Abstract
The Arabidopsis mutant defective in anther dehiscence1 (dad1) shows defects in anther dehiscence, pollen maturation, and flower opening. The defects were rescued by the exogenous application of jasmonic acid (JA) or linolenic acid, which is consistent with the reduced accumulation of JA in the dad1 flower buds. We identified the DAD1 gene by T-DNA tagging, which is characteristic to a putative N-terminal transit peptide and a conserved motif found in lipase active sites. DAD1 protein expressed in Escherichia coli hydrolyzed phospholipids in an sn-1–specific manner, and DAD1–green fluorescent protein fusion protein expressed in leaf epidermal cells localized predominantly in chloroplasts. These results indicate that the DAD1 protein is a chloroplastic phospholipase A1 that catalyzes the initial step of JA biosynthesis. DAD1 promoter::β-glucuronidase analysis revealed that the expression of DAD1 is restricted in the stamen filaments. A model is presented in which JA synthesized in the filaments regulates the water transport in stamens and petals.
INTRODUCTION
Jasmonic acid (JA) is a multifunctional growth regulator widely distributed in the plant kingdom that modulates anther dehiscence, fruit ripening, root growth, tendril coiling, and plant resistance to insects and pathogens (Creelman and Mullet, 1997). JA is derived from linolenic acid (LA), and its biosynthesis is catalyzed by several enzymes, namely, lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC), and 12-oxo-phytodienoic acid reductase (OPR), whose action is followed by three cycles of β-oxidation (Creelman and Mullet, 1997; Mueller, 1997; Schaller, 2001) (Figure 1). Some parts of this pathway are shared for the biosynthesis of oxylipin compounds such as volatile C-6 aldehydes, traumatin, cutin monomers, and α- and γ-ketols (Creelman and Mullet, 1997; Schaller, 2001) (Figure 1). All genes encoding enzymes in the JA biosynthetic pathway downstream of LA have been cloned from Arabidopsis and some other plants (Song et al., 1993; Bell et al., 1995; Laudert et al., 1996; Maucher et al., 2000; Sanders et al., 2000; Stintzi and Browse, 2000; Ziegler et al., 2000). Because LOX, AOS, and AOC were shown to be localized in chloroplasts (Bell et al., 1995; Maucher et al., 2000; Ziegler et al., 2000; Froehlich et al., 2001), it is believed that the initial steps of biosynthesis catalyzed by these enzymes are performed in chloroplasts.
Figure 1.
Biosynthetic Pathway of JA.
DAD1, the lipolytic enzyme, had long been sought and was identified in this study. The downstream steps from LA had been established (Creelman and Mullet, 1997; Mueller, 1997; Schaller, 2001). Some compounds sharing the pathway also are indicated.
In contrast to the well-investigated reactions downstream of the LA, it was not known how LA is supplied for the synthesis of JA as the need arises. Because the conversion of linoleic acid to LA is catalyzed by fatty acid desaturase only as the lipid-bound form (Browse and Somerville, 1991), the step to release the LA from cellular lipids should be essential for the biosynthesis of JA. However, the lipolytic enzyme catalyzing this reaction has not been identified.
It is thought that this lipolytic enzyme plays a critical role in plant responses to pathogen and insect attack. After treatment with elicitors or wounding of plant tissues, the amount of JA is increased drastically within 1 hr (Mueller et al., 1993; McConn et al., 1997), along with the accumulation of many free fatty acids, including LA (Mueller et al., 1993; Conconi et al., 1996). This observation strongly indicates that the lipolytic reaction is one of the major regulatory steps in the biosynthesis of JA.
The structure of JA is similar to that of animal eicosanoids, which are made from arachidonic acid, a polyunsaturated fatty acid, released from the plasma membrane phospholipids by phospholipase A2 (PLA2). Thus, the involvement of membrane phospholipids and PLA2 in JA biosynthesis has long been predicted (Mueller, 1997). Indeed, rapid induction of PLA2 activity was detected in response to wounding, systemin, and elicitors in tomato leaves (Nárvaez- Vásquez et al., 1999). However, inhibitors of animal PLA2 did not inhibit the accumulation of JA upon wounding of potato tuber tissue (Koda and Kikuta, 1992). To date, there is no direct evidence for the involvement of PLA2 in JA biosynthesis. In addition, the involvement of phospholipase D (PLD) in the wound-induced accumulation of JA was revealed in Arabidopsis (Wang et al., 2000), but this enzyme requires another lipolytic enzyme(s) to release free fatty acid. No convincing candidate for the lipolytic enzyme involved in JA biosynthesis has been identified.
The involvement of JA in anther dehiscence was revealed in genetic analyses of Arabidopsis mutants. A JA-insensitive mutant, coronatine insensitive1 (coi1), shows a male-sterile phenotype in which the anther cannot dehisce even after the flowers have opened (Feys et al., 1994; Xie et al., 1998). The triple mutant (fad3 fad7 fad8) of genes encoding fatty acid desaturases, which catalyze the desaturation of linoleic acid to make LA, also shows a dehiscence-defective phenotype (McConn and Browse, 1996). Most recently, a similar phenotype was reported in the mutant of the DELAYED DEHISCENCE 1 (DDE1)/OPR3 gene, which encodes an OPR protein (Sanders et al., 1999, 2000; Stintzi and Browse, 2000). In all of these mutants, cell organization and differentiation of anther tissues appear normal, but dehiscence does not occur at flower opening and pollen grains are inviable.
From anatomical analysis, it is known that three tissues in anthers, the endothecium, the connective, and the stomium, have important roles in anther dehiscence. The endothecium is an anther wall tissue lying between the epidermis and the tapetum, and the connective is a tissue filling the space between the vascular bundles and the locules. In Arabidopsis, after the development of the trinucleate pollen grains, a secondary thickening of cell walls or formation of “fibrous bands” occurs in the endothecium and connective cells. When the flowers begin to open, the endothecium and connective lose most of their water and shrink to cause the outward bending of the anther walls and the dehiscence of the anthers at the stomium (Keijzer, 1987; Goldberg et al., 1993; Sanders et al., 1999). The stomium is a collection of specialized epidermal cells that join the anther walls; targeted ablation of the stomium resulted in anthers that failed to dehisce, indicating that anther dehiscence requires functional stomium cells (Beals and Goldberg, 1997). However, little is known about the molecular mechanisms controlling anther dehiscence, especially the synchronous regulation of anther dehiscence, pollen maturation, and flower opening.
Here, we report the isolation and characterization of an Arabidopsis mutant, defective in anther dehiscence1 (dad1), that has defects in anther dehiscence, pollen maturation, and flower bud development. We show that the DAD1 gene encodes a particular phospholipase A1 (PLA1) lipolytic enzyme that catalyzes the initial step of JA biosynthesis. In addition, we propose a model of JA function that synchronizes pollen maturation, anther dehiscence, and flower opening.
RESULTS
dad1 Mutant Is Defective in Anther Dehiscence and Pollen Maturation
By screening 600 T-DNA mutagenized Arabidopsis lines, we identified a male-sterile mutant whose anthers could not dehisce and could not release their pollen grains at the time of flower opening, whereas the other floral organs appeared to be normal (Figures 2A to 2D). This mutant is female fertile and segregated 3:1 in the F2 generation of a backcross with wild-type plants (data not shown), indicating that it carries a recessive sporophytic mutation. We designated this mutation defective in anther dehiscence1 (dad1). Anther dehiscence could be observed on the day after flower opening, although the pollen grains were not viable (see below).
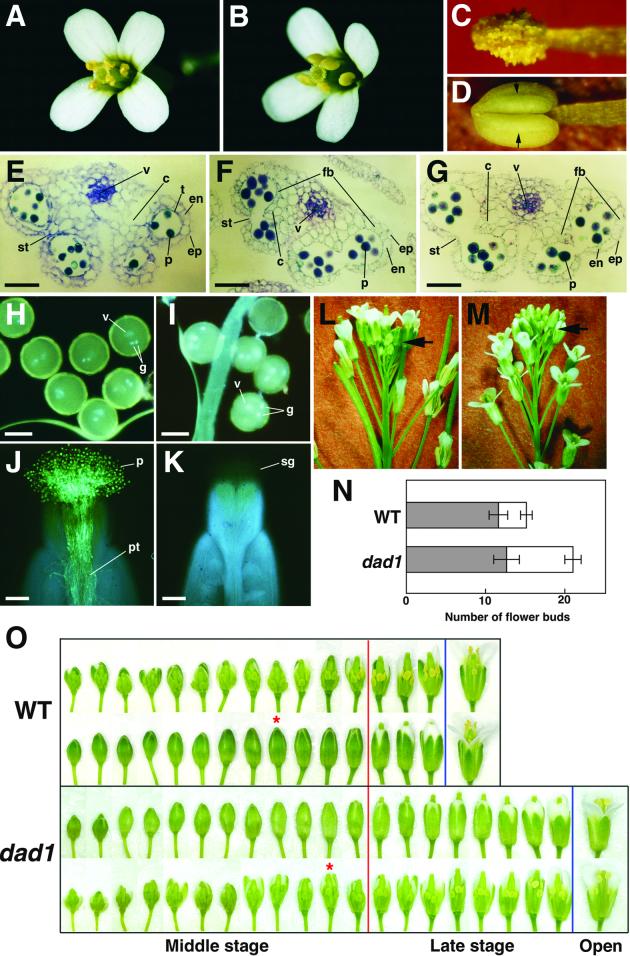
Figure 2.
