Abstract
Certain small outer envelope membrane proteins of chloroplasts are encoded by the nuclear genome without a cleavable N-terminal transit peptide. We investigated in vivo the targeting mechanism of AtOEP7, an Arabidopsis homolog of the small outer envelope membrane protein. AtOEP7 was expressed as a fusion protein with the green fluorescent protein (GFP) either transiently in protoplasts or stably in transgenic plants. In either case, fluorescence microscopy of transformed cells and protein gel blot analysis of fractionated proteins confirmed that the AtOEP7:GFP fusion protein was targeted to the chloroplast outer envelope membrane. In vivo targeting experiments revealed that two regions, the transmembrane domain (TMD) and its C-terminal neighboring seven–amino acid region, were necessary and sufficient for targeting to the chloroplast outer membrane. Substitution of aspartic acid or lysine residues with glycine residues or scrambling of the amino acid sequence of the seven–amino acid region caused mistargeting to the plasma membrane. Although the amino acid sequence of the TMD is not important for targeting, amino acid residues with large side chains inhibited targeting to the chloroplasts and resulted in the formation of large aggregates in the protoplasts. In addition, introduction of a proline residue within the TMD resulted in inhibition of targeting. Finally, a fusion protein, AtOEP7:NLS:GFP, was targeted efficiently to the chloroplast envelope membranes despite the presence of a nuclear localization signal. On the basis of these results, we conclude that the seven–amino acid region and the TMD are determinants for targeting to the chloroplast outer envelope membrane. The seven–amino acid region plays a critical role in AtOEP7 evading the endomembrane system and entering the chloroplast pathway, and the TMD plays critical roles in migration to the chloroplasts and/or subsequent insertion into the membrane.
INTRODUCTION
The majority of chloroplast proteins are encoded by the nuclear genome and synthesized in the cytosol. Transport of these nucleus-encoded proteins to the correct cellular compartment is critical for the biogenesis and function of chloroplasts (Pilgrim et al., 1998; Keegstra and Froehlich, 1999; Bauer et al., 2000). These nucleus-encoded proteins can be divided into several groups based on their final destinations, which include the stroma, the thylakoid membranes, the lumen of thylakoid membranes, and the inner and outer envelope membranes of the chloroplast (Perry and Keegstra, 1994; Kouranov et al., 1999; Sohrt and Soll, 2000). The proteins transported to the chloroplast outer envelope membrane can be divided into two groups: those with (Tranel et al., 1995; Muckel and Soll, 1996) and without (Salomon et al., 1990; Wu and Ko, 1993; Li and Chen, 1996) a cleavable N-terminal transit peptide. Most of the outer envelope membrane proteins lack the N-terminal transit peptide (Salomon et al., 1990; Wu and Ko, 1993; Li and Chen, 1996), and only a few of them, such as TOC75 and TOC159, have been shown to have this N-terminal peptide (Tranel et al., 1995; Muckel and Soll, 1996). The outer membrane proteins lacking a transit peptide, such as spinach OEP6.7, pea OEP14, and pea OEP34, all were targeted to purified chloroplasts when assayed in an in vitro import system using proteins translated in vitro (Salomon et al., 1990; Li et al., 1991; Li and Chen, 1996; Chen and Schnell, 1997). These experiments identified the first 30 amino acids of OEP14 (Li et al., 1991), the first 40 amino acids of SCE/Com70 (Wu and Ko, 1993), and the 10–amino acid hydrophobic core at the C-terminal membrane anchor of OEP34 (Li and Chen, 1996) as the outer membrane targeting signal. Unlike proteins having transit peptides, translocation of these proteins to the outer envelope membrane is not ATP dependent (Wu and Ko, 1993; Li and Chen, 1996). However, recently, Tu and Li (2000) reported that proteinaceous components of chloroplast envelope membranes are required for insertion of OEP14 into the outer envelope membrane.
In this study, we investigated the subcellular targeting of an outer envelope chloroplast protein, AtOEP7, fused to green fluorescent protein (GFP) in vivo. We monitored targeting of AtOEP7:GFP fusion proteins that were expressed either transiently in protoplasts prepared from Arabidopsis or stably in transgenic plants. Here, we present evidence that the seven-amino acid region located to the C-terminal of the transmembrane domain (TMD) of AtOEP7 determines the targeting specificity of AtOEP7 to the chloroplast outer envelope membrane and that it most likely requires a factor in the cytosol to accomplish this process.
RESULTS
Isolation and Expression of an Arabidopsis Homolog of Pea OEP14
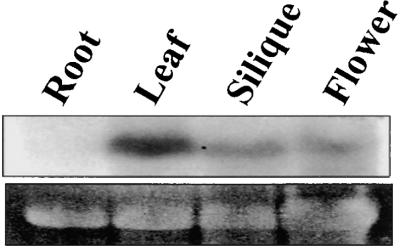
Pea OEP14 is one of several low molecular weight proteins found in the chloroplast outer envelope membrane (Li et al., 1991). A DNA sequence encoding an OEP14 homolog is present in the Arabidopsis genome with a predicted molecular mass of ∼7 kD; thus, we named the Arabidopsis homolog AtOEP7. To isolate and characterize the gene encoding AtOEP7, we performed polymerase chain reaction (PCR) amplification using gene-specific primers. The amino acid sequence of AtOEP7 showed 54 and 38% amino acid sequence identity with OEP14 of pea and OEP6.7 of spinach (Salomon et al., 1990), respectively (Figure 1). Because AtOEP7 was isolated from genomic DNA by PCR amplification, we investigated whether AtOEP7 was actually expressed in plant tissues. Therefore, RNA gel blot analysis was performed with total RNA obtained from various tissues using AtOEP7 DNA as a hybridization probe. As shown in Figure 2, an RNA transcript was clearly detected using an AtOEP7-derived probe in RNA samples obtained from green tissues but not from root tissues, supporting a role for this protein in the chloroplast.
Figure 1.
Amino Acid Sequence Alignment of AtOEP7 and Other Chloroplast Outer Membrane Proteins.
The deduced amino acid sequence of AtOEP7 was used to search GenBank using BlastP of the National Center for Biotechnology Information E-mail server. Gaps were introduced to maximize the sequence alignment. The accession numbers for pea OEP14 and spinach OEP6.7 (SpOEP6.7) are AAA63414 and AAA34035, respectively.
Figure 2.
RNA Gel Blot Analysis of AtOEP7.
Total RNA was isolated from various tissues, and 15 μg of RNA was separated on a 1.5% formaldehyde–agarose gel. RNA was transferred onto a nylon membrane and probed using 32P-labeled AtOEP7 DNA.
AtOEP7:GFP Fusion Protein Is Targeted to Chloroplast Envelope Membranes in Protoplasts
It was shown previously, using the in vitro import assay, that pea OEP14 is targeted to the outer envelope membrane by a novel pathway that does not require ATP or an N-terminal transit peptide (Li et al., 1991; Tu and Li, 2000). In this study, the GFP was fused to the N or C terminus of AtOEP7 to investigate the targeting of AtOEP7 within intact cells. Each fusion construct was introduced into protoplasts prepared from Arabidopsis tissues by the polyethylene glycol transformation method (Jin et al., 2001). As shown in Figure 3B, AtOEP7 was targeted to chloroplasts when GFP was fused to the C terminus of AtOEP7 (d and e). AtOEP7:GFP gave ring patterns surrounding the chloroplast, whereas the control, soluble modified GFP, was distributed uniformly in the cytosol (a and b), suggesting that AtOEP7:GFP is targeted to the chloroplast envelope membranes. However, when GFP was fused to the N terminus, it gave a punctate staining pattern (g and h, arrows), indicating that it was targeted to other organelles.
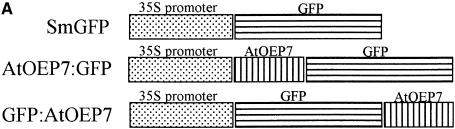
Figure 3.
