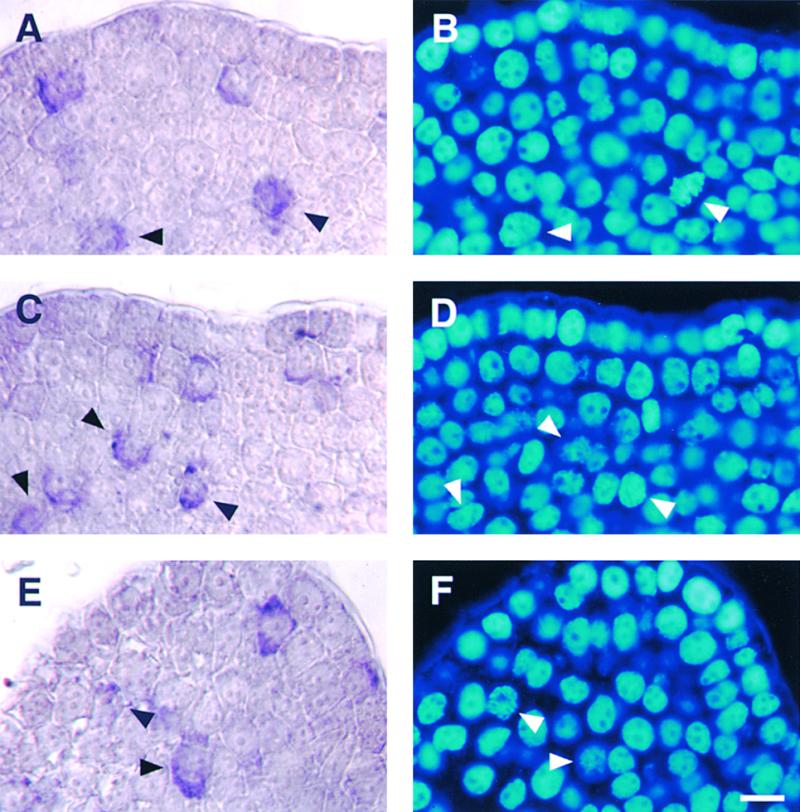
Periodic Accumulation of NtmybA1 and A2 Transcripts during the Cell Cycle in Tobacco Shoot Apices.
Longitudinal sections of shoot apices were hybridized with digoxigenin-labeled antisense RNA probes of Nicta;CycB1;3, NtmybA1, and NtmybA2. They were counterstained with DAPI and viewed with bright field or epifluorescence. Arrowheads indicate mitotic cells with hybridization signals.
(A) and (B) Nicta;CycB1;3.
(C) and (D) NtmybA1.
(E) and (F) NtmybA2.
(A), (C), and (E) show transcript localization (bright field). (B), (D), and (F) show DAPI-stained nuclei (epifluorescence). Bar in (F) = 10 μm.
Table 1.
Distribution of Cell Cycle Stages among Cells Labeled by In Situ Hybridization on Tobacco Shoot Apices
| Percent of Cells in a Particular Cell Cycle Stagea
|
|||||
|---|---|---|---|---|---|
| Cell Populations | Interphase | Prophase | Metaphase | Anaphase | Telophase |
| Overall meristemb | 85 | 11 | 2 | 1 | 1 |
| Cells labeled with following probesc | |||||
| Nicta;CycB1;3 | 23 | 43 | 23 | 6 | 5 |
| NtmybA1 | 33 | 60 | 7 | 0 | 0 |
| NtmybA2 | 30 | 66 | 4 | 0 | 0 |
Cell cycle stages were determined by microscopic observation of DAPI-stained sections.
Overall meristematic region of tobacco shoot apices.
Cells labeled with each probe by in situ hybridization on tobacco shoot apices.
NtmybA1, A2, and B Modulate the Activity of MSA-Containing Promoters
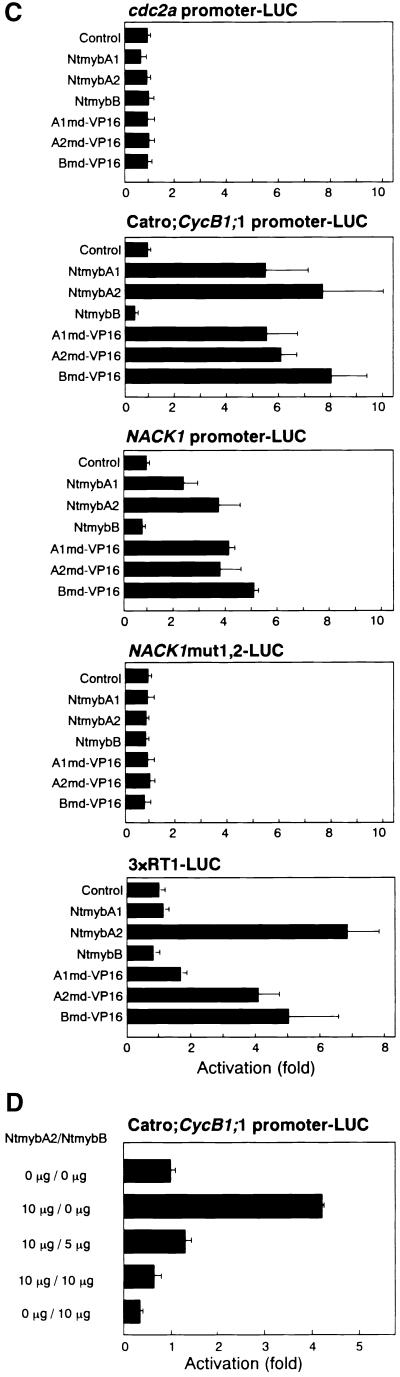
To determine whether the three Ntmyb proteins modulate the activity of promoters containing MSA elements, effector plas-mids were constructed that express full-length NtmybA1, A2, and B and fusions between Myb domains of the Ntmyb proteins and the VP16 activation domain (Figure 8A). These effector plasmids were transfected into tobacco BY2 protoplasts along with LUC reporter plasmids. As a reporter, promoters from Arabidopsis cdc2a, C. roseus Catro;CycB1;1, and tobacco NACK1 were examined (Figure 8B). The cdc2a promoter contains several putative Myb binding sequences (Imajuku et al., 1992; Chung and Parish, 1995) that differ from the consensus MSA motif (Ito, 2000). When cotransfected into BY2 protoplasts, cdc2a promoter activity was not affected by any effector (Figure 8C). Catro;CycB1;1 and NACK1 promoters were activated between fourfold and eightfold by cotransfection of the effector plasmids expressing full length NtmybA1 and A2. In contrast, cotransfection of the NtmybB effector resulted in a slight but significant decrease in reporter activity from the Catro; CycB1;1 and NACK1 promoters (Figure 8C). Both activation and repression of promoter activities were dependent on the presence of MSA elements, because neither effector affected the mutant NACK1 promoter (NACK1mut1,2) in which both of the two MSA-like sequences were mutated (Figures 8B and 8C).