Phenotypes of the dad1 Mutant.
(A) and (B) Flowers immediately after opening. (A) Wild type. (B) dad1.
(C) and (D) Anthers removed from open flowers. (C) Dehisced wild-type anther. (D) Undehisced dad1 anther. Stomium regions are indicated (arrows).
(E) to (G) Transverse sections of anthers. (E) A developing dad1 anther in a flower bud 3 days before opening. Epidermis (ep), endothecium (en), tapetum (t), stomium (st), connective (c), vascular bundle (v), and pollen grains (p) appear to be normal. (F) A wild-type anther and (G) a dad1 anther in flower buds immediately before opening. Fibrous bands (fb) are observed in endothecium and connective of both anthers. Bars = 50 μm.
(H) and (I) 4′,6-Diamidino-2-phenylindole–stained pollen grains. One vegetative (v) and two generative (g) nuclei are observed in both wild-type (H) and dad1 (I) pollen grains. Bars = 20 μm.
(J) and (K) Aniline blue–stained pistils of dad1 flowers 24 hr after pollination with wild-type pollen grains (J) and dad1 pollen grains (K), respectively. Only wild-type pollen grains (p) germinate and elongate pollen tubes (pt). Because pollen grains that did not germinate were washed away during the staining, no dad1 pollen grains are present on the transparent stigma (sg). Bars = 100 μm.
(L) and (M) Side view of wild-type (L) and dad1 (M) inflorescences. Arrows indicate the flower buds in inflorescences. Many unopened flower buds accumulate at the top of the mutant inflorescence (M).
(N) Number of flower buds on a wild-type (WT) and a dad1 inflorescence. The numbers of middle-stage flower buds (gray) and late-stage flower buds (white) on an inflorescence are shown. Error bars show the standard deviation. The data represent averages of 10 independent samples.
(O) A series of flower buds on an inflorescence of the wild type (WT) and the dad1 mutant. Only middle-stage (left of the red line) and late-stage (between the red and blue lines) flower buds are shown. The definitions of these stages are described in the text. The lower row of wild type and the upper row of dad1 are intact. In the upper row of wild type and the lower row of dad1, the front sepals and petals are removed to show the development of the floral organs. Asterisks indicate flower buds whose anthers have just begun to turn yellow.
In spite of the defect in dehiscence, the outside appearance of dad1 anthers was quite similar to that of the wild type until just before flower opening. To determine if the dehiscence defect was caused by some morphological abnormality of the anther tissues, we observed transverse sections of wild-type and dad1 anthers. At 3 days before flower opening, all cell types in the developing dad1 anthers, such as tapetum, endothecium, epidermis, and connective, as well as immature pollen grains, were normal in appearance (Figure 2E). In the later stage, just before flower opening, all structural features also were observed to be normal in the dad1 anthers, namely, the disappearance of the tapetum, breakage of the septum, differentiation of the stomium, and development of fibrous bands in the endothecium and connective cells (Figures 2F and 2G). Thus, there were no differences in structures between dad1 and wild-type anthers before dehiscence, indicating that the developmental processes in the mutant progress normally toward dehiscence but are interrupted immediately before stomium breakage.
The dad1 pollen grains in undehisced anthers of the mutant flower were indistinguishable in shape from wild-type pollen grains, although they were slightly smaller (∼90% in diameter) than those of the wild type. One vegetative nucleus and two generative nuclei were similarly observed in both wild-type and dad1 pollen grains (Figures 2H and 2I). In addition, the exine wall sculpturing was normal when we examined the mutant pollen grains by scanning electron microscopy (data not shown).
To examine pollen viability, we put the pollen grains on an agar medium. Only 2% of the dad1 pollen grains germinated, compared with more than 70% for those from dehisced wild-type anthers (data not shown). When the dad1 pollen grains were put on the stigmas of dad1 flowers, they did not germinate, whereas wild-type pollen grains germinated effectively on the stigmas and pollen tubes elongated into the ovaries (Figures 2J and 2K). Similar results were obtained when the wild-type stigmas were used in place of the dad1 stigmas (data not shown). In addition, pollen grains isolated from dehisced dad1 anthers at 1 day after flower opening did not germinate either on the medium or on the fresh stigmas (data not shown). These results indicate that the pollen grains of the dad1 mutant develop normally to the trinucleate stage but that some defect occurs at the final stage of maturation to cause inviability of the pollen grains.
Opening of Flower Buds Is Delayed in the dad1 Mutant
Another phenotype characteristic of the dad1 mutant is developmental delay of flower bud opening. We noted that there were more unopened flower buds that had accumulated above the opened flowers in the inflorescence of dad1 than in the wild-type inflorescence (Figures 2L and 2M). We divided the development of the flower buds into three stages, early, middle, and late, and counted the number of flower buds on a primary inflorescence of soil-grown plants after the first 10 to 20 flowers had opened. Three or four flowers opened every day on the inflorescences. The late stage included flower buds whose stigmas protruded from the sepals but whose petals still did not bend outward. The middle stage included flower buds younger than those of the late stage (i.e., the stigmas were covered by sepals, whereas their petals were longer than the stamens). The early stage included flower buds smaller than the middle-stage buds (i.e., from nascent flower primordia to young flower buds whose petals are still shorter than the stamens). The numbers of flower buds of the middle and early stages on a primary inflorescence were approximately the same between the wild type and the dad1 mutant (Figures 2N and 2O). However, the average number of flower buds of the late stage in the wild-type and dad1 inflorescence were 3.5 and 8.0, respectively, indicating that the number of mutant flower buds increased more than twofold compared with wild-type flower buds (Figures 2N and 2O).
To determine the effect of the dad1 mutation more precisely, we observed the development of floral organs in each stage using a series of flower buds (Figure 2O). By the end of the middle stage in wild-type anthers, tapetum degeneration, fibrous band development, and septum breakage were completed and the anther color began to be yellow. In the anther first identified as having yellowish color in the wild type, the tapetum and septum had just disappeared, suggesting that it corresponds to the beginning of anther stage 12 as defined in a previous report (Sanders et al., 1999). In the late stage, anther color turned to brilliant but opaque yellow, petals grew gradually, and finally stamens and petals grew rapidly to open the flowers. In dad1 anthers, the middle-stage events, including complete development of anther tissues, occurred as in the wild type except for a slight delay in the color change of the anthers. However, the progress of late-stage events, anther color change and stamen and petal elongation, was apparently retarded in dad1 flower buds, resulting in an extended late stage and delayed flower opening. The anther color of dad1 was paler than that of the wild type even after flower opening.
dad1 Mutant Was Rescued by the Application of LA as Well as JA
In all respects described above, the phenotype of the dad1 mutant is similar to the phenotypes of the dde1/opr3 mutant, the coi1 mutant, and the fad3 fad7 fad8 triple mutant (data not shown). dde1/opr3 and fad3 fad7 fad8 are known as JA biosynthesis mutants, and coi1 is a mutant insensitive to JA (Feys et al., 1994; McConn and Browse, 1996; Xie et al., 1998; Sanders et al., 1999, 2000; Stintzi and Browse, 2000), strongly suggesting that the DAD1 gene is involved in JA production or signaling. To examine this possibility, we applied methyl jasmonate (MeJA) to bud clusters of the dad1 mutant. Two days after treatment, we found that the anthers dehisced at the same time as flower opening in the newly opened flowers (Figure 3A). Pollen grains of these flowers germinated effectively on the pollen germination medium as well as on stigmas, and these flowers bore many seed like the wild-type flowers, suggesting that the pollen grains are completely fertile. In addition, a backlog of late-stage flower buds opened within 2 days after MeJA treatment, and the morphology of its inflorescence turned to normal (Figure 3B). These results strongly indicate that the dad1 mutation affects JA biosynthesis in flower buds.
