Abstract
Hereditary multiple exostoses (EXT) is an autosomal dominant disorder characterized by multiple cartilage-capped outgrowths from the epiphyses of long bones. In some cases, these osteochondromas progress to malignant chondrosarcomas. Alterations in at least three genes (EXT1, EXT2, and EXT3) can cause this disorder. Two of these have been isolated (EXT1 and EXT2) and encode related members of a putative tumor suppressor family. We report here the genomic structure of the human EXT2 gene consisting of 14 exons (plus 2 alternative exons) covering an estimated 108 kb of chromosome 11p11–13. We have derived the DNA sequences at all exon/intron boundaries throughout this gene—information that is important for the detailed study of mutations in EXT2. We have also characterized the mouse EXT2 cDNA and have mapped the mouse locus to chromosome 2 between D2Mit15 and Pax6. This mouse homolog should enable transgenic knockout experiments to be initiated to further elucidate gene function. Interestingly, sequence comparisons reveal that the human and mouse EXT genes have at least two homologs in the invertebrate Caenorhabditis elegans, indicating that they do not function exclusively as regulators of bone growth. This observation opens the way for a functional analysis of these genes in nematodes and other lower organisms.
[The sequence data described in this paper have been submitted to the GenBank data library under accession nos. U67353–U67368 and U67837.]
Hereditary multiple exostoses (EXT) is an autosomal dominant condition characterized by multiple cartilage-capped projections originating from the epiphyseal plates of endochondral bones (Jaffe 1943; Solomon 1961). The prevalence of this disorder has been estimated at between 1/50,000 and 1/100,000 (Hennekam 1991; Cook et al. 1993; Schmale et al. 1994). The development of exostoses appears to be correlated with growth of the epiphyseal plate, with new exostoses rarely appearing after epiphyseal plate closure (Hennekam 1991). Exostoses usually form at the ends of long bones but can also occur on the hands, feet, pelvis, ribs, and scapula. Shortened stature is sometimes seen in this disorder and is possibly attributable to these benign tumors interfering with the epiphyseal growth plates and reducing overall bone growth (Hennekam 1991). Exostoses also frequently impinge on adjacent nerves, vessels, tendons, and joints and thus require surgical removal. A serious complication of this disorder is the occasional transformation to chondrosarcoma or osteosarcoma that occurs in 0.9%–2.9% of cases (Voutsinas and Wynne-Davies 1983; Wicklund et al. 1995).
Genetic linkage analysis in EXT families has identified three loci responsible for hereditary multiple exostoses on chromosomes 8q24.1 (EXT1), 11p11–13 (EXT2), and 19p (EXT3) (Cook et al. 1993; Le Merrer et al. 1994; Wu et al. 1994; Wuyts et al. 1995). To date, the exostoses that are caused by disruption of these genes appear to be indistinguishable; no specific phenotypic differences have been observed in patients from different linkage groups (Le Merrer et al. 1994). Multiple exostoses has also been observed as one phenotype in two contiguous gene deletion syndromes that encompass EXT1 or EXT2 separately, confirming the hypothesis that haploinsufficiency for the products of these genes results in the disruption of bone growth (Ludecke et al. 1991; Parrish et al. 1991; Bartsch et al. 1996).
Loss of heterozygosity (LOH) studies using polymorphic markers that flank EXT1, EXT2, and EXT3 indicate that these chromosomal regions are lost in chondrosarcomas that develop in genetically linked families (Hecht et al. 1995, 1997; Raskind et al. 1995). In addition, LOH for loci flanking EXT1 and/or EXT2 has been detected in 7 of 17 sporadic chondrosarcomas (Raskind et al. 1995), and LOH at both EXT1 and EXT2 has been observed in a single chondrosarcoma that was derived from an EXT2-linked individual (Hecht et al. 1995). Together, these findings suggest that the EXT genes may act as tumor suppressors in developing skeletal tissues, in addition to playing a role in bone growth and modeling.
The chromosome 8 EXT1 gene was cloned by an approach involving analysis of chromosomal breakpoints (Ahn et al. 1995). The EXT2 gene was identified recently by our collaborative group using a new positional cloning strategy (Stickens et al. 1996) and subsequently was cloned independently by Wuyts et al. (1996). The two genes encode very similar, novel proteins, the functions of which are unknown. However, the presence of a putative signal sequence composed of a hydrophobic core at the amino terminus of both proteins indicates that these proteins may be either secreted or embedded within the cell membrane (Wise et al. 1997). A third member of the EXT family (named EXT-L for EXT-like) also has been identified by our group (Wise et al. 1997). This gene has been localized to chromosome 1p36.1 and is thus not a candidate for the chromosome 19-linked EXT3 gene. Currently, no evidence exists that this gene is associated with any skeletal disorder, but it lies within a chromosomal region that frequently exhibits LOH in other types of malignancies (Lothe et al. 1995; Nagai et al. 1995; Ezaki et al. 1996).