Figure 8.
NtmybA1 and A2 Function as Activators, and NtmybB Functions as a Repressor.
(A) Scheme of effector plasmids used in transient assays. NtmybA1, A2, and B effector plasmids contained the corresponding full-length cDNAs. A1md–VP16, A2md–VP16, and Bmd–VP16 effector plasmids contained fusions between Myb domains from NtmybA1, A2, and B, respectively, and the VP16 activation domain (VP16). All of these constructs were placed between a double CaMV 35S promoter (2×CaMV 35S) and the terminator signal of the CaMV 35S gene (35S ter). R1, R2, and R3 motifs in the Myb domains are shown as black boxes.
(B) Scheme of reporter plasmids used in transient assays. Promoters from cdc2a, Catro;CycB1;1, and NACK1 genes were fused to the LUC reporter gene. In NACK1mut1,2–LUC reporter plasmids, both of two MSA elements were mutated. The 3×RT1–LUC reporter contained three tandem copies of the RT1 sequence (a 9-bp MSA motif from the Catro;CycB1;1 promoter) fused to the CaMV 35S basal promoter (positions −46 to +1). Numbers indicate nucleotide positions relative to the transcriptional start site (+1). Arrowheads indicate the positions and orientations of the MSA elements. Black arrowheads, wild-type MSA sequences; white arrowheads, mutant MSA sequences.
(C) Promoter activation by NtmybA1 and A2 is dependent on the MSA element. The LUC reporter plasmids cdc2a promoter–LUC, NACK1 promoter–LUC, Catro;CycB1;1 promoter–LUC, NACK1mut1,2–LUC, and 3×RT1–LUC were cotransfected with effector plasmid expressing full- length NtmybA1, A2, and B and Myb domain–VP16 fusions (A1md–VP16, A2md–VP16, and Bmd–VP16) into BY2 protoplasts. As a control, each reporter plasmid was transfected without effectors (Control).
(D) NtmybB represses transcriptional activation mediated by NtmybA2. The Catro;CycB1;1 promoter–LUC reporter plasmid was transfected into BY2 protoplasts together with various combinations of effector plasmids expressing NtmybA2 and B.
All LUC activities are expressed relative to the reporter construct alone (value set to 1). Error bars indicate ±sd.
All Myb domain–VP16 fusions, A1md–VP16, A2md–VP16, and Bmd–VP16, activated Catro;CycB1;1 and NACK1 reporters fourfold to eightfold but did not affect the NACK1mut1,2 reporter (Figure 8C). These results indicate that NtmybB, as well as NtmybA1 and A2, can bind to MSA-containing sequences in vivo. NtmybB has similar or identical DNA binding specificity to NtmybA1 and A2; therefore, it has the potential to repress transactivation mediated by NtmybA1 and A2. To determine if NtmybB can act as a competitive inhibitor, BY2 protoplasts were transfected with LUC reporter plasmids and the NtmybA2 effector plasmid together with different amounts of the NtmybB effector plasmid (Figure 8D). Cotransfection of the NtmybA2 effector plasmid activated the Catro;CycB1;1 reporter threefold to fourfold. This activation was reduced in a dose-dependent manner by cotransfection of the NtmybB effector. These results suggest that NtmybB, lacking transcriptional activation activity, blocked NtmybA2-mediated transactivation in vivo.
We have shown previously that a synthetic promoter, 3×RT1, containing three tandem copies of a 9-bp MSA motif (RT1 from the Catro;CycB1;1 promoter) fused to the cauliflower mosaic virus (CaMV) 35S basal promoter was sufficient to direct G2/M-phase–specific reporter gene expression in vivo (Ito et al., 1998a). To determine if this synthetic promoter can be activated by the Ntmyb proteins, we cotransfected the 3×RT1 reporter into BY2 protoplasts along with effector plasmids (Figures 8B and 8C). The 3×RT1 promoter was activated by the NtmybA2 effector approximately sevenfold and was repressed by the NtmybB effector. However, the reporter activity was not activated by the NtmybA1 or the A1md–VP16 effector (Figure 8C). Therefore, we conclude that interaction between NtmybA2 and the MSA elements is sufficient for promoter activation. NtmybA1 may require additional sequences for DNA binding because the 9-bp RT1 motif does not fully cover the MSA consensus sequence that is composed of 11 bases (Ito, 2000).