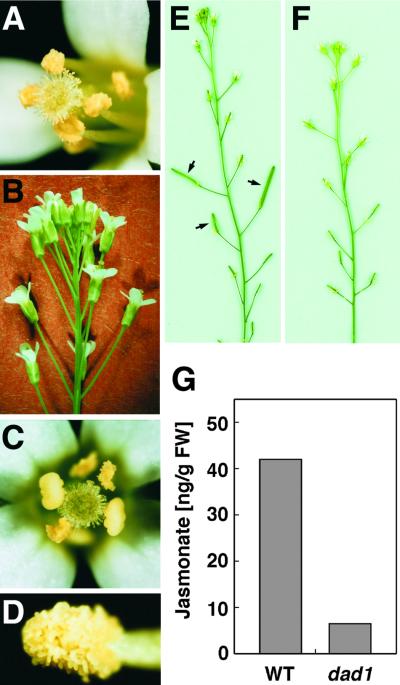
Figure 3.
Level of JA Is Decreased in the dad1 Inflorescence.
(A) and (B) Rescuing of the dad1 mutant by application of MeJA. (A) A dad1 flower opened at 2 days after application. Dehisced anthers and functional pollen grains attached to the stigma are observed. (B) Side view of the dad1 inflorescence at 2 days after application, in which there is no accumulation of unopened flower buds.
(C) to (E) Rescuing of the dad1 mutant by application of LA. (C) A dad1 flower opened at 2 days after application. Dehisced anthers and functional pollen grains attached to the stigma are observed. (D) A dehisced dad1 anther at 2 days after application. (E) A dad1 inflorescence at 7 days after application. Developing seed pods (arrows) are observed.
(F) A dad1 inflorescence at 7 days after application of linoleic acid, which cannot be converted to JA.
(G) Jasmonate levels in flower bud clusters of the wild type (WT) and the dad1 mutant. JA and MeJA were isolated, purified, and measured by gas chromatography–mass spectrometry (see Methods). The data represent the sum of both compounds. FW, fresh weight.
To determine whether the biosynthetic pathway between free LA and JA is intact in the mutant, we treated the dad1 inflorescences with LA. Two days after treatment, the newly opened flowers showed a completely restored wild-type phenotype (Figures 3C and 3D). These flowers also displayed restored fertility, that is, they bore self-pollinated seed (Figure 3E). In contrast, the flower buds treated with linoleic acid, oleic acid, or stearic acid, all of which cannot be used as JA precursors, did not restore fertility (Figure 3F and data not shown), indicating that the effect was specific to LA. In addition, the application of LA had no effect on the dde1/opr3 flower buds, in which conversion from LA to JA was blocked, although MeJA could restore their fertility (data not shown). Therefore, these data strongly suggest that the DAD1 protein acts at (or upstream of) the reaction step to release free LA from some cellular lipids to produce JA in flower buds.
Total Jasmonates Are Decreased in Flower Buds of the dad1 Mutant
JA and MeJA were extracted from flower bud clusters above the opened flowers of wild-type and dad1 mutant plants and quantified using gas chromatography–mass spectrometry. Figure 3G shows that the total amounts of both compounds in dad1 were decreased to 22% of that in the wild type. This result strongly supports our argument that the DAD1 protein is involved in JA biosynthesis.
DAD1 Encodes a Lipase-Like Protein
Because dad1 was isolated from the T-DNA insertional mutagenized population, the mutation was expected to be tagged with T-DNA. We isolated a genomic DNA fragment flanking the T-DNA left border by inverse polymerase chain reaction (PCR) and then used the isolated fragment as a probe for cosegregation analysis. We confirmed that all 110 individual dad1 plants identified from a backcrossed F2 population were homozygous for the T-DNA insertion (data not shown), suggesting that the dad1 mutation is tightly linked to the T-DNA insertion.
Genomic and cDNA cloning revealed that a concatenated T-DNA was inserted in an open reading frame assigned to the hypothetical gene T13E15.18 (At2g44810) in the Arabidopsis Genome Initiative databases. To confirm whether the predicted gene is the true DAD1 gene, we performed a complementation experiment. Transformed dad1 plants containing a T-DNA carrying a wild-type T13E15.18 gene showed restored anther dehiscence at flower opening, complete fertility, and normal inflorescence morphology (data not shown). Thus, we concluded that the T13E15.18 gene is the DAD1 gene.
Sequence comparison between the genomic and cDNA clones revealed that the DAD1 gene had no introns and encoded a polypeptide composed of 447 amino acid residues (Figure 4A), which included an N-terminal stretch missing the T13E15.18 sequence in the Arabidopsis Genome Initiative databases. After consideration of the cleavage of the putative transit peptide (described below), the molecular weight of the DAD1 protein as calculated from its transcribed sequence was 45,000. Homology searches with DAD1 amino acid sequences revealed apparent similarities of DAD1 protein to some fungal lipases, especially around their active sites (Figure 4B). The lipase active site is characterized by an active center or “catalytic triad,” composed of a serine, an aspartic acid, and a histidine residue, and by the GXSXG consensus sequence, including the serine residue of the catalytic triad, both of which are widely conserved in fungal and animal lipases (Brady et al., 1990; Winkler et al., 1990; Woolley and Petersen, 1994). The DAD1 sequence contained a GHSLG motif that fit the lipase consensus sequence and the putative catalytic triad S295, D352, and H425 (or H418) that fit the position of catalytic triad of fungal lipases (Figures 4A and 4B), indicating that DAD1 has typical features of a lipase. The dad1 T-DNA insertion was located at a site corresponding to G249 of the DAD1 amino acid sequence (Figure 4A). This insertion makes a truncated protein without the catalytic triad, suggesting that dad1 is a null mutation.
Figure 4.
Structures of DAD1 and Homologous Proteins Encoded in the Arabidopsis Genome.
(A) The deduced amino acid sequence of DAD1. The serine and aspartic acid residues and two candidates for the histidine residue, which constitute the catalytic triad, are highlighted. The lipase consensus sequence is boxed. The closed triangle indicates the putative cleavage site of the predicted transit peptide. The asterisk indicates the N-terminal residue that is fused to the C terminus of MBP in the MBP-DAD1dE fusion protein. The open triangle indicates the position corresponding to the T-DNA insertion in genomic DNA of the dad1 mutant.
(B) Amino acid alignment in the catalytic regions of the DAD1 protein, homologs identified from Arabidopsis databases, and Rhizomucor miehei lipase. Residues identical in more than half of the sequences are highlighted. The Arabidopsis sequences are classified into three classes, class I (I), class II (II), and class III (III), based on their similarities and the presence of N-terminal stretches. The serine and aspartic acid residues (closed arrowheads) and two candidates for the histidine residue (open arrowheads), which constitute the putative catalytic triad, are indicated. Boxed amino acid residues in R. miehei lipase represent the residues of the catalytic triad determined by x-ray crystallography (Brady et al., 1990). For At2g30550, we supplemented the database sequence with the in-frame 82–amino acid stretch continued on the N terminus.
(C) Scheme of DAD1 and its Arabidopsis homologs showing predicted transit peptides (closed boxes), predicted mitochondrial signal sequence (hatched box), and catalytic regions (open boxes) indicated in (B). Classes I, II, and III are indicated.
A search of the databases revealed 11 hypothetical genes in the Arabidopsis genome encoding proteins homologous with DAD1 throughout their sequences. Based on the presence of N-terminal stretches and sequence similarities in the catalytic region, we classified these proteins into three classes: class I, class II, and class III (Figures 4B and 4C). DAD1 is included in the class I proteins. The class I and class III proteins contain sequence stretches at their N-terminal portions, whereas the class II proteins lack them. Computer analyses with the ChloroP, TargetP, and PSORT programs have predicted that the class I proteins target chloroplasts and that the N-terminal stretches of class I proteins are transit peptides. The class II and class III proteins are predicted to be localized in cytosol and mitochondria, respectively. No protein is predicted to be a secretory protein like fungal lipases. In these proteins, the serine and aspartic acid residues corresponding to the catalytic triad of fungal lipases were completely conserved with their surrounding sequences. A histidine residue aligned with the catalytic triad histidine in fungal lipases also was conserved in 10 of the 12 homologous proteins, whereas another completely conserved residue was present seven residues away from it; thus, one of these two residues may join the catalytic triad. Homologies also were found between these lipases and fungal lipases at regions outside of their catalytic regions, suggesting that the entire structure of the lipases resembles that of the fungal lipase whose stereoscopic structure was highly analyzed by x-ray crystallography (Brady et al., 1990).
