Abstract
The KNAT2 (for KNOTTED-like from Arabidopsis thaliana 2) homeobox gene is expressed in the vegetative apical meristem. It is also active during flower development, suggesting a function in the structuring of flowers. To investigate its role, we used a DEXAMETHASONE (DEX)-inducible system to generate transgenic plants that overexpressed a fusion of KNAT2 with the hormone binding domain of the glucocorticoid receptor. DEX-induced plants were similar to plants overexpressing the closely related KNAT1 gene, indicating overlapping functions, although we observed differences as well. In particular, KNAT2-GR activation induced ectopic carpel features. First, KNAT2 induced the homeotic conversion of nucellus into carpel-like structures. Second, KNAT2 induced stigmatic papillae on rosette leaves in the ap2-5 background. Third, ectopic expression of the carpel identity gene AGAMOUS (AG) was observed in carpels and ovules. Interestingly, the homeotic conversion was not dependent on AG activity, because it was maintained in the ag-1 ap2-5 double mutant. Therefore, our data indicate that KNAT2 also must activate other carpel regulators. Together, these results suggest that KNAT2 plays a role in carpel development.
INTRODUCTION
Homeodomain proteins are transcription factors that have been identified in many species and that play a variety of roles in plant, animal, and fungal development (Gehring et al., 1994; Mann and Affolter, 1998; Kappen, 2000). Plant homeodomain proteins have been classified into different families according to their similarities within the homeodomain and the presence of additional motifs (Chan et al., 1998; Mayer et al., 1998; Jorgensen et al., 1999). Molecular and genetic analyses have revealed a role for these genes in various developmental processes, including meristem functioning, ovule development, and trichome elongation (reviewed in Chan et al., 1998; Mayer et al., 1998; Williams, 1998). The KNOTTED-like homeobox (KNOX) genes constitute a family in plants (Reiser et al., 2000). In maize, a subclass of the KNOTTED genes has been associated with meristem function. Gain-of-function mutations in KN1 cause knots to form along the lateral veins (Hake et al., 1989; Vollbrecht et al., 1991). Loss-of-function kn1 alleles have been isolated that lead to plants with a defect in meristem maintenance (Kerstetter et al., 1997; Vollbrecht et al., 2000).
In Arabidopsis, two classes of KNAT (for KNOTTED-like from Arabidopsis thaliana) genes have been identified. Class I comprises KNAT1, KNAT2, SHOOTMERISTEMLESS (STM), and a recently identified member, KNAT6 (Semiarti et al., 2001). Within this class, KNAT1, KNAT2, and STM are expressed mainly in the shoot apical meristem (Lincoln et al., 1994; Dockx et al., 1995; Long et al., 1996). Class II contains KNAT3, KNAT4, and KNAT5, which are expressed in all tissues (Serikawa et al., 1996). No role has been proposed yet for the class II KNAT genes. To date, only one mutant in the KNAT family has been isolated. Strong recessive alleles of STM result in plants defective in shoot meristem initiation and maintenance, whereas weak alleles exhibit a decrease in floral organ number (Barton and Poethig, 1993; Endrizzi et al., 1996). Constitutive expression of STM leads to stunted plants with a highly disorganized shoot meristem (Williams, 1998). These results suggest a role for STM in meristem maintenance. A similar function has been proposed for KNAT1. Although no loss-of-function mutant has been described for KNAT1, its overexpression induces the formation of ectopic meristems and therefore is able to activate STM, directly or indirectly (Chuck et al., 1996). In addition, the ectopic expression of both genes in young primordia in mutant or transgenic backgrounds is associated with abnormal leaf development (Ori et al., 2000; Reiser et al., 2000). In agreement with their proposed role in establishing the meristematic state, KNAT1 and STM are downregulated when cells are recruited into the incipient organs (Lincoln et al., 1994; Long et al., 1996). In Arabidopsis, two mutants, asymmetric leaves 1 (as1) and asymmetric leaves2 (as2), are altered in leaf shape and exhibit some features similar to those of plants that overexpress KNAT1 (Ori et al., 2000; Semiarti et al., 2001). These mutants show misexpression of KNAT1 and KNAT2, suggesting that AS1 and AS2 stimulate cell differentiation through the repression of KNOX genes (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001). AS1, which encodes a myb factor related to PHANTASTICA (PHAN) and ROUGH SHEAT (RS2), is downregulated by STM in stem cells (Schneeberger et al., 1998; Waites et al., 1998; Byrne et al., 2000). In organ founder cells, STM is downregulated, allowing AS1 to be expressed and to negatively regulate KNAT1 and KNAT2 (Byrne et al., 2000).
In this article, we address the function of KNAT2. Expression studies suggest that KNAT2 is expressed in the internal parts of the vegetative shoot apical meristem and is downregulated when leaf primordia are initiated (Dockx et al., 1995; Laufs et al., 1998a). In mature plants, the KNAT2 gene is expressed in the receptacle and the developing pistils of flowers and in the axillary buds of inflorescence stems (Dockx et al., 1995; Bowman et al., 1999). Nothing is known, however, concerning the function of KNAT2. To investigate its role during plant development, we used a glucocorticoid-inducible system to generate transgenic plants that overexpressed a fusion of KNAT2 and the hormone binding domain of the glucocorticoid receptor (GR) (Miesfeld et al., 1986; reviewed in Picard, 2000). The activation of the KNAT2-GR fusion with DEXAMETHASONE (DEX), a synthetic glucocorticoid, led to plants with epinastic cotyledons and curled and lobed leaves with ectopic stipules. Here, we concentrate on a particular feature induced by the activation of KNAT2-GR, a homeotic conversion of ovules into carpel-like structures. We show that this conversion was associated with an abnormal persistence of AGAMOUS (AG) expression in carpels and ovules of the MADS box gene AG, a key gene in carpel development (reviewed in Bowman et al., 1999), although the homeotic conversion as such does not depend on AG.
RESULTS
Transgenic 35S-KNAT2-GR Arabidopsis Plants Are Perturbed in Development
To gain insight into the role of KNAT2 in plant development, transgenic plants that express a KNAT2-GR fusion protein were obtained. The KNAT2 cDNA was fused translationally to a DNA fragment encoding the hormone binding domain of the glucocorticoid receptor and placed under the constitutive 35S promoter of the cauliflower mosaic virus (Miesfeld et al., 1986; Benfey et al., 1990). In the absence of hormone, DEX, the hybrid protein, is maintained in the cytoplasm and is inactive, whereas when DEX is added, the fusion moves to the nucleus and transactivates target genes (reviewed in Picard, 2000). As a result, in the absence of inducer, the plants show wild-type phenotype, whereas their development is altered upon DEX induction. Eleven and 38 independent transgenic lines of the Wassilewskija and Landsberg erecta ecotypes, respectively, were recovered and selfed to obtain homozygous lines. The overall morphology of the transgenic lines in the absence of DEX was wild type (Figure 1). In contrast, in the presence of DEX, the development of transgenic lines was altered dramatically (Figure 1). DEX-induced plants exhibited a reduced size of apical organs, lobed leaves, and distorted siliques. For a given line, the severity of the phenotype correlated with the concentration of DEX, with the maximal effect being obtained at concentrations of 1 μM and greater (Figure 1B). The most severely affected transgenic line in the Landsberg erecta ecotype was chosen for detailed examination and for subsequent genetic analyses and is referred to throughout as KNAT2-GR. Unless stated otherwise, all further experiments were performed on homozygous KNAT2-GR plants by using 10 μM DEX.
