Abstract
We reported previously that three ERF transcription factors, tobacco ERF3 (NtERF3) and Arabidopsis AtERF3 and AtERF4, which are categorized as class II ERFs, are active repressors of transcription. To clarify the roles of these repressors in transcriptional regulation in plants, we attempted to identify the functional domains of the ERF repressor that mediates the repression of transcription. Analysis of the results of a series of deletions revealed that the C-terminal 35 amino acids of NtERF3 are sufficient to confer the capacity for repression of transcription on a heterologous DNA binding domain. This repression domain suppressed the intermolecular activities of other transcriptional activators. In addition, fusion of this repression domain to the VP16 activation domain completely inhibited the transactivation function of VP16. Comparison of amino acid sequences of class II ERF repressors revealed the conservation of the sequence motif L/FDLNL/F(x)P. This motif was essential for repression because mutations within the motif eliminated the capacity for repression. We designated this motif the ERF-associated amphiphilic repression (EAR) motif, and we identified this motif in a number of zinc-finger proteins from wheat, Arabidopsis, and petunia plants. These zinc finger proteins functioned as repressors, and their repression domains were identified as regions that contained an EAR motif.
INTRODUCTION
Plant-specific transcription factors that include a DNA binding domain known as an ERF domain constitute a subfamily of the AP2/ERF domain proteins (Allen et al., 1998; Hao et al., 1998; Fujimoto et al., 2000). So-called ERF proteins were first identified as DNA binding factors that bound to the core sequence of an ethylene-responsive element (GCC box) in tobacco (Ohme-Takagi and Shinshi, 1995). Several genes that are involved in plant growth and development were found subsequently to encode ERF proteins (Wilson et al., 1996; Büttner and Singh, 1997; Stockinger et al., 1997; Zhou et al., 1997; Finkelstein et al., 1998; Liu et al., 1998; Solano et al., 1998; Menke et al., 1999; Fujimoto et al., 2000; van der Fits and Memelink, 2000). Nucleotide sequencing revealed that, in the Arabidopsis genome, 124 genes encode putative ERF proteins and form a superfamily of genes (Riechmann et al., 2000). The ERF proteins identified to date function as transcription factors that are responsive to signals induced by extracellular stress. They are involved in the induction of gene expression by stress factors, such as pathogens and cold, and by components of stress signal transduction pathways, such as ethylene, abscisic acid, and jasmonic acid (Ohme-Takagi and Shinshi, 1995; Büttner and Singh, 1997; Stockinger et al., 1997; Zhou et al., 1997; Finkelstein et al., 1998; Liu et al., 1998; Solano et al., 1998; Suzuki et al., 1998; Menke et al., 1999; Fujimoto et al., 2000; van der Fits and Memelink, 2000).
Among the members of the family of ERF proteins, tobacco ERF2 and ERF4, Arabidopsis AtERF1, AtERF2, AtERF5, ERF1, CBF1, DREB1 and DREB2, periwinkle ORCA2 and ORCA3, and tomato Pti4 have been shown to function as activators of transcription (Stockinger et al., 1997; Zhou et al., 1997; Liu et al., 1998; Solano et al., 1998; Menke et al., 1999; Fujimoto et al., 2000; Ohta et al., 2000; van der Fits and Memelink, 2000). In contrast, NtERF3, AtERF3, and AtERF4, which are categorized as class II ERFs, have been shown to be active repressors (Fujimoto et al., 2000; Ohta et al., 2000). Active repressors, in contrast to passive repressors, include independent repression domains and inhibit the initiation of transcription directly via the actions of these domains (Hanna-Rose and Hansen, 1996). Although large numbers of active repressors have been identified in yeast, mammals, and Drosophila (Hanna-Rose and Hansen, 1996), only a few transcription factors have been reported to be “active” repressors in plants (Liu et al., 1997; Mijer et al., 1997; Raventós et al., 1998; Fujimoto et al., 2000; Jin et al., 2000; Ohta et al., 2000). Moreover, the repression domains of such factors have not been defined in detail. It is important to characterize such repression domains if we are to clarify the molecular mechanisms of repression and the roles of the repressors in the regulation of transcription in plant cells.
In this study, we identified the repression domains of ERF transcriptional repressors as a first step in our attempt to characterize mechanisms of transcriptional repression in plants. We show here that the repression domains of class II ERF proteins are located in regions that contain a conserved L/FDLNL/F(x)P motif and, furthermore, that plant zinc finger proteins that contain this motif can function as repressors.