Activation of MSA-Containing Promoters by NtmybA1 and A2 Is Dominant over the Cell Cycle Position
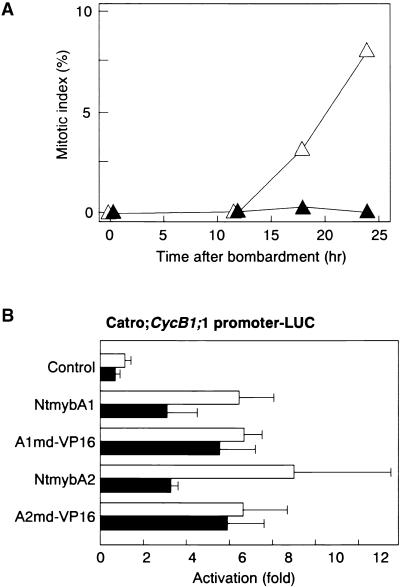
We observed significant activation of MSA-containing promoters by NtmybA1 and A2. In transient assays, this promoter activation may occur only in cells in a particular phase of the cell cycle. To determine if other G2/M-phase–specific factors are required in MSA activation by NtmybA1 and A2, we undertook transient assays in S-phase–arrested BY2 cells. BY2 cells from 8-day-old cultures, in which most of the cells are arrested in G1-phase, were bombarded with the Catro;CycB1;1 reporter and the effector plasmids expressing NtmybA1, A1md–VP16, NtmybA2, or A2md–VP16 (Figures 8A and 8B). The bombarded cells then were cultured in liquid medium with or without aphidicolin. Aphidicolin treatment leads to the arrest of BY2 cells in S-phase (Nagata et al., 1992). When cells were treated with aphidicolin, no increase of mitotic index was observed, indicating that cells were arrested in S-phase and did not enter G2- and M-phases. On the other hand, cells cultured without aphidicolin began to enter M-phase after 12 to 18 hr (Figure 9A).
Figure 9.
NtmybA1 and A2 Have Promoter Activation Abilities in S-Phase–Arrested Cells.
(A) Changes in mitotic index after bombardment. BY2 cells from 8-day-old culture were bombarded with plasmids and cultured in the presence or absence of aphidicolin. Mitotic index was measured at the times indicated after bombardment. Closed triangles, cells cultured with aphidicolin; open triangles, cells cultured without aphidicolin.
(B) Particle bombardment assay. As a reporter plasmid, Catro; CycB1;1 promoter–LUC was used. As effector plasmids, constructs expressing full length NtmybA1 and A2 and Myb domain–VP16 fusions (A1md–VP16 and A2md–VP16) were cobombarded. The bombarded cells were cultured in the presence or absence of aphidicolin for 24 hr, and LUC activity was assayed. As a control, the reporter plasmid was transfected without effector plasmids (Control). All LUC activities are expressed relative to the control (value set to 1). Error bars indicate ±sd. Closed bars, cells cultured with aphidicolin; open bars, cells cultured without aphidicolin.
All effector plasmids resulted in fourfold to sixfold activation of the Catro;CycB1;1 reporter in the presence of aphidicolin, indicating that NtmybA1 and A2 can activate the Catro;CycB1;1 promoter even in S-phase (Figure 9B), during which B-type cyclin genes normally are repressed (Figure 5A). However, the activation was significantly higher in cells cultured without aphidicolin than in aphidicolin-treated cells bombarded with NtmybA1 and A2 effectors. Such differences in activation between aphidicolin-treated and nontreated cells were not observed when A1md–VP16 and A2md–VP16 effector plasmids were bombarded (Figure 9B). These results indicate that NtmybA1 and A2 are sufficient to activate MSA-containing promoters, but the extent of activation is modulated in a phase-dependent manner. The cell cycle–dependent activation could be dependent, in part, on the C-terminal region outside of the Myb domain of NtmybA1 and A2, because the transactivation activities of Myb domain–VP16 fusions were not affected by aphidicolin treatment.
DISCUSSION
MSA Elements in Plant Promoters Provide a Common Mechanism for Directing G2/M-Phase–Specific Gene Expression
We have shown previously that MSA elements in the cyclin B1 gene Catro;CycB1;1 are necessary and sufficient for G2/M-phase–specific promoter activation (Ito et al., 1998a). Thus, the MSA element is one of the primary determinants for the timing of promoter activation of this gene. In plants, most B-type cyclin genes (B1 and B2 classes) are expressed at a similar time in the cell cycle. Their expression is induced in late G2-phase and reaches its peak in M-phase (Ito, 2000). Thus, it is likely that all B-type cyclin genes expressed at the same time are regulated by the same MSA-mediated mechanism. This idea is supported by reports that B-type cyclin promoters from Arabidopsis and C. roseus are activated at the correct time in the heterologous host, tobacco BY2 cells (Shaul et al., 1996; Ito et al., 1997). In fact, MSA-like motifs are present in upstream regions of several B-type cyclin genes from Arabidopsis, tobacco, and soybean (Ito et al., 1998a).
Our evidence demonstrates that functional MSA elements are not restricted to B-type cyclin promoters but also are found in other genes that show similar expression patterns. Transcripts for two tobacco genes, NACK1 and NACK2, accumulate with timing nearly identical to that of B-type cyclin genes during the cell cycle (Machida et al., 1998). In this study, we demonstrate that the activity of the NACK1 promoter is cell cycle regulated in a manner similar to that of the cyclin B1 gene. The G2/M-phase–specific activation of the NACK1 promoter is dependent on the presence of MSA-like motifs, because a mutant NACK1 promoter in which both of two MSA-like motifs were mutated completely lost the G2/M-phase–specific promoter activity in vivo. We conclude that the MSA-like sequences in the NACK1 promoter are functional MSA elements and suggest that MSA-dependent transcriptional control could provide a general mechanism for the G2/M-phase–specific transcription of a defined set of plant genes, analogous to the MCM1/SFF system in yeast (Breeden, 2000).
MSA Binding Factors Are Three Repeat Myb Proteins
Using the yeast one-hybrid system, we identified three distinct Myb-related proteins, NtmybA1, A2, and B, that interact specifically with the MSA sequence. Genes of the Myb family have been found in all major eukaryotic groups. In animals and yeast, the number of identified Myb genes is relatively small (Lipsick, 1996), but a much larger number of Myb genes has been identified in plants; Arabidopsis contains more than 100 Myb genes (Kranz et al., 1998). Unlike the Myb family in animals, the majority of plant Myb proteins is of the R2R3 type. All three Ntmyb proteins are of the R1R2R3 class, more similar in structure to the animal A-, B-, and c-Myb than to plant R2R3-Myb. Correspondingly, the MSA consensus sequence, TC(T/C)AACGG(T/C)(T/C)A (Ito, 2000), resembles the c-Myb binding sequence, (A/C)CCAACNG(C/T)C (Howe and Watson, 1991), which differs substantially from the consensus sequence, CC(T/C)ACC, recognized by R2R3-Myb (Grotewold et al., 1994; Solano et al., 1997).