In Vivo Targeting of AtOEP7:GFP in Protoplasts.
(A) Scheme showing the various fusion constructs used in these experiments. SmGFP, soluble modified GFP.
(B) In vivo targeting of AtOEP7:GFP in protoplasts. Arabidopsis protoplasts were transformed with SmGFP (soluble modified GFP), AtOEP7:GFP, and GFP:AtOEP7 and examined at various times after transformation. a, d, and g show GFP in the transformed protoplasts. b, e, and h show overlap of GFP (green) and chlorophyll (CH) (red). c, f, and i are bright-field images (Bright). These are representative data of protoplasts that expressed these fusion proteins 24 hr after transformation. At least three independent transformation experiments were performed with each fusion construct. Bars = 20 μm. Arrows in g indicate punctate stains.
(C) Protein gel blot analysis of AtOEP7:GFP. Total proteins were isolated from the transformed protoplasts and fractionated into soluble (S) and membrane (M) fractions by ultracentrifugation. These fractions were analyzed by protein gel blot analysis using a monoclonal anti-GFP antibody. C indicates untransformed protoplasts.
To identify the organelle that gives the punctate staining patterns, we attempted colocalization of GFP:AtOEP7 with various reporter proteins, such as sialyltransferase (ST):red fluorescent protein (RFP) for the Golgi apparatus, binding protein (BiP):RFP for the endoplasmic reticulum (ER), F1ATPase-γ:RFP for the mitochondria, and AtVTI1a:RFP for the prevacuolar compartment (Kim et al., 2001). The green fluorescent signals of GFP:AtOEP7 did not overlap the red fluorescent signals of any of these markers (data not shown). Thus, the identity of the organelle in which GFP:AtOEP7 is targeted remains to be identified. One possibility is that they may be aggregates of GFP:AtOEP7 caused by the hydrophobic TMD of AtOEP7. It has been reported that the N terminus of pea OEP14 was inserted into the intermembrane space between the two chloroplast envelope membranes (Li and Chen, 1996). Likewise, our results suggest that the N terminus of AtOEP7 is critical for targeting to the envelope outer membrane, and this function was inhibited when AtOEP7 was fused to GFP.
Next, we investigated whether the fusion protein was inserted into the chloroplast envelope membrane. Total protoplast lysates were fractionated into membrane and soluble protein fractions by ultracentrifugation (Li et al., 1991). The presence of AtOEP7:GFP in these fractions was assayed by protein gel blot analysis using a monoclonal anti-GFP antibody. The anti-GFP antibody detected a protein species with a molecular mass of 35 kD, the expected size of AtOEP7:GFP, in the membrane fraction (Figure 3C, lane M) but not in the supernatant fraction containing soluble proteins (Figure 3C, lane S). Thus, these results strongly suggest that the fusion protein was inserted into the chloroplast envelope membrane.
AtOEP7:GFP Was Inserted into the Outer Envelope Membrane of Chloroplasts in Transgenic Plants
To further confirm that AtOEP7:GFP was inserted into the outer envelope membrane, we produced transgenic plants expressing AtOEP7:GFP, because transformed protoplasts did not produce enough material for a detailed analysis. Transgenic plants expressing AtOEP7:GFP were generated using a binary vector containing the AtOEP7:GFP construct under the control of the 35S promoter of Cauliflower mosaic virus. GFP was detected in the chloroplasts of protoplasts prepared from these transgenic plants by fluorescence microscopy. The green fluorescent signals were present as rings identical to those observed in the transformed protoplasts (Figure 4A), suggesting that AtOEP7:GFP was localized at the chloroplast outer envelope membrane. Next, we further analyzed the localization of AtOEP7:GFP by protein gel blot analysis. Intact chloroplasts were isolated from protoplasts prepared from leaf tissues of transgenic plants using a Percoll gradient (Cline et al., 1985). Total chloroplast lysates were first fractionated into thylakoid membrane and soluble fractions by low-speed centrifugation. The soluble fraction that contained stromal proteins and envelope membranes then was further fractionated into high-speed membrane and soluble fractions by ultracentrifugation (Li et al., 1991). The high-speed membrane and soluble fractions contain the envelope membranes and the stromal soluble proteins, respectively. These fractions were assayed for the presence of AtOEP7:GFP using the monoclonal anti-GFP antibody. As shown in Figure 4B, a protein corresponding to AtOEP7:GFP was detected only in the high-speed membrane fraction (lane M) but not in the high-speed soluble fraction (lane S), indicating that AtOEP7:GFP is associated with membranes.
Figure 4.

Localization of AtOEP7:GFP to the Outer Envelope Membrane in Transgenic Plants.
(A) Targeting of AtOEP7:GFP in transgenic plants. Protoplasts were prepared from transgenic plants that stably overexpressed the AtOEP7:GFP gene and examined under a fluorescence microscope. GFP/CH indicates the overlap of green and red fluorescent signals of GFP and chlorophyll. Bar = 20 μm.
(B) Suborganellar localization of AtOEP7:GFP. Intact chloroplasts were purified on a Percoll gradient from the leaf tissues of transgenic plants. Total chloroplast extracts were fractionated into two fractions: low-speed soluble and pellet fractions. The low-speed soluble fraction was further fractionated into soluble (S) and membrane (M) fractions by ultracentrifugation. The high-speed soluble and membrane fractions then were probed with an anti-GFP antibody. The high-speed membrane fraction was treated with 0.1 M Na2CO3 or 1% Triton X-100. The soluble proteins were separated from the insoluble membrane fraction by ultracentrifugation and probed for the presence or absence of AtOEP7:GFP using an anti-GFP antibody. CH, chloroplast.
(C) Thermolysin treatment of intact chloroplasts. The intact chloroplasts were treated with thermolysin, and the reactions were stopped at various times. Total chloroplast extracts were prepared and probed with different antibodies as indicated.
To gain insight into the nature of the membrane association of AtOEP7:GFP, the high-speed membrane fraction was treated with either 0.1 M Na2CO3, pH 11.5, or 1.0% Triton X-100, and the proteins still associated with the treated membranes were pelleted by ultracentrifugation. The resulting soluble and membrane fractions were assayed for the presence of AtOEP7:GFP using the anti-GFP antibody. As shown in Figure 4B, AtOEP7:GFP remained in the membrane fraction after Na2CO3 treatment. In contrast, AtOEP7:GFP was detected in the soluble fraction when the high-speed membrane fraction was treated with Triton X-100. Together, these results indicate that the fusion protein was inserted into membranes. Next, we examined the thermolysin sensitivity of AtOEP7:GFP in intact chloroplasts to determine if this protein was present and, if so, to determine its orientation at the chloroplast envelope membranes (Li et al., 1991). Intact chloroplasts were isolated on a Percoll gradient and then treated with thermolysin for various times. Subsequently, total chloroplast proteins were isolated and assayed for the presence of AtOEP7:GFP by protein gel blot analysis using the anti-GFP antibody. We also performed protein gel blot analysis for several other chloroplast proteins that are already known to be localized in the chloroplast. These include the inner membrane protein Tic110 and two stromal proteins, RbcS and RbcL (Bauer et al., 2000). As shown in Figure 4C, the AtOEP7:GFP protein species at 35 kD was barely detectable after 30 min of thermolysin treatment, suggesting that at least the GFP portion of the fusion protein was accessible. In contrast, the inner membrane protein, Tic110, and the two stromal proteins, RbcL and RbcS, remained intact after 60 min of thermolysin treatment, indicating that the chloroplasts remained intact during this assay. These results strongly suggest that AtOEP7:GFP is an outer envelope membrane protein and that the C-terminal GFP domain, like the C terminus of pea OEP14 (Li and Chen, 1996), is exposed to the cytoplasm.