RESULTS
Identification of the Repression Domain of NtERF3
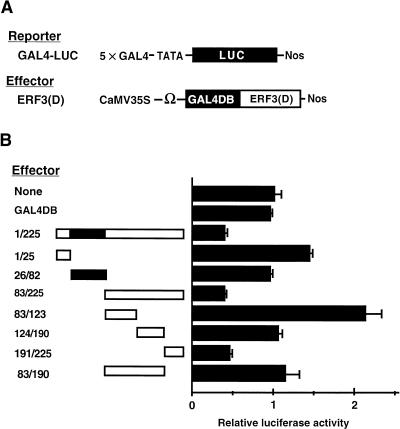
To identify the repression domain of class II ERF transcriptional repressors, we made a series of deletion constructs of the tobacco gene for NtERF3 fused to the gene for the yeast GAL4 DNA binding domain (GAL4DB) under the control of the 35S promoter of the cauliflower mosaic virus (CaMV) (GAL4DB-ERF3[D]; Figure 1A). We first divided the coding region of NtERF3 into three regions, which encoded the N-terminal region (amino acids 1 to 25 [1/25]), the ERF domain (26/82), and the C-terminal region (83/225). The effector plasmids for truncated NtERF3 (1/25, 26/82, and 83/225) and full-length NtERF3 (1/225) were used, together with a luciferase reporter gene that contained five copies of the GAL4-responsive element (GAL4-LUC), to transform into protoplasts prepared from BY-2 tobacco cultured cells by electroporation, as described by Ohta et al. (2000). As shown in Figure 1B, expression of the combination of the effector 83/225 and the reporter resulted in a 50% reduction in the expression of the reporter gene. This level of reduction of expression was similar to the level obtained with the full-length NtERF3 construct (1/225). In contrast, the effectors 1/25 and 26/82 and the GAL4 DNA binding domain alone had no effect on the expression of the reporter gene. Thus, it appeared that the repression domain of NtERF3 was located in the C-terminal region, namely 83/225.
Figure 1.
Mapping of the Repression Domain of ERF3.
(A) Scheme of the constructs used in cotransfection experiments. The GAL4-responsive reporter construct, GAL4-LUC, contained five copies of the GAL4 binding site in tandem and a minimal TATA region (starting at position −46) of the CaMV 35S promoter, the firefly gene for luciferase (LUC), and a nopaline synthase (Nos) terminator (Ohta et al., 2000). Each effector construct contained a GAL4 DNA binding domain (GAL4DB) and part of the coding region (D) of NtERF3 driven by the CaMV 35S promoter. A translational enhancer sequence from tobacco mosaic virus (Ω) was located upstream of the site of initiation of translation.
(B) Relative luciferase activities in tobacco protoplasts that had been cotransfected with effector and reporter plasmids. Schemes of the deletion mutants of NtERF3 are shown at left. The indicated portions of NtERF3 were fused to GAL4DB. Closed boxes indicate the ERF DNA binding domain. All luciferase activities are expressed relative to values obtained with the reporter construct alone (None; set arbitrarily at 1). Error bars indicate ±sd.
We further divided the C-terminal 83/225 region into three regions (83/123, 124/190, and 191/225) and examined the repression activity associated with each region to define in greater detail the repression domain of NtERF3. The effector 191/225 reduced the expression of the reporter gene by 55%, whereas effector 124/190 had no effect on the expression of the reporter gene and effector 83/123 doubled the level of expression of the reporter gene. The transactivation activity of the 83/123 region was suppressed in the presence of the 191/225 repression domain when the two regions were in the same molecule. When the 191/225 region was eliminated from effector 83/225, the resulting effector, 83/190, had no repression activity (Figure 1B). These results demonstrated that the C-terminal 35 amino acids of NtERF3 (191/225) were necessary and sufficient for repression.
Active Repression by the Repression Domain
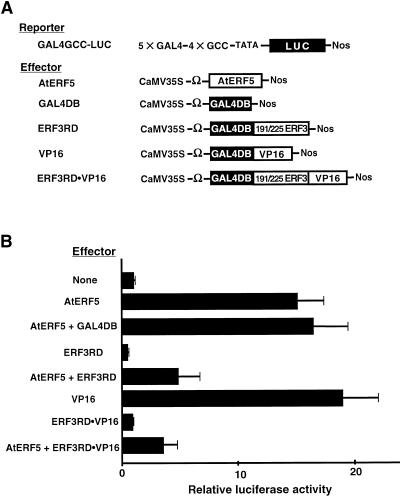
We analyzed the active repression by the 191/225ERF3 repression domain of other transcription factors using a reporter gene that contained two cis elements, the GAL4 binding site and the GCC box (GAL4GCC-LUC), and the AtERF5 effector (AtERF5), which has been shown to activate GCC box–mediated transcription (Fujimoto et al., 2000). The 191/225ERF3 repression domain in a GAL4DB fusion (ERF3RD) decreased the expression of the reporter gene by 50%. When expression of AtERF5 was driven by the CaMV 35S promoter, the effector increased the expression of the reporter gene 15-fold when introduced directly by bombardment into Arabidopsis leaves (Figure 2). When ERF3RD and AtERF5 were introduced with the reporter gene, the expression of the reporter gene increased only 4.9-fold. Because the activation of the reporter gene by AtERF5 was unaffected when the effector that contained GAL4DB alone was coexpressed with AtERF5 (Figure 2), these results indicated that ERF3RD suppressed the intermolecular transactivation activity of AtERF5, acting as an active repressor, and that ERF3RD reduced by 66% the AtERF5-activated level of expression of the reporter (ratio of extents of induction = 4.9/15.2).
Figure 2.
Active Repression by the Repression Domain of NtERF3.