Two Arabidopsis R1R2R3-type Myb genes, MYB3R-1 and MYB3R-2, have been described recently (Braun and Grotewold, 1999; Kranz et al., 2000). In addition, three other Arabidopsis R1R2R3-Myb genes can be found in currently available sequences in the database. Thus, R1R2R3-Myb genes seem to constitute a small gene family in plants. Kranz et al. (2000) also demonstrated that R1R2R3-Myb genes occurred in different plant evolutionary lineages, including mosses, ferns, and monocots. These findings showed that R1R2R3-type Myb genes predate the divergence of the animal and plant lineages and suggest that the plant R2R3-Myb genes are derived from the more ubiquitous R1R2R3-Myb genes (Kranz et al., 2000). Thus, it seems likely that the R1R2R3 type of plant Myb proteins would have a conserved function in eukaryotes, in contrast to plant R2R3-Myb, which is believed to be involved in plant-specific processes (Kranz et al., 2000).
Our results suggest that this prediction is broadly true in that both plant and animal R1R2R3-Myb proteins are involved in cell cycle control (Thompson and Ramsay, 1995; this study). However, the precise roles of Ntmyb and the animal Myb proteins in regulating cell cycle gene expression seem to have diverged. All three animal Myb proteins, A-, B-, and c-Myb, are regulated such that their maximal activity occurs during late G1- and S-phases (Weston, 1998). c-Myb is involved in the transcriptional regulation of genes such as CDC2 (Ku et al., 1993), DNA polymerase α (Venturelli et al., 1990), proliferating cell nuclear antigen (Travali et al., 1991), and cyclin A (Müller et al., 1999), all of which are important for G1/S-phase transition. Ntmyb proteins, however, do not activate the plant cdc2a promoter, which lacks any consensus MSA sequences but affects the expression of G2/M-phase–specific genes. Thus, the Ntmyb proteins appear to have a different role in cell cycle control than their animal counterparts, namely, to regulate G2/M-phase progression.
An Antagonistic Mechanism for MSA-Mediated Transcriptional Control
We have shown that NtmybA1 and A2 act as transcriptional activators for MSA-dependent transcription in BY2 cells. NtmybB also can bind to the MSA sequence in yeast and in vitro, but it lacks transactivation activity in BY2 protoplasts. Indeed, we have shown in an in vivo competition experiment that NtmybB can block NtmybA2-mediated transactivation in BY2 protoplasts. NtmybB therefore may be a repressor of transcriptional activation controlled by NtmybA2 and possibly by NtmybA1. We propose that competitive DNA binding of Ntmyb proteins with different activities for transactivation may provide a mechanism for the transcriptional regulation of plant G2/M-phase–specific genes. Indeed, a balance between activators and repressors is thought to be a common mechanism to reduce inappropriate triggering of biological responses, and such a mechanism could help to explain the unusually tight window of expression displayed by plant B-type cyclin genes.
In agreement with their similarity in structure and transactivation activity, NtmybA1 and A2 showed similar kinetics of transcript accumulation during the cell cycle of BY2 cells. The accumulation of their mRNAs was induced just before the appearance of cyclin B1 mRNA, and in situ hybridization experiments suggest that the expression of NtmybA1 and A2 declines just before that of cyclin B1. These observations are consistent with the idea that NtmybA1 and A2 are involved in the induction of B-type cyclin genes. The accumulation of NtmybB mRNA was cell cycle independent in BY2 cells and in meristems. Thus, NtmybA1 and A2 genes themselves are cell cycle regulated in a manner similar to but preceding their target gene. We have shown that NtmybA1 and A2 are sufficient for the activation of MSA-containing promoters irrespective of cell cycle position, although their transactivation activity may be modulated dependent on cell cycle position. Thus, the mechanisms governing the level of NtmybA1 and A2 proteins should be of primary importance for the transcriptional control of B-type cyclin genes. One model suggests that a constant level of NtmybB represses mitosis-specific activation of MSA elements until the levels of NtmybA1 and A2 increase above a threshold level. Increased levels of NtmybA1 and A2 then compete with NtmybB for binding to the MSA elements and permit the transcription of G2/M-phase–specific genes. Such a mechanism could explain the stringent transcriptional regulation of plant B-type cyclin genes. We are currently studying the promoters of the NtmybA1 and A2 genes with the aim of discovering the primary controls of cell cycle progression.
METHODS
Synchronization and Stable Transformation of Tobacco BY2 Cells
Maintenance, synchronization, and Agrobacterium tumefaciens–mediated transformation of tobacco (Nicotiana tabacum) BY2 cells were performed as described previously (Ito et al., 1998a). Firefly luciferase (LUC) reporter plasmids were constructed with wild-type and mutant NACK1 promoters. The wild-type NACK1 promoter fragment from −496 to +68 (relative to the transcriptional start site) was generated by polymerase chain reaction (PCR) and cloned into a BamHI–HindIII-digested pBI-LUC binary vector (Ito et al., 1998a) to generate pBI-NACK1wt. In addition, the following derivatives containing point mutations were constructed: pBI-NACK1mut1 and pBI-NACK1mut2, containing mutations in M-specific activator (MSA)-like sequences at nucleotide positions −163 to −173 and −49 to −59, respectively. In each mutant, the central core pentamer of the MSA element, AACGG, was changed to ATTGG. pBI-NACK1mut1,2 contains the combination of both mutations described above. All mutants were generated by PCR using appropriate primers, and the identities of the PCR products were confirmed by sequencing. These constructs were used to transform BY2 cells.