DAD1 Is a PLA1
To determine whether the DAD1 protein had lipase activity, we fused the whole DAD1 sequence to the C terminus of maltose binding protein (MBP) and designated it MBP-DAD1. The MBP-DAD1 protein did hydrolyze phosphatidylcholine (PC), but its enzymatic activity was low (data not shown) because the protein aggregated in solution. Then we fused the DAD1 lacking the N-terminal 72 amino acid residues, including a putative transit peptide (described below), to MBP and designated it MBP-DAD1dE. MBP-DAD1dE was more stable in solution and showed high lipolytic acyl hydrolase activity. When we removed the MBP from MBP-DAD1dE by treatment with factor Xa, the resultant DAD1dE protein showed lipolytic activity, but the activity decreased to almost one-fifth of that of the fusion protein, suggesting that the DAD1dE protein itself was not so stable in solution. Thus, we decided to use the MBP-DAD1dE fusion protein for the subsequent characterization of DAD1 activity.
The fusion protein exhibited its highest activity between pH 6.0 and 6.5, and the activity was decreased at lower or higher pH (Figure 5A). Then the substrate specificity for the DAD1 protein was examined. MBP-DAD1dE hydrolyzed PC with an activity of 42.4 nmol · min−1 · mg−1 for the first 30 min under our reaction conditions. When we used monogalactosyldiacylglycerol (MGDG) or trilinolein as a substrate, the lipolytic activity was decreased to 16 or 6%, respectively, compared with that of PC (Figure 5B). In contrast, the lipase from Rhizomucor miehei hydrolyzed MGDG and trilinolein more efficiently than it hydrolyzed the PC substrate (Figure 5B). These results indicate that the phospholipase activity of MBP-DAD1dE was obviously higher than the galactolipase or triacylglycerol lipase activities. Thus, we concluded that DAD1 was a phospholipase.
Figure 5.
PLA1 Activity of the DAD1 Protein.
(A) pH dependence of DAD1 activity. The MBP-DAD1dE protein was incubated with PC in phosphate buffers of various pH levels for 30 min at 25°C, and the released fatty acids were quantified. Data are expressed as the relative activities compared with the activity at pH 6.0, which was assigned a value of 1.0.
(B) Substrate specificity of DAD1 activity. The MBP-DAD1dE protein and R. miehei lipase were incubated with PC, MGDG, or trilinolein (TG), and the released free fatty acids were quantified. Data are expressed as the relative activities compared with the maximum activity for each enzyme (i.e., the activity for PC was assigned a value of 1.0 for MBP-DAD1dE, and the activity for MGDG was assigned a value of 1.0 for R. miehei lipase). Closed bars, MBP-DAD1dE; open bars, R. miehei lipase.
(C) and (D) Substrate specificity of DAD1 activity with respect to sn positions.
(C) The MBP-DAD1dE protein was incubated with 1-palmitoyl-2-linoleoyl-PC. After incubation for the indicated times, the amounts of palmitic acid and linoleic acid in the free fatty acid fraction and in the lysoPC fraction were measured by gas chromatography. Open circles, palmitic acid in the free fatty acid fraction; closed squares, linoleic acid in the lysoPC fraction; open squares, linoleic acid in the free fatty acid fraction; closed circles, palmitic acid in the lysoPC fraction.
(D) The MBP-DAD1dE protein was incubated with 1-palmitoyl-2-14C-linoleoyl-PC. As controls, MBP or buffer (no protein) was added in place of MBP-DAD1dE. After separation by thin layer chromatography, the radioactivity in the bands for 14C-PC (open bars), 14C-free fatty acid (hatched bars), and 14C-lysoPC (closed bars) was quantified and expressed as a proportion of the sum of the three fractions.
To examine the substrate specificity of the DAD1 protein with respect to the sn position of phospholipids, we incubated MBP-DAD1dE with 1-palmitoyl-2-linoleoyl-PC and determined the fatty acid compositions of the reaction products free fatty acid and lysoPC by gas chromatography. With increases in incubation time, the levels of palmitate and linoleate increased almost exclusively in the free fatty acid and lysoPC fractions, respectively, indicating that the MBP-DAD1dE protein exhibits sn-1–specific (or highly selective) activity (Figure 5C).
The substrate specificity of MBP-DAD1dE described above was confirmed by incubation of MBP-DAD1dE with 1-palmitoyl-2-14C-linoleoyl-PC, followed by analysis of 14C-labeled hydrolysis products by thin layer chromatography. Fifty-six percent of the labeled substrate was converted to 14C-labeled lysoPC, whereas 14C-labeled free fatty acid was detected to a negligible extent (less than 5%) (Figure 5D). In control experiment, the levels of 14C-labeled PC, lysoPC, and free fatty acid were not changed after incubation with MBP alone. Similar results were observed when 1-palmitoyl-2-14C-linoleoyl-phosphatidic acid was used as a substrate in place of 1-palmitoyl-2-14C-linoleoyl-PC (data not shown). Because these results indicated that the MBP-DAD1dE specifically hydrolyzed phospholipids at the sn-1 position, we concluded that DAD1 is a PLA1.
DAD1 Gene Expression Is Restricted in Filaments of Stamens
We prepared total RNA from flower buds, leaves, and roots of wild-type plants and performed RNA gel blot analysis using DAD1 cDNA as a probe. However, no signal was detected (data not shown), suggesting that the expression level of DAD1 was quite low. To gain higher sensitivity, we performed reverse transcriptase–mediated (RT)-PCR experiments (Figure 6A). When we used the flower bud RNA as a template, apparent amplification of the DAD1-specific signal was observed, whereas no signal was detected using leaf or root RNA. Mutant flower buds did not contain any detectable DAD1 mRNA.
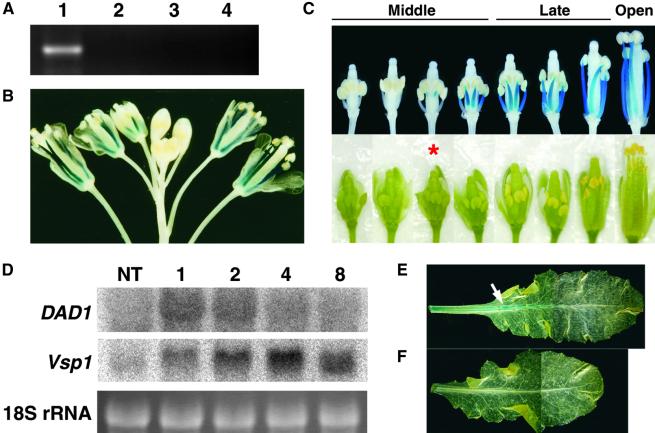
Figure 6.
Expression of the DAD1 Gene.
(A) RT-PCR analysis of DAD1 expression using total RNA isolated from Arabidopsis organs. Lane 1, wild-type flower bud clusters; lane 2, dad1 flower bud clusters; lane 3, wild-type leaves; lane 4, wild-type roots.
(B) and (C) Histochemical localization of GUS activity in DAD1::GUS plants. (B) An inflorescence. Blue signals are observed only in the stamen filaments. (C) A series of flower buds on an inflorescence. Detached flower buds were photographed (bottom row) and stained with X-Gluc solution (see Methods) after removing the sepals and petals (top row). The developmental stages of flower buds are shown above. The asterisk indicates the flower bud whose anther has begun to turn yellow.
(D) Wound-induced expression of the DAD1 gene. Gel blot analysis was performed using 10 μg of total RNA from rosette leaves harvested at the indicated times (hours) after wounding. NT, not treated. Top, RNA gel blots probed with DAD1 cDNA; middle, the same blots probed with Arabidopsis Vsp1 cDNA; bottom, ethidium bromide staining of 18S rRNA bands to confirm equal loading.
(E) and (F) Wound induction of GUS activity in rosette leaves of DAD1::GUS plants. (E) A leaf harvested and stained 1 hr after wounding showing GUS activity mainly in the midvein (arrow). (F) A leaf harvested from unwounded control plants showing no GUS activity.
To determine the temporal and spatial regulation of DAD1 gene expression, we prepared the DAD1 promoter::β-glucuronidase (GUS) construct in which the 3.5-kb 5′ upstream region of DAD1 was fused to the β-glucuronidase–coding sequence and introduced it into wild-type Arabidopsis plants. The signal of DAD1::GUS expression appeared initially at the upper part of stamen filaments in middle-stage flower buds whose anther color had just begun to turn yellow (Figures 6B and 6C). With the continued development of flower buds, DAD1::GUS expression increased gradually and extended to the base of the filaments. The highest expression was observed when the flower buds entered the late stage (Figures 6B and 6C). Consistent with the RT-PCR analysis, no signal was observed in any other organs of the transformants grown normally (data not shown). Therefore, we conclude that the expression of DAD1 was restricted to the filaments immediately before flower opening.