(A) Scheme of the constructs used in cobombardment experiments. The reporter gene GAL4GCC-LUC contained five copies of the GAL4-responsive element, four copies of the GCC box sequence in tandem, a minimal TATA region (starting at position −46) of the CaMV 35S promoter, the firefly gene for luciferase (LUC; shown as a closed box), and a nopaline synthase (Nos) terminator (Fujimoto et al., 2000). Effector constructs contained the coding sequence of AtERF5 (AtERF5), a GAL4 DNA binding domain (GAL4DB; shown as a closed box), the 191/225ERF3 repression domain fused with GAL4DB (ERF3RD), a VP16 activation domain fused with GAL4DB (VP16), and the VP16 activation domain fused with ERF3RD (ERF3RD•VP16). Each construct is driven by the CaMV 35S promoter, and an omega sequence, namely, a translational enhancer, from tobacco mosaic virus (Ω) was located upstream of the site of initiation of translation.
(B) Relative luciferase activities after cobombardment of Arabidopsis leaves with the GAL4GCC-LUC reporter gene and the GAL4DB fusion effectors with or without the AtERF5 effector. The effectors introduced into leaves with the reporter gene are shown at left. All LUC activities are expressed relative to the value obtained with the reporter construct alone (None; set arbitrarily at 1). Error bars indicate ±sd.
To determine whether ERF3RD suppressed intramolecular activation activities in addition to intermolecular activities, we generated an effector in which the strong activation domain of viral protein 16 (VP16) from herpes simplex virus (Triezenberg et al., 1988) was fused to ERF3RD (ERF3RD• VP16). The GAL4DB fusion with the activation domain of VP16 increased the expression of the GAL4GCC-LUC reporter gene 19-fold (Figure 2). However, fusion of the repression domain to the VP16 activation domain (ERF3RD• VP16) resulted in complete inhibition of the transactivation function of VP16 (Figure 2). Furthermore, ERF3RD•VP16 caused a 75% reduction in the AtERF5-activated level of expression (ratio of extents of induction = 3.6/15.2) when ERF3RD•VP16 and AtERF5 were expressed after cobombardment with the GAL4GCC-LUC reporter gene (Figure 2). These results indicated that the repression domain of NtERF3 was able to suppress transactivation activities in both an intramolecular and an intermolecular context.
Because the GAL4DB-fused ERF3RD effector did not repress the expression of reporter genes that did not have a GAL4 binding site, such as genes driven by the CaMV 35S promoter (data not shown), it seemed likely that binding to DNA was required for repression and that repression by ERF3RD was not the result of a nonspecific negative effect such as transcriptional squelching.
The Repression Domains of Class II ERFs Share a Conserved Motif
Comparison of the amino acid sequences of AtERF3 and AtERF4 with that of the 191/225 repression domain of NtERF3 revealed conservation of the sequence motif L/FDLNL/F(x)P in the C-terminal region of each protein (Figure 3A). This motif also was found in the C-terminal region of an NtERF3 homolog from rice, OsERF3, in the C-terminal region of an NtERF3 homolog from Stylosanthes hamata (GenBank accession numbers provided in Methods), in an AtERF3 homolog from tobacco, and in six ERF proteins from Arabidopsis, AtERF7, AtERF8, AtERF9, AtERF10, AtERF11, and AtERF12 (Figure 3A). These ERF proteins can be categorized as class II ERFs in view of the homology at the amino acid level among the ERF domains (Fujimoto et al., 2000).
Figure 3.
The L/FDLNL/F(x)P Motif Is Conserved in the C-Terminal Regions of Class II ERF and TFIIIA-Type Zinc Finger Proteins.
(A) Comparison of the amino acid sequences of the C-terminal regions of class II ERF proteins from various plants.
(B) Alignment of the sequences of C-terminal regions of TFIIIA-type zinc finger proteins.
Numbers in parentheses indicate the positions of the amino acid sequences. Dashes were introduced to optimize the alignment. Reverse type indicates the L/FDLNL/F(x)P motif that was found in the class II ERF and zinc finger proteins and designated EAR. Asterisks indicate the proteins whose repression activities were analyzed in the present experiments.
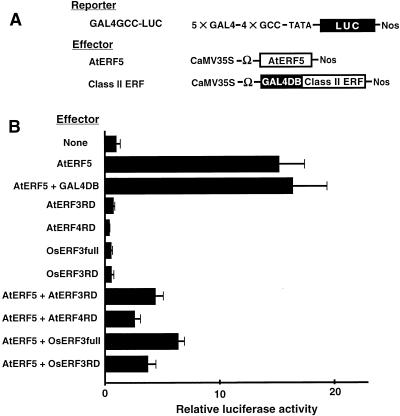
To determine whether the regions of AtERF3 and AtERF4 that contain the conserved motif might be repression domains, we fused the C-terminal 166/225 region of AtERF3 and the 183/222 region of AtERF4 to GAL4DB to generate AtERF3RD and AtERF4RD effectors, respectively. As shown in Figure 4, AtERF3RD and AtERF4RD reduced the AtERF5-activated level of expression of the GAL4GCC-LUC reporter gene by 72 and 83% (ratios of extents of induction = 4.3/15.2 and 2.6/15.2), respectively, when the effector plasmids were used to cobombard Arabidopsis leaves. Moreover, the NtERF3 homolog in rice, OsERF3, which is a polypeptide of 235 amino acids with 44% homology with NtERF3, functioned as a repressor when expressed as a GAL4DB fusion protein (OsERF3full) and reduced the AtERF5-activated level of expression of the reporter by 58% (ratio of extents of induction = 6.4/15.2). As in the case of other class II ERFs, the C-terminal 193/235 region of OsERF3, which includes the conserved motif, functioned as a repressor when expressed as a GAL4DB fusion (OsERF3RD) and reduced the AtERF5-activated level of expression by 75% (ratio of extents of induction = 3.8/15.2; Figure 4). These results indicated that the repression domains of class II ERF repressors were located in regions that contained the conserved L/FDLNL/F(x)P motif.