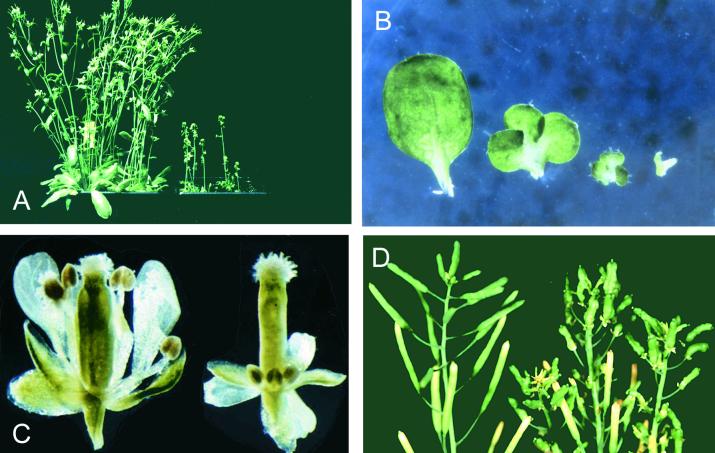
Figure 1.
Phenotypes of Noninduced and DEX-Induced KNAT2-GR Plants.
(A) Plants grown in the greenhouse in the absence (left) or presence (right) of DEX.
(B) Rosette leaves from 4-week-old plantlets grown in vitro in the presence of DEX (left to right: 0, 0.1, 0.5, and 10 μM).
(C) Flowers from noninduced (left) and DEX-induced (right) KNAT2-GR plants. DEX was applied after floral induction. The elongation of sepals, petals, stamens, and filaments was inhibited in the DEX-induced plants.
(D) Siliques from noninduced (left) and DEX-induced (right) KNAT2-GR plants. DEX was applied after floral induction. The siliques were distorted after activation of the construct.
DEX-Induced Transgenic Plants Exhibit Epinastic Cotyledons and Lobed Leaves
Nine days after sowing, in vitro KNAT2-GR seedlings grown continuously in the presence of DEX exhibited epinastic cotyledons (data not shown). No other structural abnormality was observed in the cotyledons of DEX-induced seedlings. Meristem structure was normal (data not shown). However, at 12 days after sowing, the DEX-induced KNAT2-GR plants showed a significant delay in development compared with noninduced plants. At this stage, the nontreated plants had already produced the second pair of leaves (Figure 2A), whereas the DEX-induced plants had just initiated the first pair of leaves (Figure 2B). This delay in leaf initiation suggested an alteration in shoot apical meristem function, although it also could be attributable to an inhibitory effect of KNAT2-GR on growth. Leaves from 1-month-old DEX-induced plants were curled and initiated a pair of lobes in the position of the hydathodes (Figure 2D). In wild-type or noninduced plants, lobes were visible only on young primordia (Figure 2C), whereas stipules were found at the base of the leaves. In KNAT2-GR DEX-induced plants, ectopic stipules lined the margin of the lamina (Figures 2D and 2H).
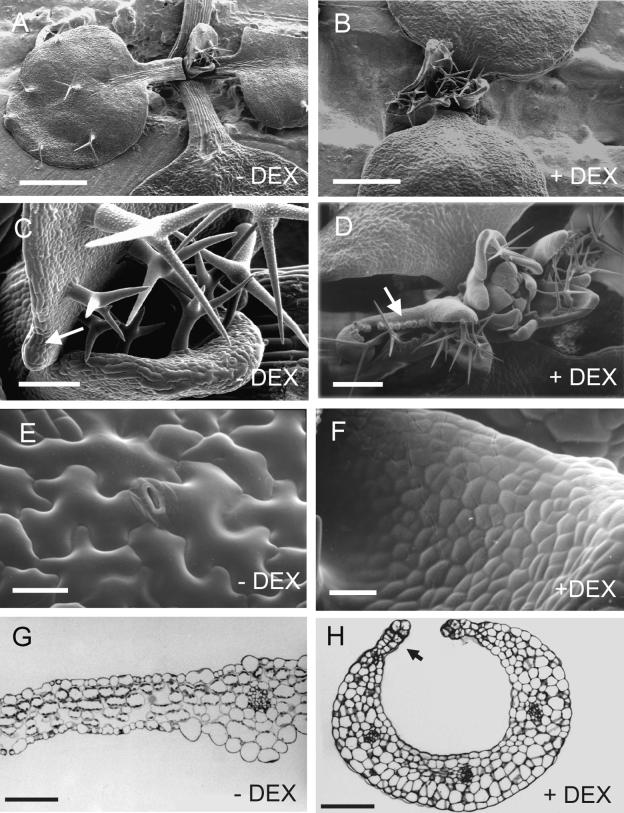
Figure 2.
Scanning Electron Microscopy and Histological Analysis of Noninduced and DEX-Induced KNAT2-GR Seedlings Grown in Vitro.
(A) Twelve-day-old noninduced KNAT2-GR seedling with its two first expanded rosette leaves. Bar = 500 μm.
(B) Twelve-day-old KNAT2-GR seedling grown in the presence of DEX. Bar = 500 μm.
(C) Closeup of a lobe from a noninduced KNAT2-GR seedling. The arrow indicates the lobe at the base of the leaf. Bar = 50 μm.
(D) Four-week-old KNAT2-GR seedling grown in the presence of DEX. The two first leaves were severely curled and developed two pairs of lobes in the position of hydathodes. The arrow indicates ectopic stipules. Bar = 250 μm.
(E) Epidermal surface of a 12-day-old noninduced rosette leaf showing irregularly shaped cells with interspersed stomata. Bar = 20 μm.
(F) Epidermal surface of a 4-week-old DEX-induced curled leaf. Bar = 20 μm.
(G) Transverse section of a noninduced leaf showing the wild-type palisade and spongy parenchyma. The section was stained in toluidine blue. Bar = 100 μm.
(H) Transverse section of a DEX-induced curled leaf showing the joined isodiametric cells. The section was stained in toluidine blue. The arrow indicates ectopic stipules. Bar = 100 μm.
Scanning electron microscopy of the epidermis of mature DEX-induced leaves revealed a defect in cell elongation and differentiation. As in the wild type, the epidermis of noninduced expanded leaves consisted of irregularly shaped cells (Figure 2E). The epidermal cell shape in DEX-induced mature leaves, however, was regular and similar to that of nonexpanded noninduced leaves (Figure 2F). Transverse sections of leaves from DEX-induced plants revealed an alteration in the internal structure. The palisade and spongy parenchyma found in the noninduced lines were normal (Figure 2G). In contrast, in DEX-induced leaves, the cells were small, isodiametric, and tightly packed with no intercellular space (Figure 2H). This structure was similar to that of the untreated young leaf or to the chlorenchyma found in the ovary wall. Thus, the activation of the KNAT2-GR fusion altered the differentiation of leaf cells.
The Nucellus Is Converted into Carpels in DEX-Induced Ovules
To examine the effects of the activation of the KNAT2-GR fusion on flower development, plants were grown in the greenhouse and treated with DEX after floral induction. An inhibition of the elongation of floral organs was observed, except for the carpels, which were a normal size (Figure 1C). In contrast to the four outer whorls, the identity of the fifth whorl was altered because ectopic carpel-like structures arose from the placental tissue in place of ovules (Figure 3B). Figures 3C and 3D show details of such carpeloid structures. The surface of these ectopic carpels was characteristic of that of the style, showing the specific cellular cuticular thickenings (data not shown). It is likely that the conversion of ovules into carpeloid structures results in the formation of more or less distorted siliques (Figures 1D and 3B).