RNA Extraction and Gel Blot Analysis
Small-scale RNA extractions were performed with TRIzol reagent (Gibco BRL, Rockville, MD) according to the manufacturer's instructions. RNA gel blot analysis and synthesis of randomly primed probes were performed as described by Ito et al. (1997). RNA gel blot hybridization was performed as described by Ito et al. (1998b). RNA signals were detected using the Fuji Imaging Analyzer (model BAS2000; Fuji Photofilm, Tokyo, Japan). Gene-specific probes for NtmybA1, A2, and B were generated by PCR amplification of the 3′ untranslated region of each cDNA. The PCR products were cloned into the pGEM-T EASY vector (Promega, Madison, WI).
Yeast One-Hybrid Screening
A yeast one-hybrid system (Clontech, Palo Alto, CA) was used to screen a BY2 cell cDNA library to identify proteins that bound to the MSA sequence. Two HIS3 reporter plasmids, CYM-HIS3 and NACK1-HIS3, were constructed by inserting MSA-containing sequences into EcoRI–XbaI intervals of the pHISi and pHISi-1 integration vectors, respectively. The CYM-HIS3 reporter construct contains six tandem copies of a 9-bp MSA motif (−66 to −56) from the Catro;CycB1;1 promoter from Catharanthus roseus, in which each motif was separated by six adenine residues. The NAKC1-HIS3 reporter construct contains three tandem repeats of the MSA sequence (−60 to −48) from the NACK1 promoter. These MSA repeat sequences were prepared by annealing complementary oligonucleotides that carry EcoR1 5′ and Xba1 3′ linkers. The reporter constructs were integrated into the genome of the yeast strain YM4271. A cDNA library was constructed from mRNA prepared from 2-day-old BY2 cells using an oligo(dT) primer and ligated downstream of the GAL4 activation domain of the pGAD10 yeast expression vector. The cDNA library was screened by transformation of the reporter strains, and the transformants were selected on medium lacking Leu and His and containing 20 mM 3-amino-1,2,4-triazole.
Electrophoretic Mobility Shift Assays
The NtmybA1 and A2 cDNA fragments, which contain internal NcoI sites at the translation initiation sites, were amplified by PCR. The NtmybB cDNA fragment was amplified by mutagenic PCR to create a NcoI site at the translation initiation site. The PCR fragments were cloned into pSPUTK vector (Stratagene, La Jolla, CA) such that translation initiated at the ATG in the NcoI site located in the Kozak sequence of the vector. Ntmyb proteins were produced by in vitro transcription/translation using a TnT-coupled reticulocyte lysate system (Promega). A 4-μL aliquot of the translation reaction was incubated in a 20-μL solution of 20 mM Hepes, pH 7.9, 20% glycerol, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 500 ng of denatured and sheared calf thymus DNA, 100 ng of poly(dI-dC), and 5 ng of 32P-labeled double-stranded oligonucleotide probe DNA at 30°C for 30 min. Synthetic DNA of the RT1 sequence (see Figure 4A), having an additional GATC sequence attached to each 5′ end, was end labeled with the Klenow fragment of DNA polymerase I and used as a DNA probe. In competition experiments, 200 ng of unlabeled oligonucleotide was added. Samples were electrophoresed on 5% acrylamide gel (acrylamide:bisacrylamide, 40:1) in 0.5 × TBE (1 × TBE is 22.5 mM Tris-borate, pH 8.0, and 0.25 mM EDTA).
In Situ Hybridization
Terminal shoot apices were isolated from 40-day-old tobacco plants and fixed immediately for 6 hr in 50 mM phosphate buffer, pH 7.0, 4% paraformaldehyde, and 0.01% Tween 20. Afterward, the tissues were dehydrated through an ethanol series and embedded in Paraplast Plus (Oxford Labware, St. Louis, MO). The tissues were cut into 8-μm sections and mounted on aminopropyltriethoxy silane–coated slides (Matsunami Glass, Osaka, Japan). RNA probes were produced from the 3′ untranslated region of each cDNA cloned in the pGEM-T EASY plasmid vector (Promega). RNA probes were synthesized and labeled by in vitro transcription using T7 or SP6 RNA polymerase (Stratagene) with a digoxigenin RNA labeling mixture (Roche Diagnostics, Mannheim, Germany). Hybridization and immunological detection were performed as described by Matsunaga et al. (1996) with the following modifications. First, the antibody was changed to anti-digoxigenin Fab fragments conjugated with alkaline phosphatase (Roche Diagnostics). Second, detection was performed using the nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate detection system (Roche Diagnostics). After the detection, preparations were stained with 1 μg·mL−1 4′,6-diamidino-2-phenylindole (DAPI) in 1% Triton X-100.
Transient Expression Assay with BY2 Protoplasts
Tobacco BY2 protoplasts were prepared from 3-day-old cells as described previously (Evans and Bravo, 1983). Transfection of the protoplasts was performed by polyethylene glycol–mediated direct gene transfer (Bilang et al., 1994), and transfected protoplasts were cultured for 24 hr at 27°C. Transfections were performed in triplicate, and at least two independent transfection experiments were performed with each construct. A dual-luciferase reporter assay was conducted according to the manufacturer's instructions (Promega). For each transfection, 10 μg of effector plasmid and 20 μg of reporter plasmid were used unless indicated otherwise. The 35S-Rluc construct, in which the Renilla reniformis luciferase reporter gene is placed downstream of the cauliflower mosaic virus (CaMV) 35S promoter, was obtained from Saeko Kitakura (Nagoya University, Nagoya, Japan). This plasmid was cotransfected as an internal control of transfection efficiency.
Plasmid Construction for Transient Expression Assays
Two parental plasmids, pUC-LUC and pCL46, were used. pUC-LUC was constructed by inserting a HindIII–SacI fragment of pDO432 (Nishiuchi et al., 1995) into the HindIII–SacI interval of pBI221 (Clontech). This plasmid contains the CaMV 35S promoter upstream of the LUC reporter gene. For cloning of the promoter fragments, the CaMV 35S promoter was removed by HindIII–BamHI digestion and the promoter fragments were cloned into the HindIII–BamHI interval of pUC-LUC. For the construction of pCL46, a fragment of the CaMV 35S basal promoter (−46 to +1) was synthesized by PCR using pDO432 as a template with a set of linker/primers (HindIII–SalI 5′ and BamHI 3′). The resulting PCR fragment was used to replace the full-length CaMV 35S promoter in pUC-LUC with the 35S basal promoter.