Figure 4.
Repression by the Repression Domains of AtERF3, AtERF4, and OsERF3.
Relative luciferase activities after cobombardment of Arabidopsis leaves with the GAL4GCC-LUC reporter gene and GAL4DB fusion effectors with or without the AtERF5 effector. The effectors were 35S-GAL4DB-AtERF3(166/225) (AtERF3RD), 35S-GAL4DB-AtERF4 (183/222) (AtERF4RD), 35S-GAL4DB-OsERF3(1/235) (OsERF3full), and 35S-GAL4DB-OsERF3(193/235) (OsERF3RD). All luciferase activities are expressed relative to the value obtained with the reporter gene alone (None; set arbitrarily at 1). Error bars indicate ±sd.
The L/FDLNL/F(x)P Motif Is Essential for Repression
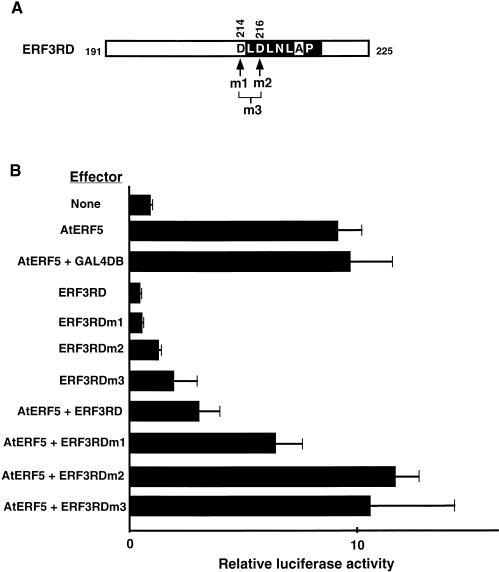
To determine whether the conserved motif is important for repression, we generated mutations in which aspartic acid residues at positions 214 (D214) and 216 (D216), located outside and inside the conserved motif of ERF3RD, respectively, were replaced by alanine. As shown in Figure 5, the ability of ERF3RD to repress transcription was abolished when both D214 and D216 together and when D216 alone were replaced by alanine residues. However, repression was unaffected when D214 alone was replaced by alanine, and the resulting mutant, ERF3RDm1, reduced the AtERF5-activated level of expression of the reporter by 33% (ratio of extents of induction = 6.5/10.2). These results showed that a mutation inside but not outside the motif abolished the capacity for repression, confirming that the motif was essential for repression. A characteristic feature of the L/FDLNL/F(x)P motif is the alternation of hydrophilic and hydrophobic amino acid residues, with the aspartic acid residue being amphiphilic. We designated this sequence motif the ERF-associated amphiphilic repression (EAR) motif.
Figure 5.
The Conserved Motif in the Repression Domain Is Essential for Repression.
(A) Scheme of the conserved motif and positions of amino acids replaced in 191/225 ERF3RD. The amino acids replaced by alanine residues are indicated by arrows; residue 214 (asparatic acid; m1), residue 216 (asparatic acid; m2), and both residues 214 and 216 (m3) were replaced by alanine residues. Reverse type indicates the EAR motif sequence.
(B) Relative luciferase activities after cobombardment of Arabidopsis leaves with the GAL4GCC-LUC reporter gene and the GAL4DB fusion effectors with or without the AtERF5 effector. The asparatic acid residues at positions 214, 216, or both 214 and 216 were replaced by alanine residues in ERF3RDm1, ERF3RDm2, and ERF3RDm3, respectively. All luciferase activities are expressed relative to the value obtained with the reporter construct alone (None; set arbitrarily at 1). Error bars indicate ±sd.
Plant Zinc Finger Proteins with an EAR Motif Are Repressors
A search of the DDBJ/EMBL/GenBank database revealed the presence of the EAR motif in the C-terminal regions of TFIIIA-type zinc finger proteins (Miller et al., 1985) from plants (Figure 3B). The proteins identified included wheat WZF1, Arabidopsis ZAT1, ZAT5, STZ/ZAT10, and ZAT11, and most members of the petunia ZPT family, with the exception of ZPT3-1 and ZPT4-1 (Sakamoto et al., 1993; Lippuner et al., 1996; Meissner and Michael, 1997; Kobayashi et al., 1998).