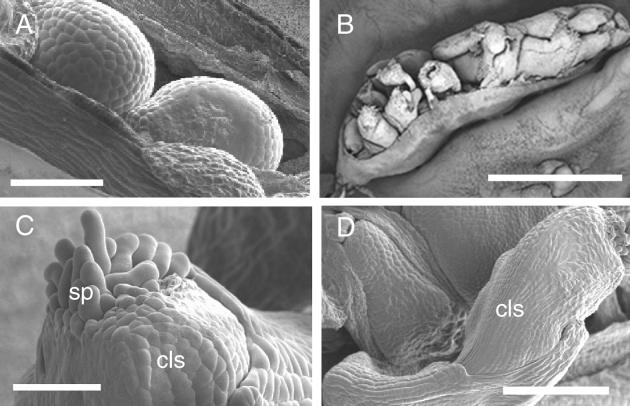
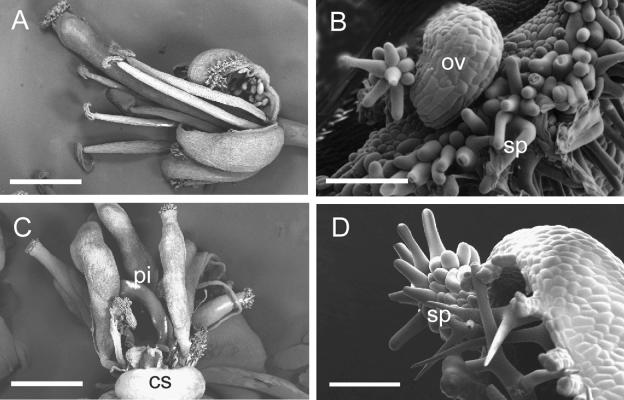
Figure 3.
Scanning Electron Micrographs of Siliques from Noninduced and DEX-Induced KNAT2-GR Plants.
Plants were watered with DEX after floral induction. Developmental stages are those defined by Smyth et al. (1990) and Robinson-Beers et al. (1992).
(A) Dissected stage 17 silique from a noninduced KNAT2-GR plant showing immature seed. Bar = 250 μm.
(B) Dissected stage 17 silique from a DEX-induced KNAT2-GR line showing carpel-like structures in place of immature seed. Bar = 2 mm.
(C) Developing carpel-like structure (cls) with stigmatic papillae (sp) from a DEX-induced silique. Bar = 50 μm.
(D) Carpel-like structure (cls) at a later stage. Bar = 200 μm.
To determine when ovules in DEX-induced KNAT2-GR plants first deviated from normal development, ovules from DEX-induced and noninduced KNAT2-GR plants were analyzed using confocal scanning laser microscopy. Optical sections of the DEX-induced ovules suggested that their development was unaltered until stage 13 as defined by Smyth et al. (1990) and Robinson-Beers et al. (1992). At this stage, the wild-type gynoecium was fully formed and comprised well-differentiated stigmatic papillae, style, and ovaries (Robinson-Beers et al., 1992). As in the noninduced ovules, the initiation and the growth of the ovule primordia were normal (Figure 4A). The inner and outer integuments of DEX-treated plants grew asymmetrically to encase the nucellus, and the micropyle was positioned near the funiculus (Figures 4B and 4C). The female gametophyte comprised seven cells, with the two polar nuclei fused (data not shown). Subsequently, the morphogenesis of the DEX-induced ovules deviated from normal development. First, the shape of the DEX-induced ovules became abnormal (Figure 4D) and the embryo sac degenerated (Figure 4E). Subsequently, the nucellus started to proliferate (Figures 4E and 4F), developing into a carpel-like structure with stigmatic papillae at the apex and a secondary carpel-like structure inside (Figures 4G and 4H).
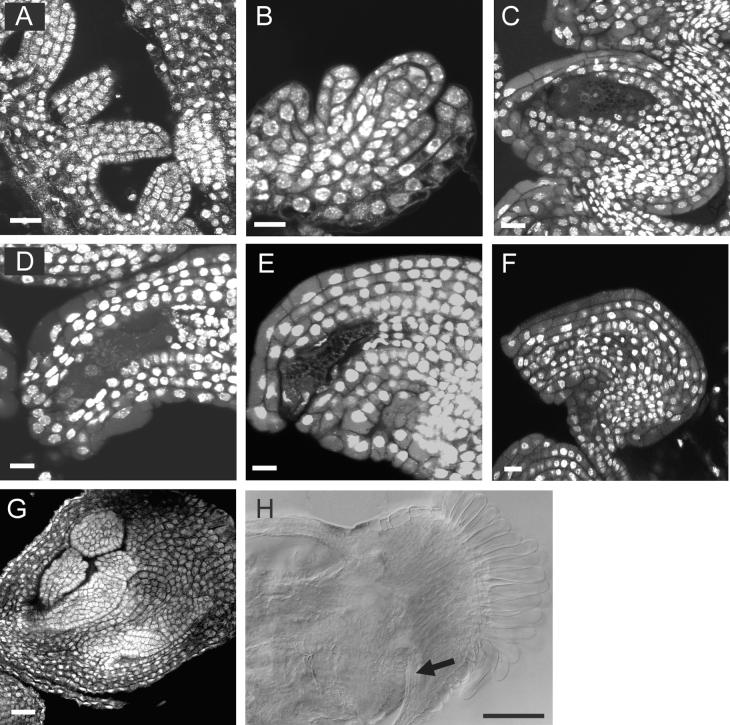
Figure 4.
Optical Sections of Developing Ovules from DEX-Induced KNAT2-GR Plants.
Pistils were stained with the nuclear stain propidium iodide and rhodamine. The cytoplasm is nonspecifically labeled by this stain. Developmental stages are those defined by Smyth et al. (1990) and Robinson-Beers et al. (1992).
(A) Stage 10 ovule from a DEX-induced KNAT2-GR plant. The ovule primordia elongated as in noninduced plants. Bar = 10 μm.
(B) Stage 12 ovule from a DEX-induced KNAT2-GR plant showing the growth of the inner and outer integuments. Bar = 10 μm.
(C) Stage 13 ovule from a DEX-induced KNAT2-GR plant with a mature embryo sac. Both integuments enveloped the nucellus. The micropyle is positioned near the funiculus. Bar = 10 μm.
(D) to (F) Developing carpel-like structures from a DEX-induced KNAT2-GR plant. The shape of the DEX-induced ovule became abnormal, the embryo sac degenerated, and the nucellus proliferated. Bar = 10 μm.
(G) Carpel-like structure from a DEX-induced KNAT2-GR plant with second order carpel-like structure. Bar = 20 μm.
(H) Cleared carpeloid structure from a KNAT2-GR plant viewed with differential interference contrast optics. The arrow indicates stigmatic papillae of the secondary carpel-like structure. Bar = 100 μm.
Ectopic Activation of KNAT2-Induced Ectopic Expression of AG in Leaves and Flowers
The conversion of the nucellus into a carpeloid structure after the ectopic activation of KNAT2 led us to examine the expression of the AG gene in DEX-induced KNAT2-GR lines. AG is a key regulatory MADS box gene that is required in the third and fourth whorls of developing flowers to specify stamen and carpel identity and to confer determinacy to the floral meristem (Bowman et al., 1989; Yanofsky et al., 1990). The homozygous KNAT2-GR line was crossed to the line carrying the AGAMOUS–β-glucuronidase (pAG-I::GUS) construct (Sieburth and Meyerowitz, 1997). A line homozygous for both transgenes was selected. We used this line in combination with in situ hybridization to study AG expression in the KNAT2-GR line. In wild-type plants, AG is not active in leaves, and its expression is detected in early stage 3 flowers and is further limited to the region of the floral meristem that gives rise to the stamens and carpels (Bowman et al., 1991a; Drews et al., 1991). In DEX-induced KNAT2-GR carrying the pAG-I::GUS construct, weak GUS activity was detected in leaves, and this expression was dependent on floral transition (data not shown). In DEX-induced flowers, in situ hybridization indicated that the early expression pattern of AG was not modified (Figures 5A and 5B). Thus, KNAT2 was not able to activate AG precociously in young floral meristems.