TMD and the Neighboring C-Terminal Seven Amino Acid Residues of AtOEP7 Are Necessary and Sufficient for Targeting to the Chloroplasts
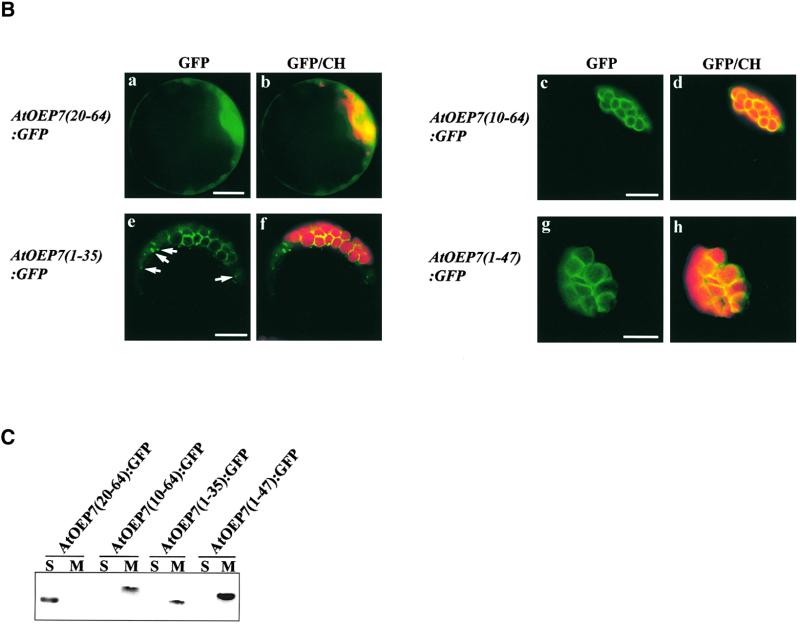
In the case of pea OEP14, the homolog of AtOEP7, it has been shown by in vitro import experiments that the N-terminal 35–amino acid region (Li et al., 1991) is necessary and sufficient for targeting of proteins into the chloroplast outer envelope membrane. We wanted to confirm whether this is true for AtOEP7 by in vivo targeting experiments. The primary structure of AtOEP7 can be divided into three regions based on hydropathy analysis: an N-terminal region with nine amino acid residues, a central hydrophobic region with 19 amino acid residues, and a C-terminal region with 35 amino acid residues (Figure 5). We generated various deletion constructs (Figure 5A), which subsequently were fused to the N terminus of GFP. These were expressed transiently in protoplasts and monitored by fluorescence microscopy. As shown in Figure 5B, the deletion fusion proteins AtOEP7(10-64): GFP (c and d), AtOEP7(1-35):GFP (e and f), and AtOEP7(1-47): GFP (g and h) were targeted to the envelope membrane of the chloroplasts. However, the targeting efficiency of AtOEP7(1-35):GFP was reduced slightly, as indicated by the presence of some punctate staining in the protoplasts (arrows in e), compared with the two other deletion fusion proteins, implying that the region from amino acid residues 36 to 47 may contribute to targeting efficiency. The punctate stains did not colocalize with ST:RFP of the Golgi apparatus, BiP:RFP of the ER, F1ATPase-γ:RFP of the mitochondria, or AtVTI1a:RFP of the prevacuolar compartment (data not shown). In contrast, the deletion mutant AtOEP7(20-64): GFP (a and b), which had a deletion of the first 19 amino acids, including 12 amino acid residues of the TMD, was not targeted to the chloroplast envelope (a and b) and instead gave a diffuse pattern throughout the cytosol.
Figure 5.
Transmembrane Region and Its Small C-Terminal Region Are Sufficient for Protein Targeting.
(A) Hydropathy plot of AtOEP7 and scheme of the various deletion mutants.
(B) In vivo targeting of various AtOEP7 deletion constructs that were expressed as GFP fusion proteins. Protoplasts were transformed with AtOEP7(20-64):GFP (a and b), AtOEP7(10-64):GFP (c and d), AtOEP7(1-35):GFP (e and f), or AtOEP7(1-47):GFP (g and h), and green fluorescent signals were examined 12 to 36 hr after transformation. At least three independent transformation experiments were performed for each fusion construct. Arrows in e indicate the punctate stains in the protoplasts. Red fluorescent signals indicate chlorophyll (CH). Bars = 20 μm.
(C) Protein gel blot analysis. Total protein extracts were prepared from the transformed protoplasts and fractionated into soluble and membrane fractions by ultracentrifugation. These fractions then were probed with an anti-GFP antibody. S and M indicate the soluble and membrane fractions, respectively.
Next, we wanted to further confirm the localization of these fusion proteins by protein gel blot analysis. Cellular extracts prepared from the transformed protoplasts were fractionated into soluble protein and membrane fractions by ultracentrifugation. These fractions were assayed for the presence of the GFP fusion proteins by protein gel blot analysis with the anti-GFP antibody. As shown in Figure 5C, all of these fusion derivatives, except for AtOEP7(20-64):GFP, were present in the membrane fractions, suggesting that they were targeted to the outer envelope membrane. Thus, these results suggest that residues 10 to 20 are necessary and residues 1 to 35 are sufficient for targeting to the chloroplast envelope membrane and that the in vivo results are in agreement with the previous results with pea OEP14 obtained in vitro (Li et al., 1991).
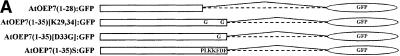
Seven–Amino Acid Region Located at the C Terminus of the TMD Is a Critical Determinant of Targeting Specificity
Although the 26–amino acid region is necessary and sufficient for chloroplast targeting, the nature of the information encoded in this region remains unclear. To further enhance our understanding of the signal sequence, we first wanted to define a minimal region that can function as a targeting signal sequence. The 26–amino acid region consists of two different regions: the 19–amino acid hydrophobic TMD and its C-terminal seven–amino acid region. We generated an additional construct without the seven–amino acid region and fused it to the N terminus of GFP (Figure 6A). As shown in Figure 6B (a and b), the deletion mutant AtOEP7(1-28):GFP was not targeted to the chloroplasts but instead gave a ring pattern, indicating that it may be targeted to the plasma membrane. To confirm this hypothesis, we attempted colocalization of AtOEP7(1-28):GFP with H+-ATPase:RFP, a marker protein for the plasma membrane. Previously, it was shown that H+-ATPase:GFP was targeted to the plasma membrane in Arabidopsis protoplasts (Kim et al., 2001). In this study, we replaced GFP with RFP. As shown in Figure 6B (c to e), the green fluorescent signals of AtOEP7(1-28):GFP closely overlapped the red fluorescent signals of H+-ATPase:RFP, confirming that AtOEP7(1-28):GFP was targeted to the plasma membrane. This result strongly suggests that the seven–amino acid region is critical for protein targeting.
Figure 6.

Seven–Amino Acid Region Is Critical for Targeting to the Chloroplast Outer Envelope Membrane.
(A) Scheme of various mutations within the seven–amino acid region.
(B) In vivo targeting. Protoplasts were transformed with AtOEP7(1-28): GFP (a and b) and AtOEP7(1-28):GFP plus H+-ATPase:RFP (c to e), and localization of green and red fluorescent signals was examined 12 to 36 hr after transformation. Green, red, and blue fluorescent signals indicate GFP, RFP, and chlorophyll (CH), respectively. Note that the autofluorescent signal of chlorophyll is colored blue.
(C) In vivo targeting of various seven–amino acid region mutants. Protoplasts were transformed with AtOEP7(1-35)[K29,34G]:GFP (a and b), AtOEP7(1-35)[D33G]:GFP (c and d), or AtOEP7(1-35)S:GFP (e and f).