The plasmid pCL46 contains HindIII and SalI sites upstream of the CaMV 35S basal promoter–LUC fusion. The promoter regions of Arabidopsis cdc2a (−1277 to −2, where the transcription initiation site has been assigned provisionally to the 5′ end of the longest cDNA; Imajuku et al., 1992), C. roseus Catro;CycB1;1 (−462 to +102), and tobacco NACK1 (−496 to +68) were amplified by PCR and cloned into BamHI–HindIII intervals of pUC-LUC to generate reporter plasmids. The mutated NACK1 promoter (NACK1mut1,2) was prepared as described above and cloned into the BamHI–HindIII interval of pUC-LUC to create the NACK1mut1,2–LUC reporter plasmid. The 3×RT1 sequence contains three tandem copies of a 9-bp MSA motif that are separated by six adenine residues (Ito et al., 1998a). This construct was prepared by annealing complementary oligonucleotides that carry HindIII 5′ and SalI 3′ linkers. They were ligated into the HindIII–SalI site of pCL46 to create reporter plasmid 3×RT1–LUC.
For construction of effector plasmids, full-length NtmybA1, A2, and B were amplified by PCR and cloned downstream of the double CaMV 35S promoter of pJIT60 (Guerineau and Mullineaux, 1993). Effector plasmids expressing fusions of Myb domains and the VP16 activation domain (A1md–VP16, A2md–VP16, and Bmd–VP16) were constructed in pGAL4/1×VP16 (Schwechheimer et al., 1998). Myb domains of NtmybA1 (amino acids 1 to 225), A2 (1 to 226), and B (1 to 230) were amplified by PCR and cloned into HindIII–BamHI intervals, replacing the GAL4 DNA binding domain. In these plasmids, the sequences encoding the Myb domains were fused in frame with the sequence encoding the VP16 activation domain downstream of the double CaMV 35S promoter. As a control plasmid, the β-glucuronidase (GUS) fragment from pBI121 (Clontech) was inserted into a HindIII–EcoRI interval of pJIT60 to create pJIT-GUS. This plasmid was added to equalize the amount of DNA in transfection experiments in which either no effector plasmid or different amounts of effector plasmids were used.
Particle Bombardment
BY2 cells from an 8-day-old culture were diluted 10 times with fresh medium, spread in a thin layer over filter papers, and subjected to particle bombardment assays. Particle gun bombardment was performed as follows: 1-μm gold biolistic particles (500 μg; Bio-Rad, Hercules, CA) were coated with 0.66 μg of reporter plasmid and 1 μg of effector plasmid or blank plasmid (pJIT-GUS). As an internal control, 0.33 μg of the 35S-Rluc plasmid was used. Bombardment was performed with a GIE-III machine (Tanaka, Sapporo, Japan). After bombardment, cells were suspended in fresh medium with or without aphidicolin (20 μg·mL−1) and cultured at 27°C. The mitotic index was measured at 0, 12, 18, and 24 hr after bombardment. Cells were harvested 24 hr after bombardment, and firefly and R. reniformis luciferase activities were assayed as described above.
Other Nucleic Acid Procedures
DNA sequences were determined with an automated DNA sequencer (model 4000; Li-Cor) using a Thermo Sequenase cycle sequencing kit (Amersham Pharmacia Biotech, Buckinghamshire, UK) and IRD41 infrared dye–labeled primers (Aloka, Tokyo, Japan). Rapid amplification of 3′ cDNA ends (RACE) was performed to obtain the 3′ region of NtmybA1 cDNA, according to the procedure of Nishida et al. (2000), using appropriate primers specific to NtmybA1. The product was cloned into pGEM-T EASY vector (Promega) and sequenced.
GenBank Accession Numbers
The GenBank accession numbers are as follows: NtmybA1, AB056122; NtmybA2, AB056123; NtmybB, AB056124; c-Myb (M15024), A-Myb (X13294), and B-Myb (X13293) from human; Drosophila melanogaster Myb (X05939); MYB3R-1 (AF176005), MYB3R-2 (AF218054), AtMYB2 (J02390), and AtGL1 (027,900) from Arabidopsis; ZmMYB1 (P20024), ZmMYB3 (P20025), and ZmMYBC (P10290) from maize; PhMYB1 (Z13996), PhMYB2 (Z13997), and PhMYB3 (Z13998) from petunia; AmMIXTA (S44926) from Antirrhinum majus; and HvMYB1 (P20026) from barley; other Arabidopsis R1R2R3-Myb genes, AF214117, T48253, and T48510.
Acknowledgments
We thank Prof. Chris Lamb (John Innes Centre, Norwich, UK) for critical reading of the manuscript. We are grateful to Atsuhiko Aoyama (University of Tokyo) for technical advice. The BY2 cDNA expression library was kindly supplied by Dr. Yosuke Takahashi (University of Tokyo). This work was supported in part by a Grant-in-Aid for Scientific Research (09740589) from the Ministry of Education, Science, and Culture, Japan, by Postdoctoral Fellowships for Research Abroad from the Japan Society for the Promotion of Science, and by the Biotechnology and Biological Sciences Research Council, UK.
References
- Albani, D., Mariconti, L., Ricagno, S., Pitto, L., Moroni, C., Helin, K., and Cella, R. (2000). DcE2F, a functional plant E2F-like transcriptional activator from Daucus carota. J. Biol. Chem. 275, 19258–19267. [DOI] [PubMed] [Google Scholar]
- Althoefer, H., Schkeiffer, A., Wassmann, K., Nordheim, A., and Ammerer, G. (1995). Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 5917–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilang, R., Klöti, A., Schrott, M., and Potrykus, I. (1994). PEG-mediated direct gene transfer and electroporation. In Plant Molecular Biology Manual, S.B. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. A1, 1–16.