DAD1 Protein Is Targeted to the Chloroplast
The ChloroP and TargetP programs predicted a chloroplastic localization of the DAD1 protein, with a putative cleavage site of the transit peptide in the DAD1 sequence between positions 46 and 47. To examine the localization experimentally, we fused green fluorescent protein (GFP) at the C terminus of DAD1 for expression of the corresponding fusion protein in living cells. Leaves of Arabidopsis and Nicotiana benthamiana were microbombarded with constructs to transiently express either GFP or DAD1-GFP under the control of the 35S promoter of cauliflower mosaic virus. Green fluorescence from DAD1-GFP colocalized with chlorophyll autofluorescence in stomatal guard cells of N. benthamiana, in which, among leaf epidermal cells, chloroplasts are well developed (Figures 7A and 7B). Similar localization also was observed in the pavement cells of N. benthamiana and Arabidopsis leaves, which gold particles coated with DAD1-GFP plasmids entered (data not shown). In contrast, fluorescence corresponding to GFP was localized in the cytoplasm as well as in the nucleus (Figures 7C and 7D). These results confirmed that the DAD1 protein targets chloroplasts in vivo, in agreement with the chloroplastic localization of some other enzymes involved in JA biosynthesis (Bell et al., 1995; Maucher et al., 2000; Ziegler et al., 2000; Froehlich et al., 2001).
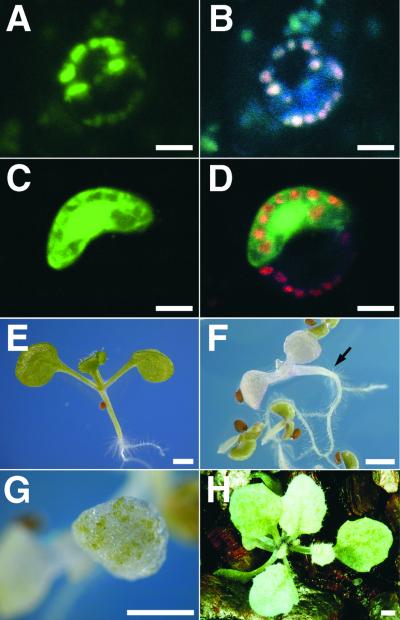
Figure 7.
Subcellular Localization and Effects of the DAD1 Protein in the Chloroplast.
(A) to (D) Subcellular localization of DAD1-GFP (A) and GFP alone (C) in guard cells of N. benthamiana leaves after particle bombardment of corresponding plasmids. (B) and (D) show the same fields as (A) and (C), respectively, illuminated by UV light to show the red autofluorescence of the chloroplasts. Bars = 10 μm.
(E) to (H) Effects of the ectopic expression of the DAD1 gene. (E) A 7-day-old wild-type seedling. (F) and (G) Seven-day-old 35S::DAD1 seedlings. The phenotypes are varied from complete pale (arrow in [F]) or green patches (G) to almost normal (data not shown). (H) A 30-day-old 35S::DAD1 plant showing pale green leaves and inflorescence. Bars = 1 mm.
Ectopic Expression of the DAD1 Gene Causes Defects in Chloroplasts
We examined the phenotype of transgenic lines expressing DAD1 under the control of the 35S promoter of cauliflower mosaic virus. T1 seedlings expressing the 35S::DAD1 gene showed a range of pale color phenotypes (Figures 7E to 7H). Some seedlings showed complete loss of green pigment and died before the formation of true leaves. Even in the seedlings of weaker phenotype, patches of bleached cells were present in their leaves, and the plants sometimes aborted their growth before flowering. These results apparently indicate that an excess amount of DAD1 protein targeted to chloroplasts hydrolyzes their membrane phospholipids to cause destruction of the chloroplasts.
DAD1 Expression Is Induced by Wounding
Plants accumulate a remarkable amount of JA immediately after wounding, when the genes involving JA biosynthesis are upregulated drastically (Bell and Mullet, 1993; Laudert et al., 1996; Heitz et al., 1997; Biesgen and Weiler, 1999; Ziegler et al., 2000). To determine whether the expression of the DAD1 gene also is induced by wounding, we isolated total RNA from wounded rosette leaves and performed RNA gel blot analysis. Figure 6D shows that the DAD1 mRNA was maximally accumulated within 1 hr after wounding and returned to the basal level by 8 hr. The time course was similar to the expression of the LOX, AOS, AOC, and OPR genes of Arabidopsis and tomato (Laudert et al., 1996; Heitz et al., 1997; Biesgen and Weiler, 1999; Wang et al., 2000; Ziegler et al., 2000) and was consistent with the time course of JA accumulation (Laudert et al., 1996; McConn et al., 1997). In contrast, the mRNA accumulation of a vegetative storage protein (VSP1) gene that was induced by JA was delayed and peaked 4 hr after wounding (Figure 6D). Thus, the DAD1 gene probably contributes to the wound induction of JA biosynthesis. The wound-induced activation of the DAD1 gene was confirmed by another experiment in which the obvious expression of the DAD1::GUS gene was observed in rosette leaves after wounding (Figures 6E and 6F).
DISCUSSION
In this study, we identified a male-sterile mutant, dad1, defective in the biosynthesis of JA, which defect caused failure in anther dehiscence simultaneous with flower opening, lack of maturation of pollen grains, and developmental delay of flower buds. The DAD1 gene was shown to encode a chloroplastic PLA1, and all defects of dad1 were rescued by the application of LA. These results indicate that the DAD1 protein is a lipolytic enzyme catalyzing the release of LA, the initial step of JA biosynthesis.
DAD1 Encodes a Novel Class of PLA1
We showed that the DAD1 protein had the remarkable property of releasing free fatty acids from phospholipids and that this activity was highly specific to the sn-1 position of the lipids. Thus, we designated DAD1 as a PLA1. The catalytic region of the DAD1 protein showed an apparent similarity to those regions of fungal triacylglycerol lipases, which show the sn-1–specific phospholipase and galactolipase activities as well (Fischer et al., 1973). Even though the identity score between the catalytic regions of DAD1 and R. miehei lipase, a fungal lipase whose three-dimensional structure is well characterized, was less than 20%, the lipase consensus sequence GXSXG and the catalytic triad were found to be highly conserved with their surrounding sequences in both lipases (Brady et al., 1990). Indeed, the DAD1 protein could hydrolyze galactolipids and triacylglycerols, but the activity was less than one-fifth and less than one-tenth, respectively, of the phospholipase activity, indicating that the enzymatic property of DAD1 is different from that of fungal lipases. To date, several types of PLA1 have been identified from bacteria, yeast, and animals. Among them, platelet-derived serine phospholipid-specific phospholipase A isolated from humans and rats, as well as venom PLA1 from hornets, showed a striking similarity to the mammalian pancreatic triacylglycerol lipase, and the hornet venom PLA1 had weak triacylglycerol lipase activity (Soldatova et al., 1993; Sato et al., 1997; Nagai et al., 1999). However, the similarity between these enzymes and fungal lipases was identified only in the narrow region surrounding the GXSXG motif. Thus, DAD1 is a novel class of PLA1. Each known PLA1, including DAD1, is structurally different from any type of PLA2 that has been characterized in animals.
Role of DAD1 in JA Biosynthesis
It was reported that the anthers of the dde1/opr3 mutant and the fad3 fad7 fad8 triple mutant, both of which have a defect in JA biosynthesis, and of coi1, which is a JA-insensitive mutant, failed or delayed their dehiscence (Feys et al., 1994; McConn and Browse, 1996; Xie et al., 1998; Sanders et al., 1999, 2000; Stintzi and Browse, 2000). Thus, these observations indicate that JA is required for anther dehiscence. The biosynthetic pathway of JA from free LA has been well investigated, and genes for all of the enzymes in this pathway have been cloned and characterized (Song et al., 1993; Bell et al., 1995; Laudert et al., 1996; Maucher et al., 2000; Sanders et al., 2000; Stintzi and Browse, 2000; Ziegler et al., 2000). In fact, the DDE1/OPR3 gene encodes an OPR protein, an enzyme of this pathway (Sanders et al., 2000; Stintzi and Browse, 2000). However, the enzyme catalyzing the initial step of JA production, which releases free LA from cellular lipids, had not been identified until now. Here, we argue that the DAD1 protein is this enzyme for the following reasons. First, the phenotype of dad1 was quite similar to that of the JA-related mutants mentioned above. Second, the amount of JA in flower buds was reduced markedly in dad1. Third, dad1 was rescued completely from lesions by the addition of LA or MeJA to the young flower buds. And fourth, DAD1 is a PLA1 that catalyzes the release of fatty acid from cellular lipids. The first three reasons clearly indicate that dad1 has a defect in JA production at the upper part of the biosynthetic pathway, and the last reason apparently shows that DAD1 has lipolytic activity to release LA, an essential reaction step to provide the LA for use in the biosynthesis of JA.