To further confirm that the seven–amino acid region plays a role in targeting, we introduced point mutations within this region (Figure 6A). For targeting of Tom20 to the mitochondrial outer membrane in animal cells, positive charges within five amino acid residues from the C terminus of the TMD have been shown to be important (Kanaji et al., 2000). The lysine or aspartic acid residues were replaced with glycine residues, and the mutant was fused to GFP. As shown in Figure 6C, AtOEP7(1-35)[K29,34G] (a and b) and AtOEP7(1-35)[D33G] (c and d) were not targeted to the chloroplasts but instead were targeted to the plasma membrane, as observed with AtOEP7(1-28), indicating that both positively and negatively charged residues play critical roles. Next, to determine the importance of the amino acid sequence of this seven–amino acid region, we generated a mutant, AtOEP7(1-35)S, that had a scrambled amino acid sequence but an identical amino acid composition within the seven–amino acid region (Figure 6A). As shown in Figure 6C (e and f), AtOEP7(1-35)S was targeted to the plasma membrane. Together, these results strongly suggest that the amino acid sequence of the seven–amino acid region is a critical signal sequence for chloroplast targeting. This is in contrast to the results obtained with Tom20 for mitochondrial targeting in animal cells (Kanaji et al., 2000).
TMD Plays an Important Role in Targeting Proteins to the Chloroplast Outer Membrane
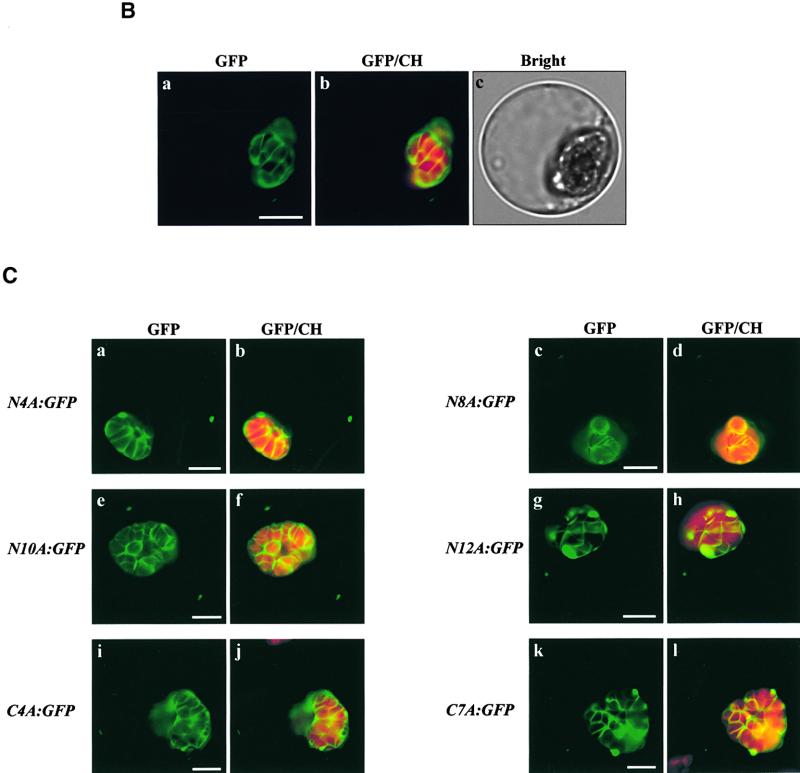
Next, we wanted to investigate whether the TMD plays a role in the targeting of AtOEP7 to the chloroplast envelope membrane. In the case of cytochrome b5, the length of the TMD has been shown to play an important role in determining its localization at the ER (Pedrazzini et al., 1996). Also, for targeting of Tom20 to the mitochondria, the TMD must have moderate hydrophobicity (Kanaji et al., 2000). To address this question, we generated various substitution mutants of the TMD (Figure 7A). First, we generated alanine scanning mutants A2-2:GFP to A2-8:GFP by simultaneously replacing two amino acid residues of the TMD with two alanine residues throughout the entire TMD. As shown in Figure 7B, all of these mutants were targeted correctly to the chloroplast, as in the case of the wild-type AtOEP7: GFP. Also, when the length of the TMD was increased by two alanine residues at the N terminus of the TMD, the resulting mutant, A2-1:GFP, was targeted to the chloroplast as efficiently as was the wild-type protein. Next, we generated additional mutants with 4, 7, 8, 10, or 12 alanine residues in a row from either the N or the C terminus of the TMD (Figure 7A). These mutants were fused to GFP and introduced into protoplasts. As shown in Figure 7C, these mutants (N4A:GFP, N8A:GFP, N10A:GFP, N12A:GFP, C4A: GFP, and C7A:GFP) were targeted correctly to the chloroplasts, suggesting that alanine can substitute for almost all of the amino acid residues in the TMD without any effect on targeting.
Figure 7.
Amino Acid Sequence of the TMD Is Not Important for Targeting.
(A) Scheme of various mutants within the TMD.
(B) Targeting of alanine scanning mutants. Protoplasts were transformed with A2-1:GFP to A2-8:GFP. A protoplast transformed with A2-1:GFP is shown. All other constructs gave identical green fluorescent patterns (data not shown).
(C) Targeting of multiple alanine substitution mutants. Protoplasts were transformed with N4A:GFP (a and b), N8A:GFP (c and d), N10A:GFP (e and f), N12A:GFP (g and h), C4A:GFP (i and j), or C7A:GFP (k and l).
Green and red signals indicate GFP and chlorophyll (CH), respectively. Bars = 20 μm.
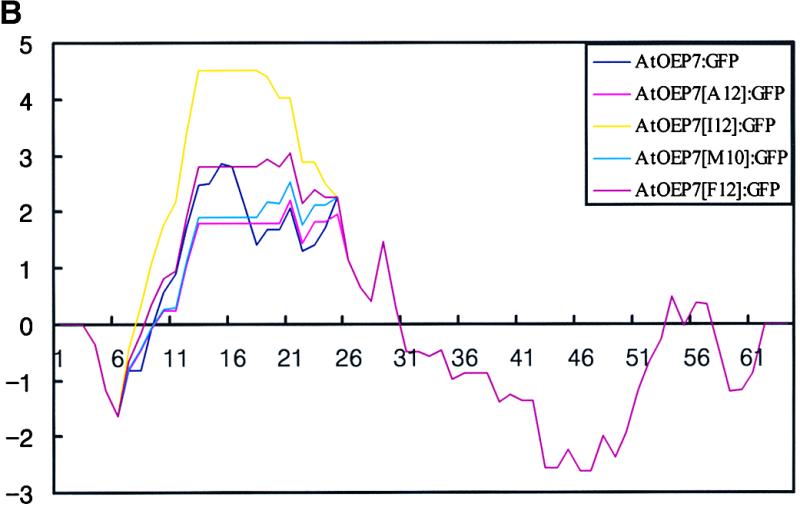
These results clearly suggest that the amino acid residues of the TMD are not important for targeting and that the TMD may function simply as an anchor for the localization of AtOEP7 at the chloroplast outer envelope membrane. However, because only alanine residue, which has the smallest side chain, was used to substitute amino acid residues in the TMD, we wanted to examine whether other hydrophobic amino acid residues having larger side chains also could be used without affecting protein targeting. This time, we selected hydrophobic amino acid residues such as isoleucine, methionine, and phenylalanine, all of which have side chains of different sizes. Again, we generated TMD mutants with 12 isoleucine, 10 methionine, or 12 phenylalanine residues in a row from the N terminus and fused these mutants to the N terminus of the GFP coding region (Figure 8A). First, we examined the hydrophobicity of these TMD mutants by the method of Kyte and Doolittle (1982). As shown in Figure 8B, the wild-type TMD had hydrophobicity ranging from 1.5 to 3. The TMDs of A12, I12, M10, and F12 had hydrophobicities ranging from 1.5 to 2, from 2 to 4.5, from 1.5 to 2, and from 1.5 to 2.8, respectively. I12 had a significantly higher hydrophobicity value compared with that of the wild type. In contrast, M10 and A12 had slightly lower hydrophobicity values. We introduced these constructs into protoplasts and examined their localization. As shown in Figure 8C, AtOEP7 [M10]:GFP (g and h) and AtOEP7[F12]:GFP (i and j) were not targeted to the chloroplasts but present as aggregates in the protoplasts, whereas AtOEP7[I12]:GFP (e and f) was efficiently targeted to chloroplasts, as was AtOEP7[A12]:GFP (c and d), indicating that targeting to the chloroplast was not dependent on hydrophobicity. Interestingly, however, AtOEP7 (1-28)[F12]:GFP (k and l) that was identical to the AtOEP7 [F12]:GFP mutant except the deletion of the C-terminal region of the TMD was targeted to the plasma membrane, suggesting that the TMD with 12 phenylalanine residues can function as a signal/anchor sequence for cotranslational translocation into the ER. This result suggests that the TMD has a preference for amino acid residues with smaller side chains.