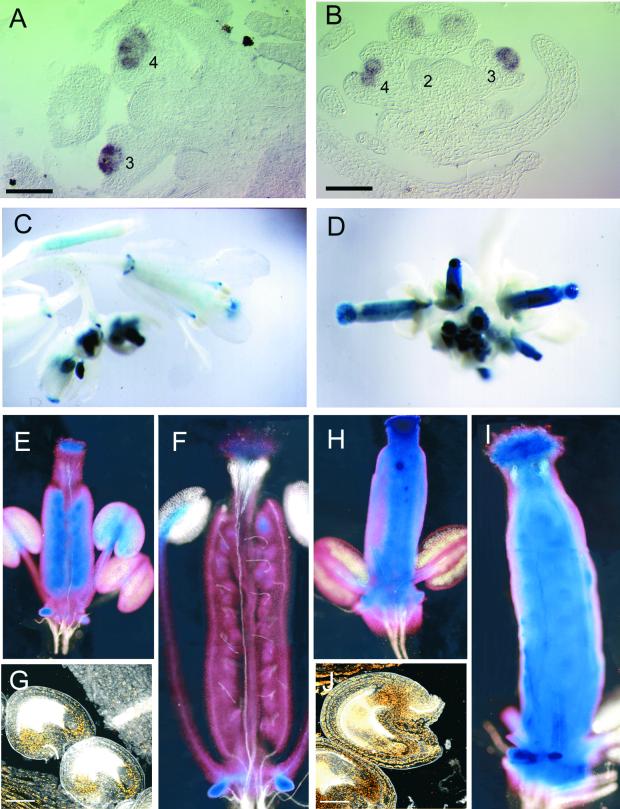
Figure 5.
AG Expression in Noninduced and DEX-Induced Inflorescences.
Numbers indicate the stages of flowers based on Smyth et al. (1990).
(A) and (B) Longitudinal sections of noninduced and DEX-induced inflorescences, respectively, showing AG mRNA expression in stage 2, 3, and 4 flowers. Bar = 50 μm.
(C) to (J) GUS expression in noninduced and DEX-induced lines homozygous for both the KNAT2-GR and pAG-I::GUS constructs.
(C) A noninduced inflorescence showing GUS activity in flowers.
(D) Inflorescence from a DEX-induced plant showing high GUS activity in carpels.
(E) and (F) Gynoecium from a noninduced plant at stages 12 and 14, respectively. The GUS activity in the carpel became progressively restricted to the stigma and the nectaries.
(G) Stage 13 ovule from a noninduced plant. GUS activity was restricted to the endothelium and the chalazal domain. The reflection of the crystalline precipitate of the GUS substrate was visualized using the confocal microscope. Bar = 50 μm.
(H) and (I) Gynoecium from a DEX-induced plant at stages 12 and 14, respectively. GUS activity was maintained in the carpel walls during development.
(J) Ovule from a DEX-induced plant. GUS activity was abnormally persistent in the ovule. Bar = 50 μm.
During flower development, strong GUS activity was present in third and fourth whorls of young flowers from noninduced inflorescences (Figure 5C). This pattern became progressively restricted to the stamens, the stigma, and the nectaries of the carpels, whereas weak staining was detected in the carpel walls (Figures 5C, 5E, and 5F). This pattern is consistent with the AG expression in wild-type plants (Bowman et al., 1991a). In contrast, high GUS activity was detected in the pistils of mature flowers after DEX treatment (Figures 5D and 5I). This high GUS activity was detected throughout the pistil: the carpel walls, the style, and the stigmatic papillae. It was maintained within the pistil even during the later stages of flower development in tissues in which AG normally is downregulated (Figures 5H and 5I). Within the wild-type ovule primordia, AG was first expressed in the region in which the integument later formed (Reiser et al., 1995). During later stages of development, the expression of AG was restricted to the endothelium and the chalazal domain (Figure 5G). This pattern was consistent with AG expression in wild-type ovules (Modrusan et al., 1994; Reiser et al., 1995; Sieburth and Meyerowitz, 1997). Figure 5J shows that GUS activity was abnormally persistent in all parts of the ovule of DEX-induced flowers. In conclusion, the ectopic activation of the KNAT2-GR fusion led to aberrant AG expression in carpels and ovules.
Interaction of KNAT2, AG, and Regulators of AG
Ectopic expression of KNAT2 led to the homeotic conversion of the nucellus into carpel-like structures and to an increase in pAG-I::GUS activity. The regulation of AG is complex and involves the transcriptional activator LEAFY (LFY) (Parcy et al., 1998; Busch et al., 1999) and several partially redundant repressors (Drews et al., 1991; Liu and Meyerowitz, 1995; Goodrich et al., 1997; Byzova et al., 1999; Krizek et al., 2000), one of which is APETALA2 (AP2) (Bomblies et al., 1999). Therefore, we introduced the KNAT2-GR construct into lfy and ap2 mutants to examine the interaction of the KNAT2-GR phenotype with these regulators. The KNAT2-GR construct was introduced into the strong allele lfy-6 to determine whether the conversion of the nucellus was dependent on LFY activity (Weigel et al., 1992). DEX-induced KNAT2-GR lfy-6 exhibited the vegetative phenotype of the DEX-induced KNAT2-GR line (data not shown). As shown in Figure 6, the noninduced terminal structure of an old KNAT2-GR lfy-6 inflorescence exhibited the lfy-6 phenotype. The inflorescence shoot consisted of fused carpeloid bracts bearing developing ovules and stigmatic papillae (Figure 6A). When plants were treated with DEX, the ovules were converted to pistils, indicating that the homeotic conversion of ovules into carpels was not dependent on LFY (Figure 6B).
Figure 6.
Terminal Flowers in the Apices of the Primary Inflorescences of KNAT2-GR lfy-6 Plants.
(A) Flowers from a noninduced KNAT2-GR lfy-6 plant with carpeloid bracts.
(B) Flowers from a DEX-induced KNAT2-GR lfy-6 plant. All ovules from carpeloid bracts were converted to pistils.
To examine the activation of KNAT2-GR in the ap2 mutant, a line homozygous for both the KNAT2-GR construct and the weak allele ap2-5 was obtained. As shown in Figures 7A and 7B, the nontreated KNAT2-GR ap2-5 line exhibited the ap2-5 phenotype (Kunst et al., 1989; Bowman et al., 1991b). Median sepals were converted to carpel-sepals, petals were converted to stamens, and the androecium and gynoecium were wild type. The carpel-sepals exhibited stigmatic papillae and ovules (Figure 7B). Occasionally, some ovules of the weak allele of ap2-5 were converted into carpel-sepal ovules (data not shown; Modrusan et al., 1994). When these plants were treated with DEX, ovules from the gynoecium were converted to carpeloid structures and ovules from median carpeloid sepals were converted to fully differentiated pistils (Figure 7C). Inside these pistils, the same homeotic conversion of the nucellus was observed as in DEX-induced KNAT2-GR plants (data not shown). Furthermore, when seed of the homozygous KNAT2-GR ap2-5 line were sown in vitro in the presence of DEX, 10% of the lines induced stigmatic tissues at the tip of the lobed leaves (Figure 7D). This carpeloid feature was observed only on the first leaves and appeared when the flowers were differentiated. Thus, an additional carpeloid feature induced by KNAT2 was revealed when the A function gene AP2 was disrupted.
Figure 7.
Scanning Electron Micrographs of Flowers from Noninduced and DEX-Induced KNAT2-GR ap2-5 Plants.
(A) Flower from a noninduced KNAT2-GR ap2-5 plant showing the ap2-5 phenotype. ap2-5 causes sepals to turn into carpel-sepal organs and petal-to-stamen transformations in the first and second whorls, respectively. Bar = 1 mm.
(B) Close-up of the margin of the noninduced KNAT2-GR ap2-5 carpeloid sepal showing ovule (ov) and stigmatic papillae (sp). Bar = 100 μm.
(C) Upper part of the flower from a DEX-induced KNAT2-GR ap2-5 plant showing pistils (pi) on the margin of the carpeloid sepal (cs). The elongation of stamen filaments from whorls two and three was inhibited completely. Bar = 1 mm.
(D) First leaf from a 2-month-old seedling grown in vitro in the presence of DEX, with stigmatic papillae (sp) at the tip. Bar = 200 μm.