- Black, A.R., and Azizkhan-Clifford, J. (1999). Regulation of E2F: A family of transcription factors involved in proliferation control. Gene 237, 281–302. [DOI] [PubMed] [Google Scholar]
- Brandeis, M., and Hunt, T. (1996). The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 15, 5280–5289. [PMC free article] [PubMed] [Google Scholar]
- Braun, E.L., and Grotewold, E. (1999). Newly discovered plant c-myb-like genes rewrite the evolution of the plant myb gene family. Plant Physiol. 121, 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden, L.L. (2000). Cyclin transcription: Timing is everything. Curr. Biol. 10, R586–R588. [DOI] [PubMed] [Google Scholar]
- Chung, S.K., and Parish, R.W. (1995). Studies on the promoter of the Arabidopsis thaliana cdc2a gene. FEBS Lett. 362, 215–219. [DOI] [PubMed] [Google Scholar]
- Cogswell, J.P., Godlevski, M.M., Bonham, M., Bisi, J., and Babiss, L. (1995). Upstream stimulatory factor regulates expression of the cell cycle-dependent cyclin B1 gene promoter. Mol. Cell. Biol. 15, 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20, 503–508. [DOI] [PubMed] [Google Scholar]
- Doerner, P., Jorgensen, J.-E., You, R., Steppuhn, J., and Lamb, C. (1996). Control of root growth and development by cyclin expression. Nature 380, 520–523. [DOI] [PubMed] [Google Scholar]
- Evans, D.A., and Bravo, J.E. (1983). Plant protoplast isolation and culture. Int. Rev. Cytol. 16 (suppl.), 33.–53. [Google Scholar]
- Fobert, P., Coen, E.S., Murphy, G.J.P., and Doonan, J.H. (1994). Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO J. 13, 616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, J., Ardelt, B., Traganos, F., and Darzynkiewicz, Z. (1994). Unscheduled expression of cyclin B1 and cyclin E in several leukemic and solid tumor lines. Cancer Res. 54, 4285–4288. [PubMed] [Google Scholar]
- Grotewold, E., Drummond, B.J., Bowen, B., and Peterson, T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76, 543–553. [DOI] [PubMed] [Google Scholar]
- Guerineau, F., and Mullineaux, P. (1993). Plant transformation and expression vectors. In Plant Molecular Biology Labfax, R.R.D. Croy, ed (Oxford, UK: Bios Scientific Publishers), pp. 121–147.
- Howe, K.M., and Watson, R.J. (1991). Nucleotide preference in sequence-specific recognition of DNA by c-myb protein. Nucleic Acids Res. 19, 3913–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, A., McKenna, W.G., and Muschel, R.J. (1998). Cell cycle dependent usage of transcriptional start sites: A novel mechanism for regulation of cyclin B1. J. Biol. Chem. 273, 31505–31509. [DOI] [PubMed] [Google Scholar]
- Imajuku, Y., Hirayama, T., Endoh, H., and Oka, A. (1992). Exon-intron organization of the Arabidopsis thaliana protein kinase genes CDC2a and CDC2b. FEBS Lett. 304, 73–77. [DOI] [PubMed] [Google Scholar]
- Ito, M. (1998). Cell cycle dependent gene expression. In Plant Cell Division, D. Francis, D. Dudits, and D. Inzé, eds (London, Portland Press), pp. 165–186.
- Ito, M. (2000). Factors controlling cyclin B expression. Plant Mol. Biol. 43, 677–690. [DOI] [PubMed] [Google Scholar]
- Ito, M., Criqui, M.-C., Sakabe, M., Ohno, T., Hata, S., Kouchi, H., Hashimoto, J., Fukuda, H., Komamine, A., and Watanabe, A. (1997). Cell-cycle-regulated transcription of A- and B-type plant cyclin genes in synchronous cultures. Plant J. 11, 983–992. [DOI] [PubMed] [Google Scholar]
- Ito, M., Iwase, M., Kodama, H., Lavisse, P., Komamine, A., Nishihama, R., Machida, Y., and Watanabe, A. (1998. a). A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, M., Kodama, H., Komamine, A., and Watanabe, A. (1998. b). Expression of extensin genes is dependent on the stage of the cell cycle and cell proliferation in suspension-cultured Catharanthus roseus cells. Plant Mol. Biol. 36, 343–351. [DOI] [PubMed] [Google Scholar]
- Jin, H., and Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. [DOI] [PubMed] [Google Scholar]
- Katula, K.S., Wright, K.L., Paul, H., Surman, D.R., Nuckolls, F.J., Smith, J.W., Ting, J.P.-Y., Yates, J., and Cogswell, J.P. (1997). Cyclin-dependent kinase activation and S-phase induction of the cyclin B1 gene are linked through the CCAAT elements. Cell Growth Differ. 8, 811–820. [PubMed] [Google Scholar]
- Koch, C., and Nasmyth, K. (1994). Cell cycle regulated transcription in yeast. Curr. Opin. Cell Biol. 6, 451–459. [DOI] [PubMed] [Google Scholar]
- Kouchi, H., Sekine, M., and Hata, S. (1995). Distinct classes of mitotic cyclins are differentially expressed in the soybean shoot apex during the cell cycle. Plant Cell 7, 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz, H.D., et al. (1998). Towards functional characterization of the member of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. [DOI] [PubMed] [Google Scholar]
- Kranz, H., Scholz, K., and Weisshaar, B. (2000). c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J. 21, 231–235. [DOI] [PubMed] [Google Scholar]
- Ku, D.H., Wen, S.C., Engelhard, A., Nicolaides, N.C., Lipson, K.E., Marino, T.A., and Calabretta, B. (1993). c-myb transactivates cdc2 expression via Myb binding sites in the 5′-flanking region of the human cdc2 gene. J. Biol. Chem. 268, 2255–2259. [PubMed] [Google Scholar]
- Lew, D.J., Dulic, V., and Reed, S.I. (1991). Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell 66, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Lipsick, J.S. (1996). One billion years of Myb. Oncogene 13, 223–235. [PubMed] [Google Scholar]