Compared with the biosynthesis pathway of animal prostaglandins, which are structurally similar to JA, many researchers believed that the precursor of JA was generated by a PLA2 (Mueller, 1997). In animal cells, the contribution of PLA2 is consistent with the positional specificity of polyunsaturated fatty acids, such as the prostaglandin precursor arachidonic acid, for the sn-2 position of phospholipids. However, at least in Arabidopsis leaves, the amount of LA at the sn-1 position in the major polar lipids is almost equal to that at the sn-2 position, and more than 30% of the sn-1 fatty acid is LA (Browse et al., 1986). Therefore, it is not surprising that the enzyme catalyzing the first step of JA biosynthesis is a PLA1. In addition, it was proposed that the DAD1 protein is localized in the chloroplasts whose envelope membranes contain the enzymes LOX, AOS, and AOC, which are involved in the downstream reaction of free LA production (Figure 1). Moreover, the outer envelope membrane of chloroplasts is a major source of PC, which is a good substrate for the DAD1 protein (Block et al., 1983; Joyard et al., 1991). Thus, we hypothesize that the DAD1 protein hydrolyzes the phospholipids of the outer envelope membrane to supply free LA to the downstream enzymes. However, recently it was shown that the tomato AOS is peripherally associated with the inner envelope membrane of chloroplasts and that the bulk of the protein faces the stroma (Froehlich et al., 2001). If the DAD1 protein also is associated with the inner envelope membrane, it may use the galactolipids, because the galactolipids account for more than 70% of the lipids of the inner envelope membrane, whereas phospholipids account for less than 20% (Block et al., 1983; Joyard et al., 1991). It is important to determine the suborganellar localization of the DAD1 protein in the chloroplast.
The amount of JA is increased markedly soon after wounding (Laudert et al., 1996; McConn et al., 1997) in many species of plants. The wound-induced accumulation of DAD1 mRNA suggests its contribution to the wound induction of JA. However, more than a 100-fold induction of JA can be detected in both dad1 and wild-type leaves after wounding (S. Ishiguro, unpublished results), indicating that some lipolytic enzyme(s) other than DAD1 must be present to participate in the wound induction of JA. Possible candidates are the DAD1-like proteins shown in Figures 4B and 4C, although no evidence for their involvement has been observed. In contrast, it was found that the levels of free fatty acid and lysophospholipid were increased markedly after wounding in tomato leaves (Conconi et al., 1996; Lee et al., 1997) and that the reaction was catalyzed by a PLA2 (Nárvaez-Vásquez et al., 1999). Thus, PLA2 also may contribute to the wound induction of JA biosynthesis, although the genes encoding this enzyme have not been identified. In addition, a wound-induced increase of phosphatidic acid in tomato leaves has been reported (Lee et al., 1997), and recently, the involvement of PLD in the wound-induced accumulation of JA was revealed experimentally by antisense suppression of a PLD gene in Arabidopsis (Wang et al., 2000). It should be noted that the involvement of PLA2 and PLD in JA accumulation has been observed only after wounding of plants. Thus, it is possible that these enzymes act only in the wound-induced accumulation of JA, whereas DAD1 and DAD1-like proteins may contribute to the biosynthesis of JA as a developmental requirement for processes such as anther dehiscence.
DAD1 Homologs in Arabidopsis
We found 12 genes encoding DAD1 and DAD1-like proteins in the Arabidopsis genome that show remarkable similarities throughout their sequences. Some other proteins, such as EDS1 and PAD4, which are involved in disease resistance, also show similarities around the lipase motif, but their overall structures are different from those of DAD1 and DAD1-like proteins (Falk et al., 1999; Jirage et al., 1999). The function of none of the DAD1-like proteins was revealed. The chloroplastic class I proteins may serve in JA biosynthesis, like the DAD1 protein. Some of them may contribute to the biosynthesis of other oxylipin compounds such as α- and γ-ketols and C6-aldehydes, which are produced by the branches of the JA biosynthesis pathway (Figure 1). The use of chloroplastic galactolipids for C6-aldehyde biosynthesis was reported recently (Matsui et al., 2000). In contrast, it was proposed that the fatty acid precursor of C6-aldehyde is supplied from outside the chloroplasts (Froehlich et al., 2001). According to this model, the contribution of the cytosolic class II proteins to C6-aldehyde biosynthesis is possible as well.
Another possible function of class II proteins is the degradation of membrane lipids at senescence. A cDNA clone encoding a carnation class II protein expressed at the onset of petal senescence has been isolated (Hong et al., 2000), and transgenic Arabidopsis plants expressing the antisense construct of an Arabidopsis homolog of this lipase showed delayed leaf senescence and enhanced seed yield (Thompson et al., 2000). Interestingly, the carnation protein had the remarkable property of hydrolyzing triacylglycerols more effectively than phospholipids (Hong et al., 2000). A cDNA clone encoding a petal-abundant lipase-like protein also has been identified from Japanese morning glory (accession number U55867). The function of the sole class III DAD1-like protein is not known.
Roles of JA in Anther Dehiscence and Opening of Flower Buds
In the flower buds of the middle stage, the typical events of anther development, namely, degeneration of the tapetum, formation of fibrous bands, breakage of the septum, and development of trinucleate pollen grains, occurred normally in both the wild type and the dad1 mutant. The only exception was the slight delay in the color change of dad1 anthers. In the late stage, when the anther color should turn yellow and petals and stamens should elongate, the remarkable defect of the dad1 mutant was identified as a developmental delay of flower buds; even after flower opening, anther dehiscence did not occur within 1 day and the pollen grains could not mature. The expression of the DAD1 gene was first observed at the upper part of the anther filaments during the middle stage and extended gradually to the base of the filaments by the end of this stage. Then it reached the maximum level when the flower buds entered the late stage. Thus, the expression profile of the DAD1 gene is consistent with the defects of the dad1 mutation. We found that exogenously applied MeJA or LA rescued the mutant from the lesion in the 6th to 17th flower buds, on average, numbered from one as the oldest in the dad1 inflorescence (data not shown); these buds correspond to the early-middle to early-late stages of flower bud development. This range seems to be consistent with the “window” in which JA is perceived by the dde1 flower buds described previously (Sanders et al., 2000). Considering the residue of applied JA in plant tissue, we conclude that the time of maximal requirement of JA is the early part of the late stage, which agrees with the maximum expression of the DAD1 gene. Sanders et al. (2000) showed that the highest DDE1/OPR3 mRNA accumulation occurred in the filaments as well as in the pistils and petals at stage 10 of their classification, which corresponds to the middle part of our middle stage. Although the expression of the DDE1/OPR3 gene was slightly earlier than what we observed for DAD1 expression and no expression of DAD1 was detected in pistils and petals, both genes may be expressed cooperatively at least in the filaments of developing flower buds to produce JA in the organs.
In angiosperms, the force to break the stomium is supplied by dehydration and shrinkage of the endothecium and connective cells surrounding the locules (Keijzer, 1987). At the same time, the liquid filling the locules is absorbed and pollen grains become desiccated. In our observations, the color change of anthers seemed to be correlated with anther desiccation, suggesting the delay of desiccation in dad1 anthers. Moreover, at the time of flower opening, the endothecium and connective cells of dad1 anthers were still fully expanded and their locules were filled with liquid. These observations indicate that the dad1 mutation blocks water transport to vascular tissues from the endothecium, connective, and locules. We observed a similar phenomenon in dde1/opr3 and coi1 mutants. Thus, it is indicated that water transport in anthers is regulated by JA. The dehydration of endothecium, connective, and locules may be attributable in part to the evaporation of water through the stomata found on the adaxial surface of anthers behind the connective tissue (Keijzer et al., 1987). In addition, Stadler et al. (1999) showed that the AtSUC1 protein, a plasma membrane H+-sucrose symporter, is accumulated in some of the connective cells surrounding the vascular tissue during the final stages of anther development and suggested that this protein accumulates sucrose in these tissues to increase water uptake. It is an attractive hypothesis that JA is required for the expression of the AtSUC1 gene as well as other genes involved in water transport in anthers.