Previously, we have described the EXT2 cDNA sequence and a limited characterization of 8 exons (Stickens et al. 1996). However, the efficient detection of mutations in EXT2-linked families requires a more thorough description of the genomic structure of EXT2, including precise exon/intron boundaries. To this end, we have subcloned a yeast artificial chromosome (YAC) clone, which contains the entire EXT2 coding region, into cosmids and have used primer-directed DNA sequencing to identify all of the exon/intron boundaries in the gene. The lengths of the introns were also determined by long-range PCR using cosmid clones and genomic DNA to define a gene of 108 kb that contains 16 exons and utilizes alternative exons and alternative polyadenylation signals.
One approach to unraveling the function of the EXT2 gene is to study its homologs in other species. Here, we describe the cDNA sequence of the mouse EXT2 gene that we have mapped to chromosome 2. We also describe two EXT homologs in Caenorhabditis elegans. The detection of EXT homologs in this nematode worm indicates that these genes do not function exclusively as regulators of bone growth and opens the way for a functional analysis of these genes in lower organisms. In addition, the description of the mouse EXT2 sequence should enable transgenic knockout experiments to be initiated to further elucidate gene function.
RESULTS
The Genomic Structure of the EXT2 Gene
In our previous study of EXT2 (Stickens et al. 1996) we identified a full-length EXT2 cDNA as well as YAC clones that spanned the EXT2 locus on chromosome 11p. PCR amplifications with primer sets from both ends of the gene demonstrated that one of the YACs (923B11 with a length of 1690 kb) contained both the 5′- and 3′-untranslated regions (UTRs) of EXT2 and thus appeared to contain the entire transcription unit (data not shown). We subcloned total genomic DNA from this YAC clone into a cosmid vector and screened ∼3000 of the resulting cosmid clones with a full-length EXT2 cDNA probe, to identify 41 positive cosmid clones. These cosmids were then picked, arrayed and stamped onto several duplicate sets of filters for subsequent rescreening. The filters were hybridized separately with 10 overlapping 300-bp fragments from the EXT2 cDNA, derived by a series of PCR amplifications across the full-length cDNA. This strategy allowed us to rapidly construct a cosmid contig of the gene and to identify a minimal cosmid tiling path of six cosmids that covered the EXT2 locus (C7, B2, C1, E1, B11, and A7; see Fig. 1).
Figure 1.
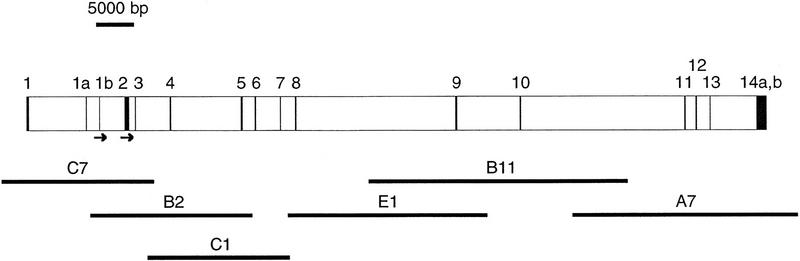
Genomic structure of the EXT2 gene. The 16 exons are represented by solid boxes. Exons 1a and 1b are alternatively spliced exons. The 3′ UTRs 14a and 14b represent alternative polyadenylation sites (see text for details). The initiation codons are indicated by arrows. The cosmids used for DNA sequencing are shown below the map. Their placements reflect their exon content, but they are not necessarily to scale.
To identify the exon/intron boundaries of EXT2, the six cosmids were then used in dye terminator DNA sequencing reactions in conjunction with primers derived from the cDNA sequence. The cDNA primers were positioned ∼150 bp apart throughout the cDNA. An exon/intron boundary was defined as the point at which the derived sequence was no longer colinear with the EXT2 cDNA. Once an exon boundary was identified, a second primer was designed that would prime in the opposite direction to confirm the junction and to identify the other exon end.
This analysis resulted in the identification of 16 exons across the EXT2 locus (Fig. 1), two of which (1a and 1b) are alternatively spliced (Stickens et al. 1996). The DNA sequences surrounding the exon/intron junctions are shown in Figure 2. All of the splice junctions conform to the GT/AG splicing rule. The 16 exon and flanking intron sequences derived in this study have been deposited in GenBank (accession nos. U67353–U67368), and PCR primers designed from these should speed up the analysis of mutations in the EXT2 gene considerably.
Figure 2.