To determine whether zinc finger proteins that contain an EAR motif might function as repressors, we constructed effectors that contained the protein-coding region of ZAT10 or ZAT11 fused to GAL4DB (ZAT10full and ZAT11full) and cobombarded Arabidopsis leaves with the effectors and the GAL4GCC-LUC reporter gene with and without AtERF5. As shown in Figure 6, both ZAT10 and ZAT11 functioned as active repressors of transcription and reduced the AtERF5-activated level of expression of the reporter by 70 and 83% (ratios of extents of induction = 4.6/15.2 and 2.5/15.2). As in the case of the repression domains of class II ERFs, we identified the C-terminal 174/227 region of ZAT10 and the C-terminal 134/178 region of ZAT11, both of which include an EAR motif, as repression domains, and they reduced the AtERF5-activated level of expression of the reporter by 89 and 81% (ratios of extents of induction = 1.6/15.2 and 2.9/15.2), respectively, as GAL4DB fusion proteins.
Figure 6.
Repression by Zinc Finger Proteins That Contain the EAR Motif.
(A) Scheme of the constructs used in cobombardment experiments. The effector constructs contained the coding sequence of full-length ZAT10 (ZAT10full), 174/227 ZAT10 (ZAT10RD), 1/173 ZAT10 (ZAT10ΔRD1), 1/189 ZAT10 (ZAT10ΔRD2), full-length ZAT11 (ZAT11full), 130/178 ZAT11 (ZAT11RD), and 1/129ZAT11 (ZAT11ΔRD1). Hatched boxes indicate EAR motifs. Nos, nopaline synthase terminator.
(B) Relative luciferase activities after cobombardment of Arabidopsis leaves with the GAL4GCC-LUC reporter gene and the GAL4DB fusion effectors with or without the AtERF5 effector. All luciferase activities are expressed relative to the value obtained with the reporter construct alone (None; set arbitrarily at 1). Error bars indicate ±sd.
To confirm that the C-terminal regions that contained the EAR motif were the only repression domains of ZAT10 and ZAT11, we constructed deletion mutants in which the 174/224 repression domain or the 189/224 region that contained the EAR motif of ZAT10 was deleted from the ZAT10full effector (ZAT10ΔRD1 and ZAT10ΔRD2, respectively) as well as a mutant in which the 134/178 repression domain of ZAT11 was deleted from the ZAT11full effector (ZAT11ΔRD), and then we examined repression by these constructs. As shown in Figure 6, transcriptional repression was abolished completely in the absence of the repression domain or the region that contained the EAR motif. The polypeptides that lacked the C-terminal region did not repress transcription; rather, they activated it. These results indicated that zinc finger proteins that contained an EAR motif functioned as repressors and that their repression domains were exclusively those regions that contained an EAR motif.
DISCUSSION
In this study, we demonstrated that polypeptides that contain the conserved L/FDLNL/F(x)P motif can confer the capacity for repression on a heterologous DNA binding domain and that the motif itself is essential for such repression. We designated the motif the EAR motif. A repression domain that contained the EAR motif (ERD) effectively suppressed not only the intermolecular transactivation activity of other transactivators when bound to DNA but also intramolecular activity, as demonstrated in the experiment with the ERF3RD•VP16 fusion protein (Figure 2).
The ERF proteins form a superfamily of transcription factors in plants. In addition to AtERF3 and AtERF4, we identified six putative ERF proteins in Arabidopsis (AtERF7, AtERF8, AtERF9, AtERF10, AtERF11, and AtERF12) that include an EAR motif and that are possible repressors. These ERF proteins can be categorized as class II ERF proteins on the basis of sequence homologies among their ERF domains (Fujimoto et al., 2000) and they are likely to have a similar DNA binding preference. AtERF4 has been shown to bind to the DRE sequence (TACCGAC), to which CBF1/DREB binds (Stockinger et al., 1997; Liu et al., 1998), as well as to the GCC box (D.-Y. Hao and M. Ohme-Takagi, unpublished data). Thus, these class II ERFs might regulate the DRE-mediated transcription of cold- and/or drought-inducible genes, in addition to the GCC box–mediated expression of genes, acting as repressors. The overall amino acid sequences of class II ERFs, outside the respective ERF domains, are very different from each other. Thus, the repression activities of class II ERF repressors might be regulated in different ways. We found that, in addition to ERF proteins, a number of TFIIIA-type zinc finger proteins from plants include an EAR motif. Wheat WZF1, which includes an EAR motif, appears to act as a transcriptional repressor (Sakamoto et al., 1993). Our results suggest that zinc finger proteins that include an EAR motif might function generally as repressors in plants.
Several ERF and zinc finger proteins that contain an EAR motif have been shown to be involved in stress responses. Both biotic and abiotic stress induce the expression of transcripts of class II ERF genes (Ohme-Takagi and Shinshi, 1995; Suzuki et al., 1998; Fujimoto et al., 2000). The promoter region of the petunia ZPT2-2 gene is responsive to wounding, cold, desiccation, and UV-B irradiation (Krol et al., 1999). The levels of STZ/ZAT10 mRNA in Arabidopsis are enhanced by increased levels of NaCl (Lippuner et al., 1996). Furthermore, the STZ gene increases the tolerance of yeast cells to both Li+ and Na+ ions (Lippuner et al., 1996). Similarly, the homolog of NtERF3 in Stylosanthes hamata increases the aluminum tolerance of yeast. Genetic analysis of Arabidopsis has revealed that both positive and negative factors are involved in signal transduction upon exposure of plants to cold, drought, and salt stress (Shitani et al., 1998; Lee et al., 1999; Xiong et al., 1999). Thus, it seems likely that repressors with an EAR motif might act as negative regulators during the transduction of stress signals.