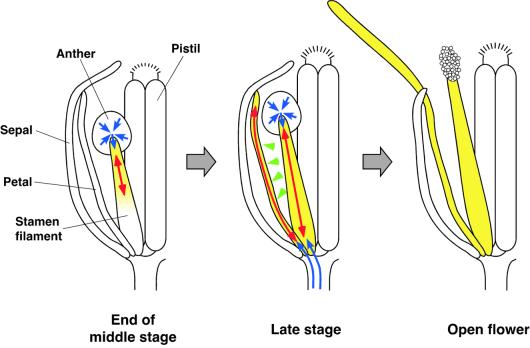
What is the significance of the predominant expression of DAD1 in stamen filaments? It was demonstrated that the dehiscence of onion anthers correlates with the extension rate of the stamen filaments (Keijzer et al., 1987). In the tomato, it was suggested that anther dehiscence is preceded by the dehydration of locules and that water is exported through the filaments to the petals along an osmotic gradient generated by starch/sugar interconversion (Bonner and Dickinson, 1990). If these activities are damaged, one might expect that the filament and petal elongation at the flower opening would be inhibited and the anther dehiscence blocked, both of which are the typical defects of the dad1 mutant. Thus, JA seems to regulate the water transport into filaments as well as into petals. Based on a putative effect of JA to promote water uptake and cell elongation, we propose the model shown in Figure 8. At the end of the middle stage, the DAD1 gene is expressed in the upper part of the filaments to produce JA. The JA induces water uptake of the cells in this region and promotes water transport from locules through the anther walls (endothecium and connective) to the filaments. The desiccation of locules promotes the maturation of pollen grains by some unknown mechanism. In the late stage, cells at both the upper and lower parts of the filaments express the DAD1 gene, produce JA, and take up water from both anther walls and pedicels, resulting in the elongation of filaments and the dehiscence of anthers. Because the JA (or its volatile derivative MeJA) produced in the filaments can be transmitted easily to petals by water flow or by diffusion, petal cells promote the water uptake and elongation to grow the petals and to open the flowers. This model could explain how the JA-promoting water transport realizes the synchronization of pollen maturation, anther dehiscence, and flower opening, which results in the effective pollination in self-pollinating Arabidopsis flowers. Easy recovery of fertility in dad1 flowers by the application of JA or LA, a nontoxic nutrient fatty acid, suggests that the DAD1 gene should be an excellent tool for the breeding of parent lines for hybrid crops and vegetables.
Figure 8.
Proposed Model of the Synchronous Regulation of Pollen Maturation, Anther Dehiscence, and Flower Opening by JA.
The yellow-colored regions represent organs that actively take up water and elongate in response to JA. Transmission of JA, elongation of organs, and transport of water are represented by green arrowheads, red arrows, and blue arrows, respectively.
METHODS
Plant Materials and Growth Conditions
The lines of Arabidopsis thaliana used here were descended from the Wassilewskija (Ws) wild type. The dad1 mutant was derived from T-DNA insertion lines that were made by the tissue culture method (Akama et al., 1992) using Agrobacterium tumefaciens harboring the binary plasmid pBIH1-IG (Ohta et al., 1990). The mutant was backcrossed twice before being used in the experiments. Seed were sown on vermiculite and allowed to imbibe for 4 days at 4°C, and plants were grown at 22°C under a 12-hr-light/12-hr-dark photoperiod.
Microscopy
For observation of anther transverse sections, flower bud clusters were fixed with 4% paraformaldehyde in PBS. Five-micrometer paraffin sections were prepared and stained with toluidine blue. Pollen grains were immersed in a solution consisting of 2 μg/mL 4′,6-diamidino-2-phenylindole and 7% sucrose and viewed by fluorescence microscopy under UV light excitation.
Pollen Germination
For in vitro pollen germination, pollen was isolated from recently fully opened flowers and placed onto plates of pollen germination medium, which consisted of 15% (w/v) sucrose, 0.4 mM Ca(NO3)2, and 0.4 mM H3BO3 solidified with 1% (w/v) agarose (Carpenter et al., 1992). In the case of the mutant, the locule was opened manually to release the pollen. Pollen was incubated for 8 hr at room temperature under moist conditions and then observed by differential interference contrast microscopy.
Aniline blue staining of pollinated pistils was performed essentially as described (Muschietti et al., 1994). Pistils were collected 24 hr after pollination and fixed in ethanol/acetic acid (3:1) for 1 hr at room temperature. After overnight softening in 8 M NaOH, the pistils were washed several times with distilled water and incubated with aniline blue solution (0.1% aniline blue in 0.1 M K2HPO4-KOH buffer, pH 11) for 3 hr in complete darkness. The stained pistils were placed in a drop of glycerol on a microscope slide and observed by fluorescence microscopy under UV light excitation.
Application of Methyl Jasmonate (MeJA) and Linolenic Acid (LA)
All opened flowers were removed from the inflorescence, and the remaining flower bud clusters were dipped into 500 μM MeJA (Wako Pure Chemical, Osaka, Japan) or 0.1% (v/v) linolenic acid (Sigma-Aldrich), both dissolved in 0.05% aqueous Tween 20.
Determination of Jasmonate
Flower bud clusters, with fully opened flowers removed, were harvested, weighed, frozen in liquid N2, and stored at −80°C. Extraction and measurement of jasmonate were performed as described (Ueda and Miyamoto, 1994). Five grams of the flower bud clusters were ground in liquid N2 with a mortar and pestle and extracted three times with 80% ethanol. After evaporation of the organic solvent in vacuo, the aqueous solution was adjusted to pH 2.0 to 3.0 with HCl and partitioned against ethyl acetate three times. Then, 5% NaHCO3 was added to the ethyl acetate phase and MeJA was partitioned into solvent phase (fraction NE). The residual 5% NaHCO3 phase was adjusted to pH 2.0 to 3.0 with HCl, and jasmonic acid (JA) was then extracted with ethyl acetate (fraction AE). Both NE and AE were partially dried with sodium sulfate and evaporated to dryness in vacuo. The residues were dissolved in a small volume of ethyl acetate, spotted onto silica gel plates (Merck), and developed with the solvent system of hexane/ethyl acetate (40:8, v/v) for NE and hexane/ethyl acetate/chloroform/acetic acid (40:40:16:1, v/v/v/v) for AE. For NE, the separated MeJA was dissolved in ethyl acetate and again purified by thin layer chromatography (TLC) developed with the solvent system of benzene/ethyl acetate (10:1, v/v). For AE, partially purified JA was dissolved in ethyl acetate and methylated to become MeJA with ethereal diazomethane. MeJA derived from both NE and AE was analyzed using a gas chromatography–selected ion monitoring system (Finnigan, Bremen, Germany). The level of MeJA was estimated from the mass-to-charge ratio 224 peak area. The results are presented as the total amounts of MeJA derived from NE and AE.
Molecular Cloning of the DAD1 Gene
To identify the genomic sequence flanking the T-DNA, we performed inverse polymerase chain reaction (PCR) as described (Deng et al., 1992). Genomic DNA prepared from dad1 leaves was digested with EcoRI, and the 2.7- to 4.3-kb fraction was isolated by agarose gel electrophoresis. The DNA was self-ligated, relinearized with BamHI, and used for PCR amplification using primers HPH-R (5′-TTTGCC-CTCGGACGAGTGCT-3′) and L2 (5′-CAAAAGGTATGCCCAAAA-ACAAC-3′). A 0.5-kb sequence flanking the T-DNA was identified in the amplified fragment and cloned into pBluescript II SK+ (Stratagene).
To confirm the cosegregation between the dad1 mutation and the T-DNA insertion, we digested genomic DNAs isolated individually from 110 F2 plants having the dad1 phenotype with EcoRI and analyzed the fragments by DNA gel blot hybridization using the 0.5-kb flanking fragment as a probe.
For isolation of genomic clones, a λ-Dash II genomic library of Arabidopsis (ecotype Ws) described previously (Wada et al., 1997) was used. The amplified library (2.0 × 105 independent clones) was screened with the 0.5-kb flanking fragment used as a probe. From the isolated clone, an 8.1-kb HindIII–SplI fragment and a 3.7-kb SpeI fragment were excised and subcloned into pBluescript II SK+.
For cDNA cloning, an amplified λ-ZAPII cDNA library of unopened flower buds of Arabidopsis (ecotype Landsberg erecta) (6.6 × 105 independent clones) was screened. The genomic 3.7-kb SpeI fragment was used as a probe.
Database searches were performed using the GenomeNet BLAST servers at the Institute for Chemical Research, Kyoto University (http://www.genome.ad.jp/), TAIR BLAST 2.0 at the Arabidopsis Information Resource Center (http://www.arabidopsis.org/home.html), and MATDB at the Munich Information Center for Protein Sequences (http://websvr.mips.biochem.mpg.de/proj/thal/). Subcellular localization was examined using ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) and TargetP (http://www.cbs.dtu.dk/services/TargetP/) at the Center for Biological Sequence Analysis and the PSORT server (http://psort.nibb.ac.jp/) at the National Institute for Basic Biology.