Figure 8.
TMD Mutants with Hydrophobic Amino Acids Having Large Side Chains Inhibit Targeting.
(A) Scheme of TMD mutants.
(B) Hydropathy analysis of the TMD mutants. Hydrophobicity of the wild-type and mutant AtOEP7s was analyzed by the method of Kyte and Doolittle (1982) with a window size of seven amino acid residues.
(C) In vivo targeting of TMD mutants. Protoplasts were transformed with AtOEP7:GFP (a and b), AtOEP7[A12]:GFP (c and d), AtOEP7[I12]:GFP (e and f), AtOEP7[M10]:GFP (g and h), AtOEP7[F12]:GFP (i and j), or AtOEP7(1-28)[F12]:GFP (k and l). Images were observed 12 to 36 hr after transformation. Green and red signals indicate GFP and chlorophyll (CH), respectively. Bars = 20 μm.
α-Helix of the TMD Is Important for Targeting to the Chloroplast Outer Envelope Membrane
It has been shown that the membrane-spanning region often has an α-helical structure. In fact, the secondary structure of the TMD region of AtOEP7 also has the α-helix (data not shown). Thus, we wanted to examine the importance of the α-helical structure of TMD for targeting. To disrupt the secondary structure, we introduced a proline residue at position 15, 19, or 23 from the N terminus, which corresponds to position 6, 10, or 14 from the N terminus of the TMD (Figure 9A). These mutants then were introduced into protoplasts as GFP fusion constructs. As shown in Figure 9B, AtOEP7[A15P]:GFP (a and b) and AtOEP7[A23P]:GFP (g and h) were targeted to the chloroplasts. At the same time, the green fluorescent signals also were present in the cytosol, indicating that these mutants were targeted to the chloroplasts with reduced efficiency. Interestingly, AtOEP7[L19P]:GFP (d and e), which has the proline residues in the middle of the TMD, was not targeted to the chloroplasts at all; instead, they were present as aggregates in the protoplast. Thus, these results strongly suggest that the α-helical structure of the TMD is important for targeting and that a proline residue in the middle of the TMD is most detrimental for targeting to the chloroplast envelope membrane.
Figure 9.
α-Helical Structure of the TMD Is Important for Targeting.
(A) Scheme of TMD mutants with proline residues.
(B) In vivo targeting of the TMD mutants. Protoplasts were transformed with AtOEP7[A15P]:GFP (a to c), AtOEP7[L19P]:GFP (d to f), and AtOEP7[A23P]:GFP (g to i). Green and red signals indicate GFP and chlorophyll (CH), respectively. Bars = 20 μm.
Targeting Signal of AtOEP7 Is Dominant over a Nuclear Localization Signal
During transport to the chloroplast, the precursors of nucleus-encoded chloroplast proteins that have a transit peptide are found to be associated with 14-3-3 and chaperone proteins (Waegemann et al., 1990; May and Soll, 2000). Therefore, in the cytosol we investigated whether AtOEP7 also might be associated with other proteins in the cytosol. We addressed this question using a fusion protein, AtOEP7: NLS:GFP, that contained the AtOEP7 TMD required for chloroplast targeting and the simian virus 40 (SV40) large T antigen nuclear localization signal (NLS) (Figure 10A) (Dingwall and Laskey, 1991). We reasoned that if AtOEP7 reaches the chloroplast as a free molecule by simple diffusion, then free AtOEP7:NLS:GFP might be transported preferentially to the nucleus despite the presence of the AtOEP7 TMD, because the number of nuclear import receptors in the cytosol is likely greater than the number of chloroplasts. We generated another fusion protein, AtOEP7(20-64):NLS, which had an identical amino acid sequence near the NLS but could not be targeted to the chloroplast. This protein was fused subsequently to GFP to yield AtOEP7(20-64):NLS:GFP (Figure 10A). Finally, the SV40 NLS was fused to the RFP and was cotransformed with each of the AtOEP7 fusion constructs described above (Pih et al., 2000). AtOEP7:NLS:GFP and NLS:RFP constructs were cotransformed into protoplasts, and the cellular localizations of these two reporter proteins were examined at various times after transformation by fluorescence microscopy. Interestingly, as shown in Figure 10B, AtOEP7:NLS:GFP was targeted efficiently to the chloroplast but was not detectable in the nucleus using fluorescence microscopy (e to h). In contrast, the control proteins AtOEP7 (20-64):NLS:GFP and NLS:RFP (a to d) were targeted efficiently to the nucleus, as observed with NLS: RFP (b and d). Together, these experiments suggest that targeting of AtOEP7 to chloroplast outer envelope membranes is an active process that can overcome nuclear targeting specified by the SV40 NLS. Because both AtOEP7: NLS:GFP and AtOEP7(20-64):NLS:GFP have a similar organization, it is likely that the nuclear targeting signal is functional in both fusion proteins. Thus, it is possible that the NLS in AtOEP7:NLS:GFP failed to deliver the protein to the nucleus because it was not accessible to the nuclear import machinery, probably because it was inhibited by a cytosolic factor(s) involved in the transport of AtOEP7:NLS:GFP to the chloroplast.
Figure 10.
In Vivo Targeting of the Fusion Protein with the NLS.
(A) Scheme of the fusion constructs.
(B) In vivo targeting of the fusion proteins. Protoplasts were transformed with AtOEP7(20-64):NLS:GFP and NLS:RFP (a to d) or AtOEP7:NLS:GFP and NLS:RFP (e to h), and the green and red fluorescent signals were examined 24 to 30 hr after transformation. The data are representative of transformed protoplasts. At least three independent transformation experiments were performed with each construct. Note that the autofluorescent signal of chlorophyll (CH) is colored blue. Bars = 20 μm.
DISCUSSION
In Vivo Targeting of AtOEP7:GFP
In this study, we investigated the targeting of an outer envelope membrane protein, AtOEP7, to the chloroplast using an in vivo system. Previous studies relied on methods using in vitro translated protein, purified chloroplasts, and protein fractionation (Salomon et al., 1990; Wu and Ko, 1993; Li and Chen, 1996). In this study, the targeting of AtOEP7 was assayed using protoplasts. The proteins under study were tagged with GFP (Davis and Vierstra, 1998), which allowed us to monitor their targeting easily within protoplasts. These data also were verified using traditional cell extract fractionation methods. First, localization of the fusion protein was established in protoplasts and transgenic plants using GFP and fluorescence microscopy. We observed that AtOEP7: GFP was targeted to the envelope of chloroplasts. This was confirmed by protein gel blot analysis, which showed that AtOEP7:GFP was present in the membrane fraction but not in the soluble protein fraction. It was further shown that AtOEP7:GFP was inserted into the membrane of transgenic plants and was sensitive to thermolysin digestion, indicating that the AtOEP7:GFP fusion protein is localized at the outer envelope membrane, with the C-terminal GFP portion exposed to the cytosol. Previous studies showed that the C-terminal portion of pea OEP14 also was exposed to the cytosol (Li et al., 1991). In addition, the in vivo targeting experiments using various deletion mutants revealed that the first 35–amino acid region was necessary and sufficient for protein targeting to the chloroplast outer envelope membrane; this is consistent with the results obtained from an in vitro import assay (Li et al., 1991).