Exon/intron boundaries of the EXT2 gene. Lowercase letters represent intron sequence; uppercase letters, exon sequence; bold letters, the conserved GT/AG splice junctions. The number of total nucleotides of each exon is indicated in parenthesis. Intron lengths are shown in parenthesis on the right. For exon 14, the length is either 783 or 1276, depending on the use of the alternative polyadenylation sites.
The EXT2 gene utilizes alternate polyadenylation signals, resulting in the production of two transcripts of 3.5 and 4.0 kb. This was determined by comparing two expressed sequence tags (ESTs, GenBank accession nos. H51339 and N22755) that overlap with the EXT2 gene but contain an additional 493 bp of 3′ UTR extending beyond the polyadenylation site described previously. This sequence is collinear with the genomic DNA sequence from the end of the gene (data not shown). A canonical polyadenylation signal [ATTAAA (Birnstiel et al. 1985)] lies 23 bp from the 3′ end of this new EST, suggesting that these ESTs were derived from transcripts that use an alternative polyadenylation signal. This interpretation was confirmed by our observation that hybridization of this more distal 3′ UTR to Northern blots results in the detection of only the larger 4.0-kb transcript (data not shown).
We next determined the approximate intron lengths within the gene, to estimate the total size of the EXT2 locus. PCR primers from the ends of adjacent exons were used to amplify complete introns within the cosmid DNAs by long-range PCR methods (Barnes 1994; Cheng et al. 1994). In two cases (between exons 7 and 8 and exons 10 and 11), a complete intron was not contained within a single cosmid (data not shown). These intron lengths were determined instead by PCR amplification of genomic DNA from the YAC clone 923B11, which contains the entire transcription unit.
To confirm that the genomic structure derived from the cosmids and the YAC was a true representation of the genome, we conducted PCRs on total human genomic DNA (from five different individuals) using exon- and intron-specific PCR primer sets. This analysis agreed completely with the exon boundaries identified from the cloned cosmids. However, discrepancies were discovered in 2 of the 15 intron lengths. In these two cases, intron lengths derived from human genomic DNAs were smaller than intron lengths derived from the genomic clones (data not shown). This indicated that the cloned cosmids and/or YAC had undergone some rearrangements. By comparing lengths of PCR products from human genomic, cosmid, and YAC DNA, we were able to determine that these rearrangements originated within the YAC and that the cosmids recapitulated these rearrangements. Rearrangements within YACs are quite frequent (Bates et al. 1992). However, in this case, the alterations appear to be insertions rather than deletions. Figure 1 diagrams the derived genomic structure of the EXT2 gene, along with the cosmid contig. The EXT2 gene extends over ∼108 kb of genomic sequence with an average intron length of 7 kb, the largest intron being 24 kb between exons 10 and 11.
The Mouse Homolog of EXT2
Mouse cDNA clones homologous to the human EXT2 gene were isolated by screening a mouse brain cDNA library with a 581-bp radiolabeled probe from exon 11 to 14 of the human gene. A cDNA contig derived from six different clones produced a cDNA sequence of 2844 bp with an open reading frame of 2154 bp (submitted to GenBank under accession no. U67837). The derived nucleotide sequence has 87.6% identity in the coding region with the human EXT2 gene. The predicted mouse EXT2 protein is 718 amino acids in length and is 94.7% identical with the human EXT2 gene product over the entire length of both proteins (Fig. 3). When comparing only the last one-third of the human and mouse proteins the identity increases to 99.5%.
Figure 3.

Alignment of the human and mouse EXT2 proteins using the gap alignment tool of the Wisconsin Package. The overall identity of the two proteins is 94.7%.
The chromosomal location of the mouse EXT2 gene was determined using SSCP analysis on an interspecific backcross mapping panel (Rowe et al. 1994) with a primer set that was shown to produce a single-strand conformational polymorphism (SSCP) variant between the two mouse species. Using this technique, the mouse homolog of the human EXT2 gene was localized to the central part of chromosome 2 between markers D2Mit15 and Pax6. This localization is consistent with synteny maps that have shown this region of mouse chromosome 2 to be syntenic with human chromosome 11p11 (the region within which the human EXT2 gene resides) (Siracusa et al. 1996).
Homologs in C. elegans
In the course of analyzing the mouse and human EXT2 sequences we conducted TBLASTN and BLASTP searches of the available nucleic acid and amino acid sequence databases. In particular, we compared the EXT family of proteins with a conceptual translation of the C. elegans EST database in a TBLASTN search (Altschul et al. 1990). This search detected four ESTs comprising three unique clones that had significant homology to the EXT proteins with P values ranging from 1.9 × 10−5 to 1.3 × 10−21. These ESTs were then compared to the C. elegans genomic DNA database that identified a single cosmid sequence (F12F6). This cosmid has been mapped to chromosome IV in C. elegans (R. Durbin and J.T. Mieg, http://www.ncbi.nlm.nih.gov) and appears to encode one of the ESTs (yk187a9.5).