A large number of transcriptional repression domains that can be transferred to heterologous DNA binding domains have been identified in eukaryotes. These domains vary in both length and sequence, and they exhibit no significant sequence homology with one another. They are loosely categorized according to their primary amino acid content, being defined as alanine rich, proline rich, and charged, for example (Hanna-Rose and Hansen, 1996). The repression domain of the protein Krüppel of Drosophila is alanine rich, and that of the YY1 transcription factor is glycine rich (Licht et al., 1990; Yang et al., 1996). The Krüppel-associated box (KRAB) is an evolutionarily conserved repression domain that is rich in acidic amino acid residues (Witzgall et al., 1994). A region of 34 amino acids in Egr-1, a small C-terminal domain of 24 amino acids that includes a leucine-proline dipeptide repeat of Mig1, a region of 52 amino acids of C/EBPɛ, and the POZ domain of BCL-6 all have been shown to be repression domains (Gashler et al., 1993; Ostling et al., 1996; Seyfert et al., 1996; Williamson et al., 1998). Furthermore, the eight amino acids of the C-terminal domain were shown recently to be the minimal repression domain of the transcription factor p53 (Hong et al., 2001). Several transcription factors from mammalian cells, such as p53, Egr-1, and C/EBPɛ, have been shown to function as both activators and repressors (Gashler et al., 1993; Williamson et al., 1998; Hong et al., 2001). In contrast, the EAR-containing class II ERFs and zinc finger proteins that we examined did not exhibit such bifunctional behavior in our transient expression assays.
In plants, several transcriptional factors have been shown to be active repressors. The N-terminal region of homeobox 1 of rice (Oshox1), the N-terminal region of a repressor of transcription from barley (HRT), and the N-terminal proline-rich region of G-box binding factor 2 from soybean (SGBF-2) are required for repression activities (Liu et al., 1997; Mijer et al., 1997; Raventós et al., 1998). AtMYB4 is a transcriptional repressor, and its repression domain has been mapped to the C-terminal region (Jin et al., 2000). However, the various repression domains, conserved sequences, or regions essential for repression have not been characterized in detail. The EAR motif is a novel motif that is essential for repression and is conserved in at least two different families of transcription factors, the ERF proteins and the TFIIIA-type zinc finger proteins. The EAR motif was found in the C-terminal regions of ERF proteins and of TFIIIA-type zinc finger proteins. However, location of the EAR motif in the C-terminal region does not seem to be a prerequisite for repression, because the ERF3RD•VP16 fusion, in which the VP16 activation domain was fused to the C-terminal region of ERF3RD, had strong repression activity (Figure 2).
It is generally accepted that the repression domain of an active repressor inhibits the activation of transcription by interacting with other cellular proteins, such as basal transcription factors, activator or coactivator proteins, and corepressors (Hanna-Rose and Hansen, 1996). The repression domain of the thyroid hormone receptor and that of the products (EVE and KR) of segmentation genes in Drosophila interact with basal transcription factors (Um et al., 1995). Several repression domains selectively repress the activities of transcriptional activators. The Krüppel protein represses transcription that is mediated by the glutamine-rich activation domain of SP1 but not by the acidic GAL4 activator, which suggests that the repression domain of the Krüppel protein might interact with some specific activator or coactivator (Licht et al., 1993). KRAB-associated protein 1 (KAP-1) is a corepressor that interacts with the KRAB domain (Friedman et al., 1996). The corepressor SMRT binds to the BTB/POZ repression domain of the LAZ3/BCL-6 oncoprotein (Dhordain et al., 1997). Some active repressors are known to act by inducing the formation of an inactive structure in the chromatin at the site of the regulated promoter. The repression domains of YY1 and Elk-1 have been shown to recruit a chromatin modification enzyme, RPD3, or the mSin3A-histone deacetylase corepressor (Yang et al., 1996, 2001). The homolog of histone deacetylase HD2 in Arabidopsis represses transcription of a reporter gene when fused with the GAL4 DNA binding domain (Wu et al., 2000), which suggests that plant repressors might recruit some chromatin-modifying enzymes for functional repression.
Efforts should now be made to isolate and characterize the proteins that interact with the EAR repression domain to clarify the mechanism of repression that is mediated by the EAR motif and the possible roles of repressors with an EAR motif in the control of gene expression. Further studies of ERF repressors should help us to characterize the finely tuned mechanisms that regulate transcription in plants.
METHODS
Construction of Reporter and Effector Plasmids
The reporter plasmids GAL4-LUC and GAL4GCC-LUC were described previously (Fujimoto et al., 2000; Ohta et al., 2000). The latter plasmid includes five repeats of the yeast GAL4 protein binding site, four repeats of the GCC box sequence, and the minimal TATA region (starting at position −46) of the 35S promoter of cauliflower mosaic virus (CaMV), located upstream of the firefly gene for luciferase.