Role of the Seven–Amino Acid Region Located at the C Terminus of the TMD
We further addressed the nature of targeting information encoded in the signaling sequence. The most important question concerning the targeting of AtOEP7 is how specificity is determined among the endomembranes. This question cannot be addressed adequately using the in vitro import assay. Using an in vivo approach, we found that the seven–amino acid region located at the C terminus of the TMD is critical. The amino acid sequence of the seven–amino acid region was especially important. Deletion mutations, substitution of positively or negatively charged amino acids with glycine, or scrambling the amino acid sequence of this region resulted in mistargeting to the plasma membrane. This result suggests that, in the absence of information encoded in this region of the TMD, the AtOEP7 protein may follow the secretory pathway (Walter and Johnson, 1994) and then be transported to the plasma membrane. In fact, the TMD of AtOEP7 is very similar to a signal/anchor sequence for cotranslational translocation into the ER. Therefore, one possible mechanism for targeting proteins to the chloroplast is for them to evade the secretory pathway during translation. This is quite similar to the targeting of a small mitochondrial outer envelope membrane protein, Tom20 (Kanaji et al., 2000). In this case, the removal of positive charges within five amino acid residues from the C terminus of the TMD caused the mutant proteins to enter the secretory pathway. As in the case of Tom20, SRP may bind to the TMD region of AtOEP7 at an early stage of translation and be released from the TMD when it encounters the seven–amino acid region.
Role of the TMD in Protein Targeting to the Chloroplasts
Next, we investigated whether the TMD plays any role in protein targeting. To address this question, we analyzed four different aspects of the TMD: the importance of specific amino acid residues, hydrophobicity, the bulkiness of the side chain of the hydrophobic amino acid residues, and the α-helical structure. From these studies, it is clear that amino acid residues within the TMD can be substituted with alanine without any detrimental effect on targeting. Even the TMD with 12 alanine residues in a row can be targeted successfully to the chloroplasts. Also, differences in the hydrophobicity of the TMD did not affect targeting significantly. This was very clear from the fact that proteins containing I12 (hydrophobicity values, 2.0 to 4.5) but not F12 (hydrophobicity values, 2.0 to 2.8) were targeted efficiently to the chloroplasts. The hydrophobicity value of I12 was significantly higher than that of the wild-type protein (which ranges from 1.5 to 3.0), whereas F12 had a similar hydrophobic range. In addition, although A12 and M12 had similar hydrophobicity values, which were slightly lower than those of the wild type, only A12 was targeted to the chloroplasts. These results suggest that hydrophobicity plays a minimal role in targeting. In the case of Tom20, the TMD with a higher hydrophobicity value was directed to the secretory pathway (Kanaji et al., 2000). In contrast, our results suggest that the size of the side chain of the hydrophobic amino acids within the TMD may be more important to targeting. The TMD for chloroplast proteins may prefer hydrophobic amino acid residues with smaller side chains.
Although an F12 domain cannot be used as the TMD for AtOEP7, it can be used to direct a protein to the plasma membrane, presumably through the secretory pathway, indicating that there is a clear difference in the preference for the amino acid residues for the TMD. In addition, the α-helical structure of the TMD appears to be important for targeting. Introduction of a proline residue into the TMD either inhibits or greatly reduces targeting efficiency, depending on the position of the proline within the TMD. Together, these results suggest that the TMD plays an important role in targeting. For the TMD of Tom20, moderate hydrophobicity was necessary for targeting. In addition, hydrophobicity is dominant over the presence of positive charges (Kanaji et al., 2000). However, for AtOEP7, it appears that the seven–amino acid region is dominant over the hydrophobicity of the TMD for chloroplast targeting. These results indicate that there are important differences between the TMD of proteins targeted to the chloroplast of plant cells and the TMD of proteins targeted to the mitochondria of animal cells. However, it is not clear whether this is attributable to differences between animal and plant cells or between mitochondria and chloroplasts.
Although we have shown that the TMD also plays an important role during targeting, we cannot answer clearly in this study why some of the mutants were not targeted to the chloroplasts, presenting instead as aggregates in the protoplasts. The lack of targeting to the chloroplast can be explained by at least two different mechanisms. One is that the TMD with 12 phenylalanines or 10 methionines cannot be inserted into the chloroplast outer envelope membrane because of either a higher energy barrier caused by the bulky side chains or an inability to interact with protein factors located at the envelope membrane (Tu and Li, 2000). Another possibility is that the TMD with 12 phenylalanines or 10 methionines may not interact with a cytosolic factor in the cytosol; thus, the mutant proteins may not be brought to the chloroplast at all. In the cytosol, the hydrophobic TMD may need to be associated with a particular cytosolic factor. Otherwise, it may have a strong tendency to be inserted into other endomembranes or to form aggregates, as observed with these M10 and F12 mutants.
Interestingly, when we introduced an NLS at the C terminus of AtOEP7, AtOEP7:NLS:GFP was targeted efficiently to the chloroplast envelope membrane despite the presence of NLS. It has been shown that the NLS of a nuclear protein is recognized by a protein import complex present in the cytosol (Mattaj and Englmeier, 1998). This result further supports the notion that a cytosolic factor has to interact with this fusion protein before it can reach the chloroplast, as has been proposed for the targeting of transit peptide–containing proteins (Waegemann et al., 1990; May and Soll, 2000). Such a protein(s) may help AtOEP7 to identify the chloroplast outer envelope membrane from a variety of different endomembranes in the cell or to guide it to interact with the proteinaceous component at the chloroplast envelope membrane (Tu and Li, 2000). In the targeting of a protein to the yeast mitochondria, it has been proposed that the nascent polypeptide-associated complex that is associated with ribosomes during translation may help guide the targeting of proteins to the mitochondria (Wiedmann et al., 1994; George et al., 1998). Thus, although the presence of a nascent polypeptide-associated complex in plant cells has not been shown, it is possible that a similar complex may operate for targeting to the chloroplasts in plant cells. In fact, we identified an ankyrin repeat protein that interacts specifically with the TMD of AtOEP7 (Y.J. Lee and I. Hwang, unpublished results), thus supporting the second possibility. Further studies are necessary to determine which of these two hypotheses is correct.
METHODS
Growth of Plants
Arabidopsis thaliana (ecotype Columbia) was grown on Murashige and Skoog (1962) (MS) plates at 20°C in a culture room with a 16/8-hr light/dark cycle. Arabidopsis seedlings were grown in MS liquid medium with constant shaking (160 rpm) at 20°C under a 16/8-hr light/dark cycle and then used to prepare protoplasts.
RNA Gel Blot Analysis
For RNA gel blot analysis, total RNA was isolated from various tissues from Arabidopsis grown on MS plates and soil. Fifteen micrograms of total RNA was used for RNA gel blot analysis using AtOEP7 as a hybridization probe (Ausubel et al., 1989).
Transformation of Arabidopsis
To generate transgenic Arabidopsis, we used the in planta transformation method (Clough and Bent, 1998). A transformation vector was constructed by introducing the gene encoding the various fusion proteins downstream of the 35S cauliflower mosaic virus promoter of pBI121 in place of the β-glucuronidase coding region. The transformants were selected on an MS agar plate supplemented with 50 μg/mL kanamycin.