To identify additional ESTs that might overlap with yk187a9.5 and possibly extend the cDNA sequence, this EST was compared to the C. elegans EST database. An overlapping EST (yk173d8.5) was identified that also overlaps with sequences in cosmid F12F6. The Genefinder gene identification program (Favello et al. 1995) predicted a total of nine transcripts from this cosmid, two of which overlap with these ESTs. Sequence alignments indicate that this set of ESTs is most similar to EXT1. The combined BLAST alignments comparing the EXT1 protein to the conceptual translation products of F12F6 are shown in Figure 4A and show that this cosmid is homologous to only the amino terminus of the EXT1 protein. Presumably the carboxy-terminal portion of this gene is also contained within the cDNAs from which the ESTs were derived, but these cDNAs have not yet been sequenced in their entirety. The conceptual translation of the cosmid, rather than the ESTs, is shown in Figure 4A because more EXT1 homologous sequences are present in the cosmid DNA sequence than are contained within the relatively short ESTs.
Figure 4.
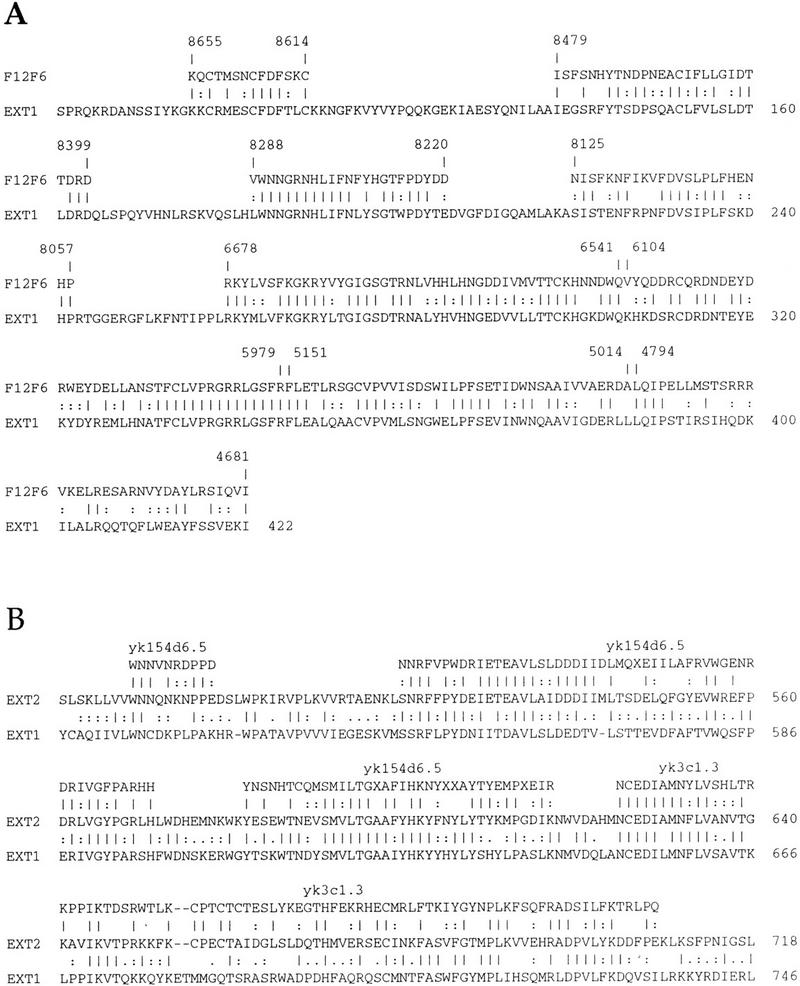
Alignments of human EXT1 and EXT2 proteins with C. elegans proteins using combined TBLASTN results. (A) A conceptual translation of the C. elegans cosmid F12F6 is aligned with the human EXT1 protein. The numbering above corresponds to positions within the nucleotide sequence of F12F6. (B) A conceptual translation of two overlapping C. elegans ESTs aligned with the human EXT2 protein. The alignment of EXT1 with EXT2 is also shown.
The remaining three ESTs (yk154d6.3, yk154d6.5, and yk3c1.3) identified in the TBLASTN search overlap with each other and are most similar to the carboxy-terminal end of EXT2. A search of these ESTs in the C. elegans genomic DNA sequence database using BLAST did not reveal any homologies. However, these ESTs have been mapped to YAC Y40C2 on C. elegans chromosome III (R. Durbin and J.T. Mieg, http://www.ncbi.nlm.nih.gov). Therefore, it appears that these ESTs are distinct from those that map to cosmid F12F6 and that two EXT homologous genes exist in C. elegans. The combined BLAST alignments comparing the EXT2 protein and the conceptual translation products of the three ESTs are shown in Figure 4B.