We next examined whether the KNAT2-GR phenotype also depended on ectopic AG activity. To this purpose, a line homozygous for both the 35S-KNAT2-GR construct and the strong mutant allele ag-1 was obtained (Yanofsky et al., 1990). DEX-induced KNAT2-GR ag-1 plants exhibited the phenotypic vegetative features of the DEX-induced KNAT2-GR line (data not shown). Flowers from the DEX-induced KNAT2-GR ag-1 plants consisted of an indeterminate number of whorls of sepals and petals in the pattern: (sepals, petals, petals)n as in ag single mutants. The size of sepals and petals was reduced (data not shown). These data indicate that these phenotypic features observed in the DEX-induced KNAT2-GR lines did not depend on AG activity. Because no carpels and consequently no ovules were produced in ag-1 mutant flowers, we could not determine if the homeotic conversion of the nucellus depended on AG. Therefore, a line carrying the KNAT2-GR construct in an ap2-5 ag-1 background in which ovules form on carpeloid sepals was obtained (Bowman et al., 1991b; Western and Haughn, 1999). As shown in Figure 8A, the noninduced KNAT2-GR ap2-5 ag-1 flower exhibited the ap2-5 ag-1 phenotype. The flower consisted of an indeterminate number of whorls of leafy carpeloid organs, and petaloid stamens in the pattern: (leafy carpeloid organs, petaloid stamens, petaloid stamens)n. The median carpeloid leaves had stigmatic tissue and structures resembling ovules (Figure 8B). When plants were treated with DEX, young inflorescences exhibited flowers with organs of reduced size (Figure 8C). Ovules from older DEX-induced KNAT2-GR ap2-5 ag-1 flowers were converted to pistils, indicating that the homeotic conversion of ovules into carpeloid structures was not dependent on AG in the ap2-5 background (Figures 8C and 8D).
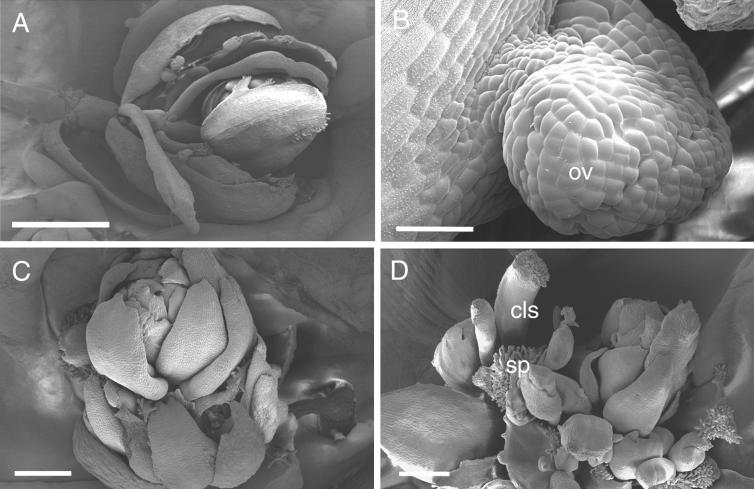
Figure 8.
Scanning Electron Micrographs of Flowers from Noninduced and DEX-Induced KNAT2-GR ap2-5 ag-1 Plants.
Plants were watered with DEX after floral induction.
(A) Flower from a noninduced KNAT2-GR ap2-5 ag-1 plant showing the ap2-5 ag-1 phenotype. The flower consisted of an indeterminate number of carpeloid leaves and petaloid stamens. Bar = 1 mm.
(B) Closeup of the noninduced carpeloid leaf showing the ovule (ov). Bar = 50 μm.
(C) A young flower from a DEX-induced KNAT2-GR ap2-5 ag-1 plant. The elongation of organs was inhibited. Bar = 250 μm.
(D) An old flower from a DEX-induced KNAT2-GR ap2-5 ag-1 plant showing stigmatic papillae (sp) and carpel-like structures (cls). Bar = 250 μm.
KNAT2 Expression during Inflorescence Carpel Development
Mature plants expressing a KNAT2-GUS fusion showed that the KNAT2 promoter was active in the receptacle and the developing pistils of flowers (Dockx et al., 1995; Bowman et al., 1999). Because the previous experiments indicated a major effect on carpel development, we refined these studies in the developing flower by using the KNAT2-GUS line in combination with in situ hybridization. During vegetative development, the expression of KNAT2 was restricted to the inner parts of the meristem (Figure 9A). In the inflorescence meristem and in stage 1 and 2 flowers, no GUS activity was detected (Figure 9D). GUS activity was barely detected in floral buds at stage 3 (Figure 9D). This activity increased in floral buds at stage 4 (Figure 9E). Later, high GUS activity was detected at the apical limit of the pedicel and in the developing fourth whorl (Figure 9F). This GUS activity became restricted to the placental tissue from which ovule initiation occurs (Figure 9G). These results were confirmed by in situ hybridization (Figures 9B and 9C). Thus, the KNAT2 gene, which is expressed in the vegetative shoot apical meristem, is not active in the inflorescence meristem. It is reactivated subsequently in stage 3 floral meristems and finally becomes restricted to the carpel.
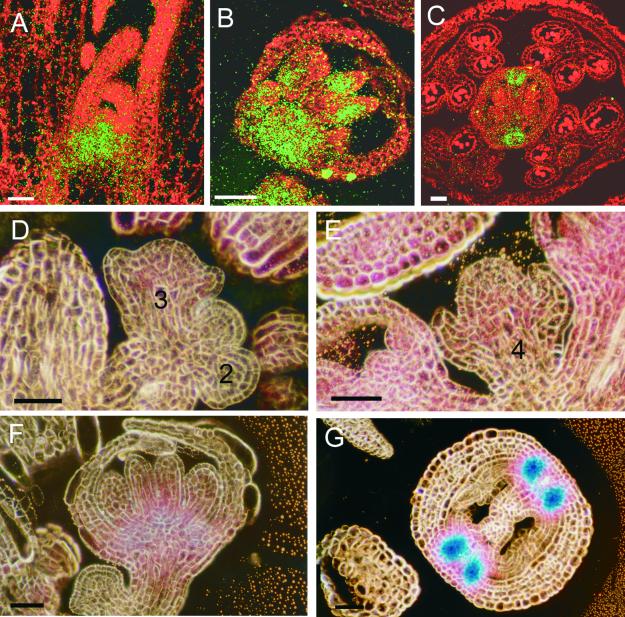
Figure 9.
In Situ Hybridization of a KNAT2 Anti-mRNA Probe and Histochemical Localization of KNAT2-GUS Activity during Flower Development.
Numbers indicate the stages of flowers based on Smyth et al. (1990).
(A) Median longitudinal section through a vegetative meristem. The expression of KNAT2 is restricted to the inner part of the meristem. Bar = 50 μm.
(B) Median longitudinal section through a stage 8 flower showing KNAT2 expression in young carpel primordia. Bar = 50 μm.
(C) Cross-sections through pistils from a flower at stage 12. The expression of KNAT2 became restricted to the replum between the two carpels that form the pistil. Bar = 50 μm.
(D) Longitudinal section through an inflorescence meristem containing stage 2 and 3 flowers. Weak GUS activity was detected only in the stage 3 flowers. Bar = 50 μm.
(E) Longitudinal section through a stage 4 floral meristem showing GUS activity throughout the meristem.
(F) The stage 7 flower shows abundant GUS staining at the base of the floral meristem and in the developing carpels. Bar = 50 μm.
(G) Cross-section of a carpel showing GUS activity restricted to the replum between the two carpels that form the pistil. Bar = 50 μm.