Construction of Fusion Proteins
To construct the AtOEP7:GFP gene, the AtOEP7 coding region was amplified by polymerase chain reaction (PCR) using primers OEP7-F (5′-GACGACGACGCAGCGATG-3′) and OEP7-R (5′-GGATCCCCA-AACCCTCTTTGGATGT-3′) that removed the natural termination codon. This was ligated in frame to the 5′ end of GFP. To construct GFP: AtOEP7, the GFP coding region was amplified by PCR using primers GFP-F (5′-CCCGGGCGATGGGAAAAACTTCGG-3′) and GFP-R (5′-TTACAAACCATCTTTGGA-3′) that removed the natural termination codon and ligated in frame to the 5′ end of the AtOEP7 gene. Various deletion mutations and gene fusion constructs were generated by PCR amplification. The primers were as follows: (1) 5′-ATGGCGACTGTGGTGGTCGCA-3′ and OEP7-R for AtOEP7(10-64); (2) 5′-ATGGGATGGTTAGCCATAGAG-3′ and OEP7-R for AtOEP7 (20-64); (3) OEP7-F and 5′-GGATCCGGAATTTATCGAGGAAAG-3′ for AtOEP7(1-35); (4) OEP7-F and 5′-GGATCCGGTCTTTGG-TTGGGTCAG-3′ for AtOEP7(1-47); (5) OEP7F and 5′-GGATCC-TGAAAGCGATCTCTATGGC-3′ for AtOEP7(1-28); (6) OEP7F and 5′-GGATCCGGAAACCATCGAGGAAAGGACCGAAAGCGATCTCTT-GG-3′ for AtOEP7 (1-35)K29,34G; (7) OEP7F and 5′-GGATCCGGA-ATTTACCGAGGAAAGGCTTGAAAG-3′ for AtOEP7(1-35)D33G; (8) OEP7F and 5′-GGATCCGGAAATCGAACTTTTTGAGAGGGAAAG-CGATCTCTATGG-3′ for AtOEP7(1-35)S29-35; (9) OEP7F, OEP7R and 5′-GGAAAAACTTCGGGAGCGGCGGCGGCGACTGTGGTGGTC-GCA-3′ for AtOEP7 (A2-1); (10) OEP7F, OEP7R, 5′-GCGAAACAGGCGGCGGCGGTGGTCGCAGCGATGGCG-3′, and 5′-CGCCGCCGCCTGTTTCGCTCCCGA-3′ for AtOEP7(A2-2); (11) OEP7F, OEP7R, 5′-CAGGCGACTGTGGCGGCGGCAGCGATGGCGTTAGGA-3′, and 5′-CGCCGCCACAGTCGCCTGTTTCGC-3′ for AtOEP7(A2-3); (12) OEP7F, OEP7R, 5′-GTGGTCGCAGCGGCGGCGTTAGGATGGTTA-GCCATA-3′, and 5′-CGCCGCCGCTGCGACCACCACAGT-3′ for AtOEP7(A2-4); (13) OEP7F, OEP7R, 5′-GCGGCGTGGTTAGCCATAGAGATC-3′, and 5′-TATGGCTAACCACGCCGCCGCCATCGCTGCGACCAC-3′ for AtOEP7 (A2-5); (14) OEP7F, OEP7R, 5′-GCG-GCGGCCATAGAGATCGCTTTC-3′, and 5′-GATCTCTATGGCCGCCGCTCCTAACGCCATCGCTGC-3′ for AtOEP7(A2-6); (15) OEP7F, OEP7R, 5′-GCGGCGATCGCTTTCAAGCCTTTC-3′, and 5′-CTTGAA-AGCGATCGCCGCGGCTAACCATCCTAACGC-3′ for AtOEP7(A2-7); (16) OEP7F, OEP7R, 5′-GCGGCTTTCAAGCCTTTCCTC-3′, and 5′-GAAAGGCTTGAAAGCCGCCTCTATGGCTAACCATCC-3′ for AtOEP7 (A2-8); (17) OEP7F, OEP7R, 5′-GCGGCGGCAGCGATGGCGTTA-GGA-3′, and 5′-CGCCATCGCTGCCGCCGCCGCCGCCGCCTG-TTTCGC-3′ for AtOEP7(NA4); (18) OEP7F, OEP7R, 5′-GCGGCG-TTAGGATGGTTAGCCATA-3′, and 5′-TAACCATCCTAACGCCGCCGC-CGCCGCCGCCGCCGC-3′ for AtOEP7(NA8); (19) OEP7F, OEP7R, 5′-GCAGCGGCGGCGGCTGCATGGTTAGCCATAGAGATC-3′, and 5′-TGCAGCCGCCGCCGCTGCCGCCGCCGCCGCCGC-3′ for AtOEP7 (NA10); (20) OEP7F, OEP7R, 5′-GCGGCGGCTGCAGCTGCAGCC-ATAGAGATCGCTTTC-3′, and 5′-TGCAGCTGCAGCCGCCGCCGC-TGC-3′ for AtOEP7(A12); (21) OEP7F, OEP7R, 5′-GCGGCGGCGGCTTTCAAGCCTTTC-3′, and 5′-CTTGAAAGCCGCCGCCGCGGC-TAACCATCCTAACGC-3′ for AtOEP7(CA4); (22) OEP7F, OEP7R, 5′-GCGGCGGCCGCGGCGGCGGCTTTCAAG-3′, and 5′-CGCCGCCGCGGCCGCCGCTCCTAACGCCATCGCTGC-3′ for AtOEP7(CA7); (23) OEP7F, OEP7R, 5′-CCCGCGATGGCGTTAGGATGG-3′ and 5′-TCCTAACGCCATCGCGGGGACCACCACAGTCGC-3′ for AtOEP7 (A15P), (24) OEP7F, OEP7R, 5′-CCCGGATGGTTAGCCATAGAG-3′ and 5′-TATGGCTAACCATCCGGGCGCCATCGCTGCGAC-3′ for AtOEP7 (L19P); (25) OEP7F, OEP7R, 5′-CCCATAGAGATCGCT-TTCAAG-3′ and 5′-GAAAGCGATCTCTATGGGTAACCATCCTAA-CGC-3′ for AtOEP7(A23P); (26) OEP7F, OEP7R, 5′-TTTTTCTTT-TTCTTTTTCTTAGCCATAGAGATCGCT-3′, 5′-AAAGAAGAAAAAAAA-GAACTGTTTCGCTCCCGAAGT-3′ and 5′-GAAAAAGAAAAAGAA-AAAAAAGAAGAAAAAAAAGAA-3′ for AtOEP7(F12); (27) OEP7F, OEP7R, 5′-ATGATGGCAATGATGATGTTAGCCATAGAGATCGCT-3′, 5′-CATCATCATCATTGCCATCTGTTTCGCTCCCGAAGT-3′ and 5′-CATCATCATTGCCATCATCATCATCATCATTGCCAT-3′ for AtOEP7 (M10); and (28) OEP7F, OEP7R, 5′-ATAATCATTATAATTATCTTAGCC-ATAGAGATCGCT-3′, 5′-TATAATGATAATTATGATCTGTTTCGC-TCCCGAAGT-3′ and 5′-GCTAAGATAATTATAATGATTATTATAATGATAATTATGATCTG-3′ for AtOEP7(I12). The PCR-amplified DNA products were confirmed by nucleotide sequencing and subsequently introduced into an expression vector with the 35S cauliflower mosaic virus promoter and nos terminator. AtOEP7:NLS:GFP and AtOEP7(20-64):NLS:GFP were constructed using BamHI sites of AtOEP7 and NLS:GFP.
Polyethylene Glycol–Mediated Protoplast Transformation
The plasmid DNAs were purified using a Qiagen column (Qiagen, Valencia, CA) according to the manufacturer's instructions. The fusion constructs were introduced into Arabidopsis protoplasts prepared from whole seedlings by the polyethylene glycol–mediated transformation procedure (Jin et al., 2001). Expression of the fusion constructs was monitored at various times after transformation by fluorescence microscopy using a Zeiss (Jena, Germany) Axioplan fluorescence microscope, and the images were captured with a cooled charge-coupled device camera. The filter sets used were XF116 (exciter, 474AF20; dichroic, 500DRLP; emitter, 510AF23), XF33/E (exciter, 535DF35; dichroic, 570DRLP; emitter, 605DF50), and XF137 (exciter, 540AF30; dichroic, 570DRLP; emitter, 585ALP) (Omega, Inc., Brattleboro, VT) for green fluorescent protein, red fluorescent protein, and autofluorescence of chlorophyll, respectively. The data were processed using Adobe (Mountain View, CA) Photoshop software and presented in pseudocolor format.