DISCUSSION
In this study we have determined the exon/intron structure, exon/intron boundary DNA sequences, and size of the human EXT2 gene. This gene is comprised of 16 exons, including 2 alternatively spliced exons, has alternate polyadenylation sites, and covers ∼108 kb of genomic sequence. All of this information has been deposited in, and is accessible through, GenBank. This is clinically important for two reasons: (1) It defines the exon sequences and PCR primers that are necessary for a more thorough mutational analysis of familial and sporadic cases of multiple exostoses; and (2) it defines the genomic borders of the gene that are important for investigating deletions accurately in this region and for future LOH studies in EXT-linked and sporadic tumors (Hecht et al. 1995, 1997; Raskind et al. 1995). Only one intragenic marker (D11S903) is currently known within EXT2 (Stickens et al. 1996). Therefore, the cosmid contig generated in this work should be useful as a source of new intragenic and flanking polymorphic markers that can be used for the detection of deletions in exostoses-derived chondrosarcomas and sporadic chondrosarcomas.
We have also identified the mouse homolog of EXT2 and derived its entire coding sequence. The identity between the mouse EXT2 and the mouse EXT1 proteins identified previously is 34.7%—a figure that is very similar to the 35.2% identity observed between the human EXT1 and EXT2 proteins (Stickens et al. 1996). The identity in the last one-third of EXT1 and EXT2 increases to 44.5% in both the human and mouse proteins and suggests that this region has functional significance. This increased homology at the carboxyl terminus has also been observed in the comparison of the EXT-L protein with the EXT1 and EXT2 gene products (Wise et al. 1997). We have mapped the mouse EXT2 gene within mouse chromosome 2 between D2Mit15 and Pax6 to a region that was known previously to contain a conserved linkage group with human chromosome 11p11 (Siracusa et al. 1996). However, a search of known mouse mutations mapped to this region does not reveal any that have an observed alteration in bone growth or modeling. Nevertheless, the small size of the mouse may not lend itself easily to finding this type of bone malformation, and it is possible that this phenotype may have been overlooked. In any case, our definition of the entire coding sequence of the mouse EXT2 homolog should certainly help in the derivation of transgenic knockouts and further insights into EXT2 function.
A search of the available sequence databases with the EXT proteins surprisingly found two compelling homologs in the nematode worm C. elegans. One of these appears to be the C. elegans EXT1 homolog and maps to chromosome IV. A comparison of amino acids 260–422 from the human EXT1 protein with the corresponding region of this C. elegans putative gene demonstrates a 55.2% identity, whereas a comparison of these same amino acids of EXT1 with the human EXT-L and EXT2 proteins shows only 40.5% and 35.0% identity, respectively. As was mentioned above, the human and mouse EXT proteins show most similarity at their carboxy-terminal ends. The second worm homolog (comprised of three ESTs) does show significant homology to other EXT proteins at the carboxyl terminus and is most homologous to the human EXT2 protein. This gene appears to map to chromosome III of C. elegans.
A previous study that we participated in (Stickens et al. 1996) commented on the apparent presence of EXT genes only in vertebrates and speculated that this might indicate their exclusive involvement in bone growth. The identification of at least two homologs in the nematode worm C. elegans suggests that these EXT proteins play a broader role. Many C. elegans mutants have been roughly mapped to the regions in which the two EXT homologs are located, and several of these mutants cause defects in cell migration and differentiation in early development (R. Durbin and J.T. Mieg, http://www.ncbi.nlm.nih.gov). It is possible that exostoses develop in vertebrates because of aberrant chondrocyte migration within the epiphyseal plate or excessive chondrocyte proliferation at the periphery of the bone. The known C. elegans mutants are thus attractive candidates for future functional analysis. However, the sheer number of roughly mapped mutations in these regions means that assigning one of them as a candidate for a disrupted EXT worm homolog will be difficult. Nevertheless, this study lays the foundations for future functional analyses of the EXT2 gene by complementation experiments in C. elegans and by gene disruption methods in both C. elegans and mouse.
METHODS
Construction and Screening of EXT2 Cosmids
High molecular weight DNA from the Centre d’Etude du Polymorphisme Humain (CEPH) Mega-YAC 923B11 was isolated according to the method of Sambrook et al. (1989). Twelve micrograms of total 923B11 yeast DNA was digested partially with 0.1 unit of Sau3A1 for 10 min and phosphatased. A portion of the DNA (2.5 μg) was ligated to 1 μg of sCos-1 (Stratagene) vector arms in ligase buffer (66 mm Tris (pH 7.5), 5 mm MgCl2, 1 mm DTT, and 1 mm ATP) using 5 units of T4 DNA ligase (Boehringer Mannheim). The reaction was incubated at 14°C for 18 hr.