The effector plasmids 35S-GAL4DB, 35S-GAL4DB-ERF3, 35S-AtERF5, and 35S-GAL4DB-VP16 were constructed previously (Fujimoto et al., 2000; Ohta et al., 2000). Effector plasmids containing the truncated coding region of ERF3 (35S-GAL4BD-ERF3[D]) were generated by insertion of DNA fragments of NtERF3, generated by polymerase chain reaction (PCR), into the SmaI and SalI sites of 35S-GAL4DB (Ohta et al., 2000). The rice expressed sequence tag (EST) clone for OsERF3 was sequenced in its entirety. The effector plasmid GAL4-ERF3RD•VP16 was constructed by inserting part of the coding region of NtERF3 (191/225) downstream of GAL4DB in the GAL4DB-VP16 plasmid (Fujimoto et al., 2000). Point mutations in the repression domain of NtERF3 were created by PCR-based site-directed mutagenesis, and the resulting DNA fragments were inserted into the SmaI and SalI sites of 35S-GAL4DB. Fragments encoding ZAT10 and ZAT11 of Arabidopsis thaliana were generated by PCR from the corresponding ESTs and were inserted into the SmaI and SalI sites of 35S-GAL4DB.
Transient Expression
Preparation of tobacco (Nicotiana tabacum) protoplasts, electroporation, and analysis of expression were performed as described previously (Ohta et al., 2000). Analysis of transient expression in Arabidopsis leaves after particle bombardment was as described previously (Fujimoto et al., 2000). In cotransfection assays, we used 1.6 μg of reporter constructs and 1.2 μg of effector constructs for each bombardment. Luciferase assays were performed with the Dual-Luciferase Reporter Assay System and a luminescence reader (TD-20/20; Promega, Madison, WI). To normalize values after each transfection, 0.4 μg of plasmid pPTRL, which included a luciferase gene from Renilla under the control of the CaMV 35S promoter, was used as an internal control (Ohta et al., 2000). The values cited are averages, with standard deviations, of results from a minimum of three independent experiments. Normalized luciferase activity recorded after transfection with the GAL4-LUC plasmid or GAL4GCC-LUC alone was set arbitrarily at 1.
GenBank Accession Numbers
The GenBank accession numbers are as follows: NtERF3 homolog from Stylosanthes hamata, U91857; AtERF3 homolog from tobacco, AB02475; AtERF7, AB032201; AtERF8, AB036884; AtERF9, AB047648; AtERF10, AB047649; AtERF11, AB055882; AtERF12, AB055883; OsERF3, AB036883; rice EST clone, C26399; ZAT10 EST, Z29828; ZAT11, T44892.
Acknowledgments
The authors thank Dr. Kaoru Suzuki for helpful discussions, the National Institute of Agrobiological Resources (Tsukuba, Japan) for the rice EST clone, and the Arabidopsis Biological Resource Center (Ohio State University, Columbus) for Arabidopsis EST clones.
References
- Allen, M.D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., and Suzuki, M. (1998). A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17, 5485–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, M., and Singh, K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhordain, P., Albagli, O., Lin, R.J., Ansieau, S., Quief, S., Leutz, A., Kerckaert, J.P., Evans, R.M., and Leprince, D. (1997). Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA 94, 10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J.R., Fredericks, W.J., Jensen, D.E., Speicher, D.W., Huang, X.P., Neilson, E.G., and Rauscher III, F.J. (1996). KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10, 2067–2078. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gashler, A.L., Swaminathan, S., and Sukhatme, V.P. (1993). A novel repression module, an extensive activation domain, and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Mol. Cell. Biol. 13, 4556–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna-Rose, W., and Hansen, U. (1996). Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 12, 229–234. [DOI] [PubMed] [Google Scholar]
- Hao, D., Ohme-Takagi, M., and Sarai, A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273, 26857–26861. [DOI] [PubMed] [Google Scholar]
- Hong, T.M., Chen, J.J., Peck, K., Yang, P.C., and Wu, C.W. (2001). p53 amino acids 339–346 represent the minimal p53 repression domain. J. Biol. Chem. 276, 1510–1515. [DOI] [PubMed] [Google Scholar]
- Jin, H., Cominelli, E., Baily, P., Parr, A., Mehrtens, F., Jones, J., Toneilli, C., Weisshaar, B., and Martin, C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, A., Sakamoto, A., Kubo, K., Rybka, Z., Kanno, Y., and Takatsuji, H. (1998). Seven zinc-finger transcription factors are expressed sequentially during the development of anthers in petunia. Plant J. 13, 571–576. [DOI] [PubMed] [Google Scholar]
- Krol, A.R., Poecke, R.M.P., Vorst, O.F.J., Voogt, C., Leeuwen, W., Borst-Vrensen, T.W.M., Takatsuji, H., and Plas, L.H.W. (1999). Developmental and wound-, cold-, desiccation-, ultraviolet-B-stress-induced modulations in the expression of the petunia zinc-finger transcription factor gene ZPT2–2. Plant Physiol. 121, 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., Xiong, L., Ishitani, M., Stevenson, B., and Zhu, J.-K. (1999). Cold-regulated gene expression and freezing tolerance in an Arabidopsis thaliana mutant. Plant J. 17, 301–308. [DOI] [PubMed] [Google Scholar]