DISCUSSION
KNAT2 Induces the Homeotic Conversion of Nucellus into Carpel-like Structures
KNAT2 overexpression induced a number of features also found in plants that overexpress KNAT1, such as reduced organ size and lobed leaves with ectopic stipules. DEX-induced KNAT2-GR lines, however, also show differences, and we focused our analysis on the most striking feature that distinguishes the effects of KNAT2 overexpression: the ectopic induction of carpel features in leaves and the homeotic conversion of the nucellus into a carpel-like structure. This relation with organ identity was probably limited to the carpels, because we could not detect any ectopic expression of genes involved in other floral organs, such as APETALA1 and APETALA3 (data not shown). It must be noted that the homeotic conversion was observed only in the Landsberg erecta ecotype. The activation of the KNAT2-GR fusion in the Wassilewskija ecotype led to a degeneration of ovules, smaller siliques, and the premature termination of the inflorescence meristem (data not shown). This is in agreement with the observation of Modrusan et al. (1994) that Landsberg erecta is one of the most appropriate ecotypes for the promotion of carpel-like structures. It must be noted that the most severely affected 35S::KNAT1 transgenic lines displayed the same distorted siliques, although no homeotic conversion was illustrated (Chuck et al., 1996).
Mutations in two other genes, AP2 and BEL1, also led to the conversion of ovules into carpeloid structures. We do not think, however, that KNAT2 acts via the inactivation of one of these genes. Mutations in BELL1 (BEL1) result in abnormal bell-shaped ovules (bel1 ovules) that lack the inner integument and develop an abnormal integument-like structure (Robinson-Beers et al., 1992). Some of these abnormal integuments are converted homeotically into carpeloid structures (Modrusan et al., 1994; Ray et al., 1994). The homeotic conversion of bel1 ovules is related to a persistent accumulation of AG mRNA during later stages of ovule development and depends on AG activity (Modrusan et al., 1994; Reiser et al., 1995; Western and Haughn, 1999). In contrast to bel1 ovules, the carpeloid structures induced by KNAT2-GR derived from the nucellus. Thus, KNAT2 acts via a different pathway. The strong allele ap2-6 exhibits normal ovules, filamentous ovules, and carpel-sepal ovules (Bowman et al., 1991b; Modrusan et al., 1994). These carpel-sepal ovules develop earlier than bel1 ovules and are not derived from the integument region or from the nucellus (Modrusan et al., 1994; Western and Haughn, 1999). Although these results suggest that KNAT2 does not interfere with BEL1 and AP2 in ovules, it cannot be ruled out that the gene ectopically inactivates other inhibitory functions.
KNAT2, AG, and Regulators of AG
Because AG is a crucial factor involved in carpel development, we further investigated a possible link with KNAT2. The identity of the four whorls in DEX-induced KNAT2-GR flowers was not altered, suggesting that KNAT2 was not able to overcome the regulators of AG in the early flower. This was confirmed by in situ hybridization, which indicated that KNAT2 did not activate AG precociously in young floral buds. Therefore, KNAT2 was not able to overcome the mechanisms that restrict AG to the third and fourth whorls, a process that involves several factors such as AP2, CURLY LEAF, LEUNIG, STERILE APETALA, and AINTEGUMENTA (Drews et al., 1991; Liu and Meyerowitz, 1995; Goodrich et al., 1997; Byzova et al., 1999; Krizek et al., 2000). In contrast, the expression of pAG-I::GUS was maintained over longer periods in the pistil and in the ovules.
It was initially tempting to propose that the homeotic conversion observed in DEX-induced KNAT2-GR plants was caused by the persistent expression of AG in ovules. However, the homeotic conversion did not depend on AG activity, because carpeloid structures were maintained in the ap2-5 ag-1 double mutant. In this context, it is worth mentioning that the most obvious carpel features induced by KNAT2 (i.e., the formation of stigmatic tissue on leaves in the ap2-5 background) and the homeotic conversion of the nucellus into a carpeloid structure are not mimicked by AG overexpression (Ray et al., 1994; Goodrich et al., 1997; Mizukami and Ma, 1997). Therefore, our results imply that the effects of KNAT2 probably are not limited to AG and that the gene also could activate, in an AG-independent pathway, other carpel regulators. Such a pathway is defined by the CRABS CLAW and SPATULA genes, which specify a number of carpel characteristics, including style, stigma, and ovule production (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Heisler et al., 2001). Alternatively, KNAT2 could activate downstream targets usually induced by AG. AG-like genes involved in carpel development represent such putative targets of KNAT2 (Bowman et al., 1999; Pelaz et al., 2000). The homeotic conversion also was observed in a lfy background, indicating that KNAT2-GR could act via a LFY-independent pathway. Alternatively, KNAT2 could operate downstream of LFY. In either case, however, we cannot exclude that in the wild type, KNAT2 interacts with LFY.
A Role of KNAT2 in Carpel Development?
Although our data were obtained from overexpression studies, several observations suggest a potential activity of KNAT2 in carpel development. This activity does not concern only the carpeloid features induced in ovules and leaves or the ectopic expression of AG. KNAT2 also is reactivated at the right moment. It is possible that the related KNAT1 and STM genes also could play a role in carpel development. It is noteworthy, in this context, that STM is expressed strongly in the apex of the floral meristem, where AG is activated (Long et al., 1996). It should be stressed that the carpel constitutes a specialized meristematic tissue that produces ovules (Sessions and Zambryski, 1995). Its meristematic nature is illustrated not only by its layered structure, which corresponds to the characteristic L1, L2, and L3 structure present in the apical meristem, but by other meristem genes such as KNAT1 and STM, which are active in the carpel as well.
In this context, it seems likely that the KNAT proteins have to interact at some level with the factors that more specifically control carpel development. This is in contrast to other organs, such as leaves, petals, and sepals, in which the KNOTTED-like homeobox genes must be inactivated. Although we have focused our attention on the carpel, the function of the gene probably is not limited to the flower, because it is expressed in the inner layers of the vegetative apical meristem. At this stage, the role of KNAT2 in the meristem is unclear, although an overlap in function between KNAT2 and its closely related family members has been suggested. The evidence is not limited to the partially overlapping expression patterns. As mentioned above, the lobed leaves and their reduced size also are characteristics of KNAT1 overexpression. In addition, KNAT2, when overexpressed, is able to rescue, at least partially, a stm mutant (our unpublished results; see also Byrne et al., 2000), again suggesting potential common targets.
METHODS
Construction of the Chimeric 35S-KNAT2-GR Gene, Plant Transformation, and Dexamethasone Treatments
To make a translational fusion between the KNAT2 gene and the DNA fragment encoding the glucocorticoid receptor, a BamHI site was introduced at the stop codon of KNAT2. A XbaI-BamHI fragment containing the KNAT2 coding sequence was fused to a DNA fragment encoding the glucocorticoid receptor domain (amino acids 519 to 795; Miesfeld et al., 1986) into the pBI-DGR plasmid (based on the pBI121 plasmid; Clontech, Palo Alto, CA) and transcribed from the cauliflower mosaic virus 35S promoter. The new binary vector, called pTK1-6, harboring the gene cassette 35S-KNAT2-GR was introduced into Agrobacterium tumefaciens LBA4404 (Gelvin and Schilperoort, 1988). Two different Arabidopsis thaliana ecotypes (Wassilewskija and Landsberg erecta) were transformed with the construct by using the vacuum infiltration method (Bechtold and Pelletier, 1998). Transformants were selected on medium containing 50 mg/L kanamycin. Plants were grown either in vitro as described by Santoni et al. (1994) or in a greenhouse.
Dexamethasone (DEX; Sigma) was dissolved in ethanol at 10 mM. Unless stated otherwise, all experiments were performed using 10 μM DEX. DEX was added either to the in vitro medium or to the nutritive solution (Coïc and Lesaint, 1971). Plants were grown continuously in the presence of DEX. To test the effect of the induction of the KNAT2-GR fusion on flower development, greenhouse-grown plants were watered continuously with DEX after floral induction.