The ligated DNA was packaged using Gigapack III Gold packaging extract (Stratagene) according to the manufacturer’s directions and infected into XL-1 Blue MR cells. The cells were titered, and ∼3000 colonies were screened (Sambrook et al. 1989) using the full-length EXT2 cDNA as a radiolabeled probe. Forty-one cosmid clones were detected, and these were arrayed into a 96-well microtiter dish, stamped onto nylon filter-covered LB-ampicillin plates, and processed for rescreening with fragments of the EXT2 cDNA (Sambrook et al. 1989). Ten fragments of the EXT2 cDNA were hybridized to the arrayed colony filters to determine an approximate cosmid contig and to select a minimal tiling path for directed DNA sequencing. Six cosmids that covered the entire EXT2 genomic region were purified for sequencing using a QIAfilter plasmid midi kit (Qiagen).
DNA Sequencing of Exon/Intron Boundaries
Cosmid DNAs were sequenced using 1 μg of the template. For the identification of exon boundaries, primers were designed in both directions that were separated by ∼150 bp. These primers were used in conjunction with the ABI dye terminator FS+ kit and an ABI 377 automated DNA sequencer to derive sequences from the cosmid templates. When sequence analysis revealed a divergence between the known cDNA sequence and the genomic DNA sequence (an exon/intron boundary), a secondary primer was constructed in the opposite direction. This was then used in a sequencing reaction to reconfirm the boundary and to identify the other end of the exon. The exon, as well as the partial flanking intron sequences, has been submitted to GenBank (accession nos. U67353–U67368).
Genomic Reconstruction and Estimation of Intron Lengths
Intron lengths were estimated by long-range PCR with primers from adjacent exons using the Expand Long Template PCR System (Boehringer Mannheim). The template for these reactions was 10 pg of cosmid DNA that contained the flanking exons of the intron being analyzed. The length of the introns between exons 7–8 and 10–11 could not be determined by this method, as no cosmids contained both of the respective exons. In these two cases, 1 ng of total 923B11 yeast DNA was used as the genomic DNA template. These intron lengths were confirmed using 250 ng of high molecular weight total human genomic DNA in long PCR reactions.
Mouse cDNA Library Screening and cDNA Sequencing
A PCR product from nucleotide 2097 to 2677 of the human EXT2 cDNA sequence (GenBank accession no. U62740) was generated and radiolabeled using a Rediprime random prime labeling kit (Amersham). This was used as a hybridization probe to screen filter lifts from a mouse brain Lambda ZAP cDNA library (Stratagene). Approximately 600,000 mouse cDNA clones were screened, resulting in the identification of 26 positive clones. Eight of these were picked and excised in vivo using M13K07 helper phage (GIBCO-BRL) according to the manufacturer’s instructions. The clones were amplified by PCR using T3/T7 primers to determine the lengths of the inserts. The PCR products were then cut with Sau3A1 for restriction fragment fingerprint analysis. By this criterion six of the inserts were related. These six DNAs were sequenced using dye terminator cycle sequencing reactions (ABI) on an ABI377 automated DNA sequencer. The mouse cDNA sequence was submitted to GenBank (GenBank accession no. U67837).
Mouse Chromosomal Localization
Primers to the mouse EXT2 3′ UTR (TAGAGGAGTCAGCTGTCCAG and CTTTCAGTAGACATCTTGGC) were found to detect a polymorphism in C57BL/6J and Mus spretus genomic DNA by SSCP analysis (Orita et al. 1989a,b) using mutation detection enhancement (MDE) acrylamide gel solution (FMC BioProducts) for gel electrophoresis. These primers were typed through the Jackson Laboratory BSB [(C57BL/6J × M. spretus)F1 × C57BL/6J] backcross mapping panel resource (Rowe et al. 1994) to determine the chromosomal position. Segregation patterns were analyzed by the Jackson Laboratory and placed the mouse EXT2 gene within the central part of chromosome 2. The map position relative to other markers is as follow: proximal–Hoxd11, Acra, D2Mit11–4.25 cM +/−2.08 (SE)–D2Mit15–1.06 +/− 1.06–Ext2–4.25 +/−2.08–Pax6–distal.
Sequence Analysis
The gap alignment tool of the Wisconsin Package (Genetics Computer Group) (Devereux et al. 1984) was used to align the human and mouse EXT2 conceptual translation products. The C. elegans EXT homologs were identified using the TBLASTN and BLASTP (Altschul et al. 1990) programs; searching the EXT1, EXT2, and EXTL protein sequence against the conceptual translations of the C. elegans genomic and EST databases, and also the GeneFinder (Favello et al. 1995) predicted proteins at the Sanger Center C. elegans database.