- Licht, J.D., Grossel, M.J., Figge, J., and Hansen, U.M. (1990). Drosophila Krüppel protein is a transcriptional repressor. Nature 346, 76–79. [DOI] [PubMed] [Google Scholar]
- Licht, J.D., Ro, M., English, M.A., Grossel, M., and Hansen, U. (1993). Selective repression of transcriptional activators at a distance by the Drosophila Kruppel protein. Proc. Natl. Acad. Sci. USA 90, 11361–11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner, V., Cyert, M.S., and Gasser, C.S. (1996). Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J. Biol. Chem. 271, 12859–12866. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Hagen, G., and Guilfoyle, T.J. (1997). A G-box-binding protein from soybean binds to the E1 auxin-responsive element in the soybean GF3 promoter and contains a proline-rich repression domain. Plant Physiol. 115, 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner, R., and Michael, A.J. (1997). Isolation and characterization of a diverse family of Arabidopsis two- and three-fingered C2H2 zinc-finger protein genes and cDNAs. Plant Mol. Biol. 33, 615–624. [DOI] [PubMed] [Google Scholar]
- Menke, F.L., Champion, A., Kijne, J.W., and Memelink, J. (1999). A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 16, 4455–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijer, A.H., Scarpella, E., van Dijk, E.L., Oin, L., Taal, A.J.C., Rueb, S., Harrington, S.E., McCouch, S.R., Schilperoot, R.A., and Hoge, J.H. (1997). Transcriptional repression by Oshox1, a novel homeodomain leucine zipper protein from rice. Plant J. 11, 263–276. [DOI] [PubMed] [Google Scholar]
- Miller, J., McLahlan, A.D., and Klug, A. (1985). Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4, 1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M., Ohme-Takagi, M., and Shinshi, H. (2000). Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J. 22, 29–38. [DOI] [PubMed] [Google Scholar]
- Ostling, J., Carlberg, M., and Ronne, H. (1996). Functional domains in the Mig1 repressor. Mol. Cell. Biol. 16, 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raventós, D., Skriver, K., Schlein, M., Karnahl, K., Rogers, S.W., Rogers, J.C., and Mundy, J. (1998). HRT, a novel zinc finger, transcriptional repressor from barley. J. Biol. Chem. 273, 23313–23320. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- Sakamoto, A., Minami, M., Huh, G.H., and Iwabuchi, M. (1993). The putative zinc-finger protein WZF1 interacts with a cis-acting element of wheat histone genes. Eur. J. Biochem. 217, 1049–1056. [DOI] [PubMed] [Google Scholar]
- Seyfert, V.L., Allman, D., He, Y., and Staudt, L.M. (1996). Transcriptional repression by the proto-oncogene BCL-6. Oncogene 12, 2331–2342. [PubMed] [Google Scholar]
- Shitani, M., Xiong, L., Lee, H., Stevenson, B., and Zhu, J.K. (1998). HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 10, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Suzuki, N., Ohme-Takagi, M., and Shinshi, H. (1998). Immediate early induction of mRNAs for ethylene-responsive transcription factors in tobacco leaf strips after cutting. Plant J. 15, 657–665. [DOI] [PubMed] [Google Scholar]
- Triezenberg, S.J., Kingsbury, R.C., and McKnight, S.L. (1988). Functional dissection of VP16, the transactivator of herpes simplex virus immediate early gene expression. Genes Dev. 2, 718–729. [DOI] [PubMed] [Google Scholar]
- Um, M., Li, C., and Manley, J.L. (1995). The transcriptional repressor even-skipped interacts directly with TATA-binding protein. Mol. Cell. Biol. 15, 5007–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits, L., and Memelink, J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 14, 295–297. [DOI] [PubMed] [Google Scholar]
- Williamson, E.A., Xu, H.N., Gombart, A.F., Verbeek, W., Chumakov, A.M., Friedman, A.D., and Koeffler, H.P. (1998). Identification of transcriptional activation and repression domains in human CCAAT/enhancer-binding protein ɛ. J. Biol. Chem. 273, 14796–14804. [DOI] [PubMed] [Google Scholar]
- Wilson, K., Long, D., Swinburne, J., and Coupland, G. (1996). A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzgall, R., O'Leary, E., Leaf, A., Onaldi, D., and Bonventre, J.V. (1994). The Krüppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc. Natl. Acad. Sci. USA 91, 4514–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Tian, L., Malik, K., Brown, D., and Miki, B. (2000). Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22, 19–27. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Ishitani, M., Lee, H., and Zhu, J.-K. (1999). HOS5: A negative regulator of osmotic stress-induced gene expression in Arabidopsis thaliana. Plant J. 19, 569–578. [DOI] [PubMed] [Google Scholar]
- Yang, S.H., Vickers, E., Brehm, A., Kouzarides, T., and Sharrocks, A.D. (2001). Temporal recruitment of the mSin3A-histone deacety-lase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol. 21, 2802–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.M., Inouye, C., Zeng, Y., Bearss, D., and Seto, E. (1996). Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. USA 93, 12845–12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Tang, X., and Martin, G.B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16, 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]