Protein Fractionation and Protein Gel Blot Analysis
To prepare cell extracts from protoplasts, transformed protoplasts were subjected to repeated freeze and thaw cycles and then centrifuged at 9800g at 4°C for 5 min in a microfuge to remove cell debris. The cell extracts then were fractionated into soluble and membrane fractions by ultracentrifugation at 100,000g for 30 min. Each fraction was assayed for the presence of the AtOEP7:GFP fusion protein by protein gel blot analysis using a monoclonal anti-GFP antibody (Clontech, Palo Alto, CA). The protein gel blot was developed with an enhanced chemiluminescence kit (Amersham).
Homogenates of leaf tissues of transgenic plants were prepared and separated into top, broken chloroplast, and intact chloroplast fractions on a Percoll step gradient (Cline et al., 1985). The intact chloroplasts were lysed to prepare chloroplast extracts. Chloroplast proteins were centrifuged by low-speed centrifugation at 1000g for 10 min, and the supernatant of the low-speed centrifugation was fractionated into membrane and soluble fractions by ultracentrifugation at 100,000g for 30 min (Park et al., 1998). The membrane fraction was treated with 0.1 M Na2CO3, pH 11.5, or 1% Triton X-100 in a buffer (25 mM Hepes, pH 7.5, 1 mM DTT, 1 mM MgCl2, 250 mM sucrose, 1 μg/mL aprotinin, 1 μg/mL leupeptin, and 100 μg/mL phenylmethylsulfonyl fluoride) and then pelleted by ultracentrifugation at 100,000g for 30 min. The membrane and soluble fractions were assayed by protein gel blot analysis for the presence of AtOEP7:GFP. To examine thermolysin sensitivity, the intact chloroplasts were treated with thermolysin as described previously (Li et al., 1991). Total chloroplast proteins were prepared from thermolysin-treated chloroplasts and probed with anti-GFP antibody by protein gel blot analysis.
GenBank Accession Number
The GenBank accession number for the AtOEP7 DNA sequence is CAB43440.
Acknowledgments
The polyclonal anti-Tic110 and anti-RbcS complex antibodies were provided by F. Kessler (Swiss Federal Institute of Technology, Zurich, Switzerland) and J.H. Kim (Kyeonghee University, Seoul, Korea), respectively. This work was supported by a grant from the National Creative Research Initiatives of the Ministry of Science and Technology (Korea).
References
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1989). Current Protocols in Molecular Biology. (New York: Greene Publishing Associates/Wiley-Interscience).
- Bauer, J., Chen, K., Hiltbunner, A., Wehrli, E., Eugster, M., Schnell, D., and Kessler, F. (2000). The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403, 203–207. [DOI] [PubMed] [Google Scholar]
- Chen, D., and Schnell, D.J. (1997). Insertion of the 34-kDa chloroplast protein import component, IAP34, into the chloroplast outer membrane is dependent on its intrinsic GTP-binding capacity. J. Biol. Chem. 272, 6614–6620. [DOI] [PubMed] [Google Scholar]
- Cline, K., Werner-Washburne, M., Lubben, T.H., and Keegstra, K. (1985). Precursors to two nuclear-encoded chloroplast proteins bind to the outer envelope membrane before being imported into chloroplasts. J. Biol. Chem. 260, 3691–3696. [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, 521–528. [DOI] [PubMed] [Google Scholar]
- Dingwall, C., and Laskey, R.A. (1991). Nuclear targeting sequences: A consensus? Trends Biochem. Sci. 16, 478–481. [DOI] [PubMed] [Google Scholar]
- George, R., Beddoe, T., Landl, K., and Lithgow, T. (1998). The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria. Proc. Natl. Acad. Sci. USA 95, 2296–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J.B., Kim, Y.A., Kim, S.J., Lee, S.H., Kim, D.H., Cheong, G.-W., and Hwang, I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13, 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaji, S., Iwahashi, J., Kida, Y., Sakaguchi, M., and Mihara, K. (2000). Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151, 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra, K., and Froehlich, J.E. (1999). Protein import into chloroplasts. Curr. Opin. Plant Biol. 2, 471–476. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Eu, Y.-J., Yoo, C.M., Kim, Y.W., Pih, K.T., Jin, J.B., Kim, S.J., Stenmark, H., and Hwang, I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov, A., Wang, H., and Schnell, D.J. (1999). Tic22 is targeted to the intermembrane space of chloroplasts by a novel pathway. J. Biol. Chem. 274, 25181–25186. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Li, H.M., and Chen, L.J. (1996). Protein targeting and integration signal for the chloroplastic outer envelope membrane. Plant Cell 8, 2117–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.M., Moore, T., and Keegstra, K. (1991). Targeting of proteins to the outer envelope membrane uses a different pathway than transport into chloroplasts. Plant Cell 3, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj, I.W., and Englmeier, L. (1998). Nucleocytoplasmic transport: The soluble phase. Annu. Rev. Biochem. 67, 265–306. [DOI] [PubMed] [Google Scholar]
- May, T., and Soll, J. (2000). 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12, 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckel, E., and Soll, J. (1996). A protein import receptor of chloroplast is inserted into the outer envelope membrane by a novel pathway. J. Biol. Chem. 271, 23846–23852. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Park, J.M., Cho, J.H., Kang, S.G., Jang, H.J., Pih, K.T., Piao, H.L., Cho, M.J., and Hwang, I. (1998). A dynamin-like protein in Arabidopsis thaliana is involved in biogenesis of thylakoid membranes. EMBO J. 17, 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini, E., Villa, A., and Borgese, N. (1996). A mutant cytochrome b5 with a lengthened membrane anchor escapes from the endoplasmic reticulum and reaches the plasma membrane. Proc. Natl. Acad. Sci. USA 93, 4207–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, S.E., and Keegstra, K. (1994). Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell 6, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pih, K.T., Yi, M.J., Liang, Y.S., Shin, B.J., Cho, M.J., Hwang, I., and Son, D. (2000). Molecular cloning and targeting of a fibrillarin homolog from Arabidopsis. Plant Physiol. 123, 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim, M.L., van Wijk, K.J., Parry, D.H., Sy, D.A., and Hoffman, N.E. (1998). Expression of a dominant negative form of cpSRP54 inhibits chloroplast biogenesis in Arabidopsis. Plant J. 13, 177–186. [DOI] [PubMed] [Google Scholar]
- Salomon, M., Fischer, K., Flugge, U.I., and Soll, J. (1990). Sequence analysis and protein import studies of an outer chloroplast envelope polypeptide. Proc. Natl. Acad. Sci. USA 87, 5778–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrt, K., and Soll, J. (2000). Toc64, a new component of the protein translocon of chloroplasts. J. Cell Biol. 148, 1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel, P.J., Froehlich, J., Goyal, A., and Keegstra, K. (1995). A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 14, 2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, S.-L., and Li, H. (2000). Insertion of OEP14 into the outer envelope membrane is mediated by proteinaceous components of chloroplasts. Plant Cell 12, 1951–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann, K., Paulsen, H., and Soll, J. (1990). Translocation of proteins into chloroplasts requires cytosolic factors to obtain import competence. FEBS Lett. 261, 89–92. [Google Scholar]
- Walter, P., and Johnson, A.E. (1994). Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA 77, 7112–7116. [DOI] [PubMed] [Google Scholar]
- Wiedmann, B., Sakai, H., Davis, T.A., and Wiedmann, M. (1994). A protein complex required for signal sequence–specific sorting and translocation. Nature 370, 434–440. [DOI] [PubMed] [Google Scholar]
- Wu, C., and Ko, K. (1993). Identification of an uncleavable targeting signal in the 70-kilodalton spinach chloroplast outer envelope membrane protein. J. Biol. Chem. 268, 19384–19391. [PubMed] [Google Scholar]