Acknowledgments
We are grateful to Dr. Anne Bowcock for critical reading of this manuscript and to Dr. Leon Avery for helpful advice and discussion during the course of this work. G.C. is the recipient of a Perot Scholar Award. This work was supported by grant HG00368 to M.L. from the National Center for Human Genome Research and by a collaborative agreement (project TSRHC 11-95-345) funded by Texas Scottish Rite Hospital to M.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lovett@ryburn.swmed.edu; FAX (214) 648-1666.
REFERENCES
- Ahn J, Ludecke HJ, Lindow S, Horton WA, Lee B, Wagner MJ, Horsthemke B, Wells DE. Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1) Nature Genet. 1995;11:137–143. doi: 10.1038/ng1095-137. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barnes WM. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch O, Wuyts W, Van Hul W, Hecht JT, Meinecke P, Hogue D, Werner W, Zabel B, Hinkel GK, Powell CM, Shaffer LG, Willems PJ. Delineation of a contiguous gene syndrome with multiple exostoses, enlarged parietal foramina, craniofacial dysostosis, and mental retardation, caused by deletions in the short arm of chromosome 11. Am J Hum Genet. 1996;58:734–742. [PMC free article] [PubMed] [Google Scholar]
- Bates GP, Valdes J, Hummerich H, Baxendale S, Le Paslier DL, Monaco AP, Tagle D, MacDonald ME, Altherr M, Ross M, et al. Characterization of a yeast artificial chromosome contig spanning the Huntington’s disease gene candidate region. Nature Genet. 1992;1:180–187. doi: 10.1038/ng0692-180. [DOI] [PubMed] [Google Scholar]
- Birnstiel ML, Busslinger M, Strub K. Transcription termination and 3′ processing: The end is in site! Cell. 1985;41:349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Cheng S, Fockler C, Barnes WM, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A, Raskind W, Blanton SH, Pauli RM, Gregg RG, Francomano CA, Puffenberger E, Conrad EU, Schmale G, Schellenberg G, et al. Genetic heterogeneity in families with hereditary multiple exostoses. Am J Hum Genet. 1993;53:71–79. [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki T, Yanagisawa A, Ohta K, Aiso S, Watanabe M, Hibi T, Kato Y, Nakajima T, Ariyama T, Inazawa J, Nakamura Y, Horii A. Deletion mapping on chromosome 1p in well-differentiated gastric cancer. Br J Cancer. 1996;73:424–428. doi: 10.1038/bjc.1996.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favello A, Hillier L, Wilson RK. Genomic DNA sequencing methods. Methods Cell Biol. 1995;48:551–569. doi: 10.1016/s0091-679x(08)61403-x. [DOI] [PubMed] [Google Scholar]
- Hecht JT, Hogue D, Strong LC, Hansen MF, Blanton SH, Wagner M. Hereditary multiple exostosis and chondrosarcoma: Linkage to chromosome 11 and loss of heterozygosity for EXT-linked markers on chromosomes 11 and 8. Am J Hum Genet. 1995;56:1125–1131. [PMC free article] [PubMed] [Google Scholar]
- Hecht JT, Hogue D, Wang Y, Blanton SH, Wagner M, Strong LC, Raskind W, Hansen MF, Wells D. Hereditary multiple exostoses: Mutational studies of familial EXT1 cases and EXT associated malignancies. Am J Hum Genet. 1997;60:80–86. [PMC free article] [PubMed] [Google Scholar]
- Hennekam RC. Hereditary multiple exostoses. J Med Genet. 1991;28:262–266. doi: 10.1136/jmg.28.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe HL. Hereditary multiple exostosis. Arch Pathol. 1943;36:335–357. [Google Scholar]
- Le Merrer M, Legeai-Mallet L, Jeannin PM, Horsthemke B, Schinzel A, Plauchu H, Toutain A, Achard F, Munnich A, Maroteaux P. A gene for hereditary multiple exostoses maps to chromosome 19p. Hum Mol Genet. 1994;3:717–722. doi: 10.1093/hmg/3.5.717. [DOI] [PubMed] [Google Scholar]
- Lothe RA, Andersen SN, Hofstad B, Meling GI, Peltomaki P, Heim S, Brogger A, Vatn M, Rognum TO, Borresen AL. Deletion of 1p loci and microsatellite instability in colorectal polyps. Genes Chromosomes Cancer. 1995;14:182–188. doi: 10.1002/gcc.2870140305. [DOI] [PubMed] [Google Scholar]