Genetic Analysis
All lines used were in the Landsberg erecta background. The line carrying the ap2-5 mutation was obtained from the Nottingham Arabidopsis Stock Centre, UK. ag-1 lfy-6 mutants and the AGAMOUS–β-glucuronidase (pAG-I::GUS) lines were kindly supplied by François Parcy. The translational KNAT2-GUS fusion and the pAG-I::GUS constructs have been described (Dockx et al., 1995; Sieburth and Meyerowitz, 1997). To obtain plants homozygous for both the 35S-KNAT2-GR construct and the ap2-5 mutation, an ap2-5 homozygote was crossed to the homozygous KNAT2-GR line. F2 plants were sown in vitro in the presence of 10 μM DEX. Homozygotes for the 35S-KNAT2-GR construct were selected on the basis of the epinasty of the cotyledons; this phenotype was not observed in hemizygotes. Seedlings were then transferred to the greenhouse and selected for the ap2-5 phenotype. To introduce the 35S-KNAT2-GR construct in the lfy-6 mutant, lfy-6/+ heterozygous plants were crossed to the homozygous KNAT2-GR line and a plant homozygous for both KNAT2-GR and lfy-6 was selected. To introduce the 35S-KNAT2-GR construct in the ag-1 mutant, ag-1/+ heterozygous plants were crossed to the homozygous KNAT2-GR line and a plant homozygous for both KNAT2-GR and ag-1 was selected. To obtain ap2-5 ag-1 double mutants carrying the KNAT2-GR construct, homozygous plants for KNAT2-GR ap2-5 were crossed to a plant homozygous for KNAT2-GR and heterozygous for ag-1.
In Situ Localization of GUS Activity
Plant tissues were collected, fixed in 80% acetone at −20°C for 1 hr, and stained for 2 to 16 hr at 37°C in a solution containing 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (Duchefa, Roubaix, France) dissolved in n-DMSO, 0.1% Triton X-100, 0.5 mM K4Fe(CN)6, and 50 mM sodium phosphate buffer, pH 7.2. After staining and extraction in ethanol, stained tissue samples were visualized directly with a binocular microscope. Alternatively, stained tissues were cleared in Herr's liquid overnight (Herr, 1971). Subsequently, the crystalline precipitate of the GUS substrate was visualized using either dark-field optics or the confocal microscope, as described below. To localize the GUS activity in sections, tissues were fixed after 5-bromo-4-chloro-3-indolyl-β-glucuronic acid staining for 2 hr in 4% formaldehyde (fresh from paraformaldehyde) in PBS, pH 7.5, under vacuum for 20 min and left in fixative overnight. Subsequently, plants were dehydrated in gradual steps (10, 30, 50, 70, 90, and 100% ethanol) and embedded in Technovit 7100 (Kulzer, Heraus, Wehrheim, Germany) according to the manufacturer's instructions. Ten-micro-meter sections were made on a Leica-Jung (Leica, Rueil-Malmaison, France) RM2055 rotary microtome carrying a disposable Adamas steel knife. Sections were photographed on a Zeiss Axioskop microscope (Jena, Germany) using Kodak Ektar 25 film.
Histological Analysis
Seedlings were embedded in historesin (Leica) according to the manufacturer's instructions. Three-micrometer sections were cut on a Reichert-Jung RM 2055 microtome, stained in 0.05% toluidine blue, and examined with a Nikon Microphot FXA microscope (Garden City, NY).
Nomarski Microscopy
Pistils were fixed in a mixture of formaldehyde, propionic acid, and 50% ethanol (5:5:90 [v/v]) for 3 hr at room temperature and then cleared in a mixture of 85% lactic acid and phenol (2:1 [w/w]) for 30 min according to Motamayor et al. (2000). Then, pistils of different stages were dissected with a needle in a drop of the lactic acid:phenol solution and mounted in a drop of the same mixture. Pistils and ovules were photographed with differential interference contrast optics by using a Nikon Microphot FXA microscope with Fujicolor Superia 200 ASA film (Fuji, Tilburg, The Netherlands).
Confocal Microscopy and Scanning Electron Microscopy
Inflorescences were fixed and stained with propidium iodide to visualize DNA as described by Laufs et al. (1998b). Inflorescences were then transferred to a graded glycerol series (10, 30, 50, 70, and 80%). To visualize the cells, inflorescences were incubated for 15 min in Rhodamine 123 (0.1 mg/mL; Molecular Probes Europe, Leiden, The Netherlands). This stain nonspecifically labels the cytoplasm. Pistils were mounted in a drop of citifluor glycerol/PBS (Oxford Institute, Orsay, France) and viewed directly with a Leica TCS-NT confocal laser scanning microscope (Leica Microsystems, Heidelberg, Germany) equipped with an argon/krypton laser (Omnichrome, Chino, CA). Rhodamine fluorescence was collected through a fluorescein isothiocyanate filter set (BP530/30) using the 488-nm laser line (450 lines/sec; 1024 × 1024 pixels) with a 40× 0.5 to 1.0 long-working-distance FLUOTAR objective. To visualize the propidium iodide, the 568-nm laser line was used in combination with a tetramethyl rhodamine β-isothyocyanate filter set (LP590). To collect rhodamine and propidium iodide signals simultaneously, a reflect short pass filter (RSP580) was used to separate the emission beam into two parts. Optical sections were generated and printed on a Kodak Digital Science 8650 PS dye sublimation printer.
To visualize the crystalline precipitate of the GUS substrate, pistils were treated as described above except for the rhodamine step. The reflection of the crystalline precipitate of the GUS substrate was visualized using the 568-nm laser line and a 590-nm long-pass filter. A RSP580 beam splitter was used to separate the two channels. Six optical sections (spaced 1 μm apart) were generated per sample. Scanning electron microscopy samples were prepared as described by Traas et al. (1995).
In Situ Hybridization
Plants were fixed in 4% formaldehyde (fresh from paraformaldehyde) in PBS under vacuum twice for 20 min and left in fixative overnight. After fixation, plants were washed, dehydrated, and embedded in paraffin essentially as described by Jackson (1992). Paraffin sections (8 to 10 μm thick) were cut with a disposable metal knife and attached to precoated glass slides (Fisher). Sections were hybridized with 35S-labeled antisense or sense KNAT2 RNAs. The KNAT2 antisense RNA probe was generated from the plasmid pCK130 that contained the 1.3-kb KNAT2 cDNA (GenBank accession number X81353) cloned into the XbaI site of pGEM vector (Promega). The control KNAT2 sense RNA was generated from pCK130 by using the SP6 RNA polymerase. An XbaI-XhoI subclone of the KNAT2 cDNA lacking the homeobox pCK131 gave expression patterns identical to the full-length KNAT2 cDNA.
Acknowledgments
We thank F. Parcy for providing the pTC51565 plasmid, ag-1, lfy-6, and pAG-I::GUS and pLFY::GUS lines and for his helpful discussions. We thank Christophe Reuzeau, Patrick Laufs, Christine Horlow, and Helen North for helpful comments. We thank Anne-Marie Jaumet and Patricia Kock for their skillful assistance. Part of this work was supported by the EFC REGIA program.