- Machida, Y., Nakashima, M., Morikiyo, K., Banno, H., Ishikawa, M., Soyano, T., and Nishihama, R. (1998). MAPKKK-related protein kinase NPK1: Regulation of the M phase of the plant cell cycle. J. Plant Res. 111, 243–246. [Google Scholar]
- Magyar, Z., Atanassova, A., De Veylder, L., Rombauts, S., and Inze, D. (2000). Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Lett. 486, 79–87. [DOI] [PubMed] [Google Scholar]
- Maher, M., Cong, F., Kindelberger, D., Nasmyth, K., and Dalton, S. (1995). Cell cycle-regulated transcription of the CLB2 gene is dependent on Mcm1 and Ternary complex factor. Mol. Cell. Biol. 15, 3129–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga, S., Kawano, S., Takano, H., Uchida, H., Sakai, A., and Kuroiwa, T. (1996). Isolation and developmental expression of male reproductive organ-specific genes in a dioecious campion, Melandrium album (Silene latifolia). Plant J. 10, 679–689. [DOI] [PubMed] [Google Scholar]
- McKinney, J.D., and Heintz, N. (1991). Transcriptional regulation in the eukaryotic cell cycle. Trends Biochem. Sci. 16, 430–435. [DOI] [PubMed] [Google Scholar]
- Mironov, V., De Veylder, L., Van Montagu, M., and Inzé, D. (1999). Cyclin-dependent kinases and cell division in plants: The nexus. Plant Cell 11, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, C., Yang, R., Idos, G., Tidow, N., Diederichs, S., Koch, O.M., Verbeek, W., Bender, T.P., and Koeffler, H.P. (1999). c-myb transactivates the human cyclin A1 promoter and induces cyclin A1 gene expression. Blood 94, 4255–4262. [PubMed] [Google Scholar]
- Müller, R. (1995). Transcriptional regulation during the mammalian cell cycle. Trends Genet. 11, 173–178. [DOI] [PubMed] [Google Scholar]
- Nagata, T., Nemoto, Y., and Hasezawa, S. (1992). Tobacco BY2 cell line as the “HeLa” cell in cell biology of higher plants. Int. Rev. Cytol. 132, 1–30. [Google Scholar]
- Nishida, I., Sugiura, M., Enju, A., and Nakamura, M. (2000). A second gene for acyl-(acyl-carrier-protein):glycerol-3-phosphate acyltransferase in squash, Cucurbita moschata cv. Shirogikuza, codes for an oleate-selective isozyme: Molecular cloning and protein purification studies. Plant Cell Physiol. 41, 1381–1391. [DOI] [PubMed] [Google Scholar]
- Nishiuchi, T., Nakamura, T., Abe, T., Kodama, H., Nishumura, M., and Iba, K. (1995). Tissue-specific and light-responsive regulation of the promoter region of the Arabidopsis thaliana chloroplast ω-3 fatty acid desaturase gene (FAD7). Plant Mol. Biol. 29, 599–609. [DOI] [PubMed] [Google Scholar]
- Piaggio, G., Farina, A., Perrotti, D., Manni, I., Fuschi, P., Sacchi, A., and Gaetano, C. (1995). Structure and growth-dependent regulation of the human cyclin B1 promoter. Exp. Cell Res. 216, 396–402. [DOI] [PubMed] [Google Scholar]
- Pines, J., and Hunter, T. (1990). p34cdc2: The S and M kinase? New Biol. 2, 389–401. [PubMed] [Google Scholar]
- Ramirez-Parra, E., and Gutierrez, C. (2000). Characterization of wheat DP, a heterodimerization partner of the plant E2F transcription factor which stimulates E2F-DNA binding. FEBS Lett. 486, 73–78. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra, E., Xie, Q., Boniotti, M.B., and Gutierrez, C. (1999). The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G(1)/S regulators. Nucleic Acids Res. 27, 3527–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, H.E., Lew, D.J., Henze, M., Sugimoto, K., and Reed, S.I. (1992). Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 6, 2021–2034. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Smith, C., and Bevan, M.W. (1998). The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: Design of modular transcription factors for high-level expression. Plant Mol. Biol. 36, 195–204. [DOI] [PubMed] [Google Scholar]
- Sekine, M., Ito, M., Uemukai, K., Maeda, Y., Nakagami, H., and Shinmyo, A. (1999). Isolation and characterization of the E2F-like gene in plants. FEBS Lett. 460, 117–122. [DOI] [PubMed] [Google Scholar]
- Shaul, O., Mironov, V., Burssens, S., Van Montagu, M., and Inzé, D. (1996). Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc. Natl. Acad. Sci. USA 93, 4868–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano, R., Fuertes, A., Sanchez-Pulido, L., Valencia, A., and Paz-Ares, J. (1997). A single residue substitution causes a switch from the dual DNA binding specificity of plant transcription factor MYB.Ph3 to the animal c-MYB specificity. J. Biol. Chem. 272, 2889–2895. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D., and Futcher, B. (1998). Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, M.A., and Ramsay, R.G. (1995). Myb: An old oncoprotein with new roles. BioEssays 17, 341–350. [DOI] [PubMed] [Google Scholar]
- Travali, S., Ferber, A., Reiss, K., Sell, C., Koniecki, J., Calabretta, B., and Baserga, R. (1991). Effect of the myb gene product on expression of the PCNA gene in fibroblasts. Oncogene 6, 887–894. [PubMed] [Google Scholar]
- Tréhin, C., Glab, N., Perennes, C., Planchais, S., and Bergounioux, C. (1999). M phase-specific activation of the Nicotiana sylvestris cyclin B1 promoter involves multiple regulatory elements. Plant J. 17, 263–273. [Google Scholar]
- Venturelli, D., Travali, S., and Calabretta, B. (1990). Inhibition of T-cell proliferation by a MYB antisense oligomer is accompanied by selective down-regulation of DNA polymerase α expression. Proc. Natl. Acad. Sci. USA 87, 5963–5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston, K. (1998). Myb proteins in life, death and differentiation. Curr. Opin. Genet. Dev. 8, 76–81. [DOI] [PubMed] [Google Scholar]