- Ludecke HJ, Johnson C, Wagner MJ, Wells DE, Turleau C, Tommerup N, Latos-Bielenska A, Sandig KR, Meinecke P, Zabel B, et al. Molecular definition of the shortest region of deletion overlap in the Langer-Giedion syndrome. Am J Hum Genet. 1991;49:1197–1206. [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Negrini M, Carter SL, Gillum DR, Rosenberg AL, Schwartz GF, Croce CM. Detection and cloning of a common region of loss of heterozygosity at chromosome 1p in breast cancer. Cancer Res. 1995;55:1752–1757. [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci. 1989a;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M, Suzuki Y, Sekiya T, Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989b;5:874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- Parrish JE, Wagner MJ, Hecht JT, Scott Jr CI, Wells DE. Molecular analysis of overlapping chromosomal deletions in patients with Langer-Giedion syndrome. Genomics. 1991;11:54–61. doi: 10.1016/0888-7543(91)90101-j. [DOI] [PubMed] [Google Scholar]
- Raskind WH, Conrad EU, Chansky H, Matsushita M. Loss of heterozygosity in chondrosarcomas for markers linked to hereditary multiple exostoses loci on chromosomes 8 and 11. Am J Hum Genet. 1995;56:1132–1139. [PMC free article] [PubMed] [Google Scholar]
- Rowe LB, Nadeau JH, Turner R, Frankel WN, Letts VA, Eppig JT, Ko MS, Thurston SJ, Birkenmeier EH. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmale GA, Conrad E, Raskind WH. The natural history of hereditary multiple exostoses. J Bone Jt Surg Am Vol. 1994;76:986–992. doi: 10.2106/00004623-199407000-00005. [DOI] [PubMed] [Google Scholar]
- Siracusa LD, Morgan JL, Fisher JK, Abbott CM, Peters J. Mouse Chromosome 2. Mamm Genome. 1996;6:S51–S63. [PubMed] [Google Scholar]
- Solomon L. Bone growth in diaphyseal aclasis. J Bone J Surg Br Vol. 1961;43:700–716. doi: 10.1302/0301-620X.43B4.700. [DOI] [PubMed] [Google Scholar]
- Stickens D, Clines G, Burbee D, Ramos P, Thomas S, Hogue D, Hecht JT, Lovett M, Evans GA. The EXT2 multiple exostoses gene defines a family of putative tumour suppressor genes. Nature Genet. 1996;14:25–32. doi: 10.1038/ng0996-25. [DOI] [PubMed] [Google Scholar]
- Voutsinas S, Wynne-Davies R. The infrequency of malignant disease in diaphyseal aclasis and neurofibromatosis. J Med Genet. 1983;20:345–349. doi: 10.1136/jmg.20.5.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicklund CL, Pauli RM, Johnston D, Hecht JT. Natural history study of hereditary multiple exostoses. Am J Med Genet. 1995;55:43–46. doi: 10.1002/ajmg.1320550113. [DOI] [PubMed] [Google Scholar]
- Wise CA, Clines GA, Massa H, Trask BJ, Lovett M. Identification and localization of the gene for EXTL, a third member of the multiple exostoses gene family. Genome Res. 1997;7:10–16. doi: 10.1101/gr.7.1.10. [DOI] [PubMed] [Google Scholar]
- Wu YQ, Heutink P, de Vries BB, Sandkuijl LA, van den Ouweland AM, Niermeijer MF, Galjaard H, Reyniers E, Willems PJ, Halley DJ. Assignment of a second locus for multiple exostoses to the pericentromeric region of chromosome 11. Hum Mol Genet. 1994;3:167–171. doi: 10.1093/hmg/3.1.167. [DOI] [PubMed] [Google Scholar]
- Wuyts W, Ramlakhan S, Van Hul W, Hecht JT, van den Ouweland AM, Raskind WH, Hofstede FC, Reyniers E, Wells DE, de Vries B, et al. Refinement of the multiple exostoses locus (EXT2) to a 3-cM interval on chromosome 11. Am J Hum Genet. 1995;57:382–387. [PMC free article] [PubMed] [Google Scholar]
- Wuyts W, Vanhul W, Wauters J, Nemtsova M, Reyniers E, Vanhul E, Deboulle K, Devries BBA, Hendrickx J, Herrygers I, Bossuyt P, Balemans W, Fransen E, Vits L, Coucke P, Nowak NJ, Shows TB, Mallet L, Vandenouweland AMW, McGaughran J, Halley DJJ, Willems PJ. Positional cloning of a gene involved in hereditary multiple exostoses. Hum Mol Genet. 1996;5:1547–1557. doi: 10.1093/hmg/5.10.1547. [DOI] [PubMed] [Google Scholar]