References
- Alvarez, J., and Smyth, D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386. [DOI] [PubMed] [Google Scholar]
- Barton, K.M., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild-type and in the shoot meristemless mutant. Development 119, 823–831. [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Benfey, P.N., Ren, L., and Chua, N.H. (1990). Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J. 9, 1677–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies, K., Dagenais, N., and Weigel, D. (1999). Redundant enhancers mediate transcriptional repression of AGAMOUS by APETALA2. Dev. Biol. 216, 260–264. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., and Smyth, D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Drews, G.N., and Meyerowitz, E.M. (1991. a). Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991. b). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Baum, S.F., Eshed, Y.E., Putterill, J., and Alvarez, J. (1999). Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Dev. Biol. 45, 155–205. [DOI] [PubMed] [Google Scholar]
- Busch, M.A., Bomblies, K., and Weigel, D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science. 285, 585–587. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Byzova, M.V., Franken, J., Aarts, M.G., de Almeida-Engler, J., Engler, G., Mariani, C., Van Lookeren Campagne, M.M., and Angenent, G.C. (1999). Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev. 13, 1002–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, R.L., Gago, G.M., Palena, C.M., and Gonzalez, D.H. (1998). Homeoboxes in plant development. Biochim. Biophys. Acta 1442, 1–19. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1-induced lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coïc, Y., and Lesaint, C. (1971). Comment assurer une bonne nutrition en eau et en minéraux en horticulture. Horticulture 8, 11–14. [Google Scholar]
- Dockx, J., Quaedvlieg, N., Keultjes, G., Kock, P., Weisbeek, P., and Smeekens, S. (1995). The homeobox gene ATK1 of Arabidopsis thaliana is expressed in the shoot apex of the seedling and in flowers and inflorescence stems of mature plants. Plant Mol. Biol. 28, 723–737. [DOI] [PubMed] [Google Scholar]
- Drews, G.N., Bowman, J.L., and Meyerowitz, E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the apetala 2 product. Cell 65, 991–1002. [DOI] [PubMed] [Google Scholar]
- Endrizzi, K., Moussian, B., Haecker, A., Levin, J.Z., and Laux, T. (1996). The shoot meristemless gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10, 967–979. [DOI] [PubMed] [Google Scholar]
- Gehring, W.J., Affolter, M., and Burglin, T. (1994). Homeodomain proteins. Annu. Rev. Biochem. 63, 487–526. [DOI] [PubMed] [Google Scholar]
- Gelvin, S.B., and Schilperoort, R.A. (1988). Plant Molecular Biology Manual. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51. [DOI] [PubMed] [Google Scholar]
- Hake, S., Vollbrecht, E., and Freeling, M. (1989). Cloning Knotted, the dominant morphological mutant in maize, using Ds2 as a transposon tag. EMBO J. 8, 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler, M.G., Atkinson, A., Bylstra, Y.H., Walsh, R., and Smyth, D.R. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Herr, J.M. (1971). A new clearing technique for the study of ovule development in angiosperms. Am. J. Bot. 58, 785–790. [Google Scholar]
- Jackson, D.P. (1992). In situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach, D.J. Bowles, S.J. Gurr, and M. McPhereson, eds (Oxford, UK: Oxford University Press), pp. 163–174.
- Jorgensen, J.E., Grounlund, M., Pallisgaard, N., Larsen, K., Marcker, K.A., and Jensen, E.O. (1999). A new class of plant homeobox is expressed in specific regions of determinate symbiotic root nodules. Plant Mol. Biol. 40, 65–77. [DOI] [PubMed] [Google Scholar]
- Kappen, C. (2000). Analysis of a complete homeobox gene repertoire: Implications for the evolution of diversity. Proc. Natl. Acad. Sci. USA 97, 4481–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter, R.A., Laudencia-Chingcuanco, D., Smith, L.G., and Hake, S. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124, 3045–3054. [DOI] [PubMed] [Google Scholar]
- Krizek, B.A., Prost, V., and Macias, A. (2000). AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell 12, 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst, L., Klenz, J.E., Martinez-Zapater, J., and Haughn, G.W. (1989). AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1, 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs, P., Dockx, J., Kronenberger, J., and Traas, J. (1998. a). MGOUN1 and MGOUN2: Two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development 125, 1253–1260. [DOI] [PubMed] [Google Scholar]
- Laufs, P., Grandjean, O., Jonak, C., Kieu, K., and Traas, J. (1998. b). Cellular parameters of the shoot apical meristem in Arabidopsis. Plant Cell 10, 1375–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leave morphology when overexpressed in transgenic plants. Plant Cell 6, 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., and Meyerowitz, E.M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121, 975–991. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Mann, R.S., and Affolter, M. (1998). Hox proteins meet more partners. Curr. Opin. Genet. Dev. 8, 423–429. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 11, 805–815. [DOI] [PubMed] [Google Scholar]
- Miesfeld, R., Rusconi, S., Godowski, P.J., Maler, B.A., Okret, S., Wikstrom, A.C., Gustafsson, J.A., and Yamamoto, K.R. (1986). Genetic complementation of a glucocorticoid receptor deficiency by expression of cloned receptor cDNA. Cell 46, 389–399. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y., and Ma, H. (1997). Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell 9, 393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrusan, Z., Reiser, L., Feldmann, K.A., Fisher, R.L., and Haughn, G.W. (1994). Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamayor, J.C., Vezon, D., Bajon, C., Sauvanet, A., Grandjean, O., Marchand, M., Bechtold, N., Pelletier, G., and Horlow, C. (2000). Switch (swi1), an Arabidopsis thaliana mutant affected in the female meiotic switch. Sex. Plant Reprod. 12, 209–218. [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, G., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532. [DOI] [PubMed] [Google Scholar]
- Parcy, F., Nilsson, O., Busch, M.A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395, 561–566. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Picard, D. (2000). Posttranslational regulation of proteins by fusions to steroid-binding domains. Methods Enzymol. 327, 385–401. [DOI] [PubMed] [Google Scholar]
- Ray, A., Robinson-Beers, K., Ray, S., Baker, S.C., Lang, J.D., Preuss, D., Milligan, S.B., and Gasser, C.S. (1994). Arabidopsis floral homeotic gene BELL (Bel1) controls ovule development through negative regulation of AGAMOUS gene (AG). Proc. Natl. Acad. Sci. USA 91, 5761–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, L., Modrusan, Z., Margossian, L., Samach, A., Ohad, N., Haughn, G.W., and Fischer, R.L. (1995). The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83, 735–742. [DOI] [PubMed] [Google Scholar]
- Reiser, L., Sanchez-Baracaldo, P., and Hake, S. (2000). Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol. 42, 151–166. [PubMed] [Google Scholar]
- Robinson-Beers, K., Pruitt, R.E., and Gasser, C.S. (1992). Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4, 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni, V., Bellini, C., and Caboche, M. (1994). Use of two-dimensional protein-pattern analysis for the characterization of Arabidopsis thaliana mutants. Planta 192, 557–566. [Google Scholar]
- Schneeberger, R., Tsiantis, M., Freeling, M., and Langdale, J.A. (1998). The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125, 2857–2865. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 10, 1771–1783. [DOI] [PubMed] [Google Scholar]
- Serikawa, K.A., Martinez-Laborda, A., and Zambryski, P. (1996). Three knotted1-like homeobox genes in Arabidopsis. Plant Mol. Biol. 32, 673–683. [DOI] [PubMed] [Google Scholar]
- Sessions, R.A., and Zambryski, P.C. (1995). Arabidopsis gynoecium structure in the wild and in ettin mutants. Development 121, 1519–1532. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., and Meyerowitz, E.M. (1997). Molecular dissection of the Agamous control region shows that cis elements spatial regulation are located intragenically. Plant Cell 9, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas, J., Laufs, P., Jullien, M., and Caboche, M. (1995). A mutation affecting etiolation and cell elongation in Nicotiana plumbaginifolia causes abnormal division plane alignment and pattern formation in the root meristem. Plant J. 7, 785–796. [Google Scholar]
- Vollbrecht, E., Veit, B., Sinha, N., and Hake, S. (1991). The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350, 241–243. [DOI] [PubMed] [Google Scholar]
- Vollbrecht, E., Reiser, R., and Hake, S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene KNOTTED1. Development 127, 3161–3172. [DOI] [PubMed] [Google Scholar]
- Waites, R., Selvadurai, H.R., Oliver, I.R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93, 779–789. [DOI] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Western, T.L., and Haughn, G.W. (1999). BELL1 and AGAMOUS genes promote ovule identity in Arabidopsis thaliana. Plant J. 18, 329–336. [DOI] [PubMed] [Google Scholar]
- Williams, R.W. (1998). Plant homeobox genes: Many functions stem from a common motif. Bioessays 20, 280–282. [DOI] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]