Complementation Experiment
The 3.7-kb SpeI fragment covering the entire DAD1 coding region, the 1.5-kb 5′ upstream region, and the 0.7-kb 3′ downstream region was subcloned into the SmaI site of the binary vector pARK5-MCS and introduced into Agrobacterium. Transformation of the dad1 mutant was performed using a tissue culture method with 10 mg/L bialaphos to select for transformants (Akama et al., 1992).
Examination of DAD1 Gene Expression
Total RNA was isolated from flower bud clusters, rosette leaves, or roots using ISOGEN reagent (Nippon Gene, Tokyo, Japan). We used the SuperScript Preamplification System for First Strand cDNA Synthesis (Gibco BRL) to prepare the first-strand cDNA for reverse transcriptase–mediated (RT)-PCR, according to the manufacturer's protocol. Forty cycles of PCR were performed using the DAD1 gene-specific primers FP3 (5′-CTCCTTGAGAAGCAAGGCACGAAG-3′) and RP9 (5′-CCGAAGCTCCTTACCGATTTCAG-3′).
Wounding was applied with a pair of forceps by pinching 10 times across the midvein at the distal half of a rosette leaf. Before wounding or at 1, 2, 4, and 8 hr after wounding, leaves were harvested and used for total RNA isolation as described above. RNA samples were separated on a 1% agarose/formaldehyde denaturing gel, transferred to a Hybond N+ membrane (Amersham Pharmacia), and probed with DAD1 cDNA and VSP1 cDNA. The VSP1 cDNA (715 bp long) was amplified by RT-PCR using total RNA isolated from wounded rosette leaves. Primers VSP1-133F (5′-GAACTCTTAGAG-AAAGAGGGACTG-3′) and VSP1-848R (5′-GGGACAATGCCATGA-AGATAGATG-3′) were used for amplification.
DAD1::GUS Transgenic Plants
The 3.5-kb HindIII–SplI fragment covering the 5′ upstream region of the DAD1 gene was isolated and subcloned into the binary vector pBI101.2 (Clontech, Palo Alto, CA) between HindIII and SmaI to make a translational fusion of the DAD1 and β-glucuronidase (GUS) genes. Transformation of Arabidopsis (Ws) was performed using the vacuum infiltration method (Bechtold et al., 1993). For GUS histochemical staining, organs were incubated overnight with X-Gluc solution (1.9 mM 5-bromo-4-chloro-3-indolyl-β-glucuronide, 0.5 mM K3Fe[CN]6, 0.5 mM K4Fe[CN]6, 0.1% Triton X-100, and 50 mM Na-phosphate buffer, pH 7.0) at 37°C.
Ectopic Expression of the DAD1 Gene
The coding sequence of the DAD1 gene was amplified from its genomic clone by PCR using DAD1-CF1 (5′-CATACTCCATTCTCT-AGATACAACC-3′) and DAD1-CR1 (5′-TTCTCCCGGGAAGTTAGA-GATATGTCAC-3′) as primers, and the amplified fragment was introduced into the EcoRV site of pBluescript II SK+ to make the plasmid pscDAD1. This plasmid was digested at XbaI and SmaI sites on the PCR primers described above, respectively, and the insert including the entire DAD1 coding sequence was introduced between the XbaI and SacI sites of the vector pMAT137-AGI (Matsuoka and Nakamura, 1991) downstream of the 35S promoter. Transformation of Arabidopsis (Ws) was performed by the vacuum infiltration method (Bechtold et al., 1993).
Particle Bombardment of the DAD1–Green Fluorescent Protein (GFP) Fusion Gene
To construct the plasmid carrying the 35S::DAD1-GFP gene, we digested pscDAD1 with XbaI and SacI and inserted the DAD1-coding fragment between the 35S promoter and GFP (XbaI and BamHI sites) of the pUC18-based plasmid pTH-2, which carries a synthetic GFP gene, sGFP(S65T) (Chiu et al., 1996). At the ligation step, we used annealed synthetic oligonucleotides consisting of 5′-CCACGTGTC-CATTCAGAGAAACAGCTCGGAGAGTTCTCCATAGATCTGGAGGAG-GAG-3′ and 5′-GATCCTCCTCCTCCAGATCTATGGAGAACTCTC-CGAGCTGTTTCTCTGAATGGACACGTGGAGCT-3′ as a SacI–BamHI adapter to make a fusion of full-length DAD1 and GFP. The pTH-2 was used as the 35S::GFP control.
The abaxial surfaces of Nicotiana benthamiana leaves were bombarded by the Bio-Rad particle delivery system (PDS-1000/He) with a 1350-p.s.i. rupture disk and 1.0-μm gold microcarriers. Bombarded leaves were incubated in the dark at 22°C for 36 hr and observed by fluorescence microscopy under blue light excitation. For detection of chloroplast autofluorescence, UV light excitation was used.
Expression and Purification of the DAD1 Fusion Protein
To construct plasmids expressing the maltose binding protein (MBP)-DAD1 protein, pscDAD1 was digested with SplI and SalI, and the DAD1-coding fragment was inserted between the EcoRI and SalI sites of the pMAL-c2 vector (New England Biolabs, Beverly, MA). For the ligation, an annealed oligonucleotide adapter for EcoRI and SplI sites consisting of 5′-AATTCATGAGATTCTCTCTTTCTCCC-3′ and 5′-GTACGGGAGAAAGAGAGAATCTCATG-3′ was used. For MBP-DAD1dE, which lacked the putative transit peptide, the same plasmid was digested with EcoRI and self-ligated. The resultant constructs were expressed in Escherichia coli BL21-CodonPlus (DE3) RIL (Stratagene), yielding a fusion protein consisting of DAD1 (or DAD1dE) linked through a factor Xa cleavage site to MBP. The proteins were purified from E. coli extracts by amylose column chromatography. In some experiments, the fusion protein was treated with factor Xa to release the DAD1 (or DAD1dE). Protein was assayed as described (Bradford, 1976) using BSA as a standard.
Assays of Phospholipase Activity
Phosphatidylcholine (PC) from soybean (Sigma-Aldrich), monogalactosyldiacylglycerol (MGDG) from wheat (galactosyl diglyceride; Sigma-Aldrich), and trilinolein (Sigma-Aldrich) were used as substrates for measuring lipase activities. The reaction mixture contained 50 mM K-phosphate buffer, pH 6.0, 0.8 mg/mL substrate, 0.2% Triton X-100, and enzyme protein (14 μg) in a final volume of 50 μL. The substrates were emulsified in 5% gum arabic by sonication for 30 sec before being added to the reaction mixture. The reaction was performed at 25°C for various periods of time up to 30 min. The reaction mixture then was extracted with chloroform, and the extract was dried under a N2 stream. The level of fatty acid included in the residue was assayed enzymatically with a diagnostic kit for the measurement of free fatty acid (NEFA C-Test; Wako Pure Chemical), with linoleic acid (Sigma-Aldrich) as a standard. For comparison, lipase from Rhizomucor miehei (Sigma-Aldrich) was incubated with substrates in 50 mM Tris-HCl, pH 8.0, at 37°C.
Examination of the sn position specificity of phospholipase activity was performed as described (Gatt and Barenholz, 1969) with some modifications. A 0.8-mL reaction mixture containing 1.3 μM 1-palmitoyl-2-linoleoyl-PC, 50 mM K-phosphate buffer, pH 6.0, 0.2% Triton X-100, and 220 μg of MBP-DAD1dE was incubated at 25°C. At appropriate time intervals, 100-μL aliquots of reaction mixture were withdrawn for lipid extraction, and the reaction products free fatty acid and lysoPC were purified by TLC. Fatty acid methyl esters were prepared from the reaction products by methanolysis at 80°C for 3 hr in 5% HCl/methanol, extracted with hexane, and then quantified by gas chromatography (GC18A; Shimadzu, Kyoto, Japan) on an apparatus equipped with a fused silica capillary column (HR-SS-10, 0.25 mm × 50 m; Shinwa Chemical Industries, Kyoto, Japan).
In another experiment, 15 nmol of 1-palmitoyl-2-[1-14C]-linoleoyl-PC (200 GBq/mmol; American Radiolabeled Chemicals, St. Louis, MO) or 1-palmitoyl-2-[1-14C]-linoleoyl-phosphatidic acid (200 GBq/mmol), which was derived from the radiolabeled PC with phospholipase D (Kates and Sastry, 1969), was incubated with 92 μg of MBP-DAD1dE under standard conditions for 30 min. Lipids were extracted, dried, and separated by TLC using chloroform/methanol/28% ammonia (65:25:5, v/v/v). Radioactivities corresponding to free fatty acid, lysoPC (or lysophosphatidic acid), and PC (or phosphatidic acid) were quantified with the Bio-Imaging Analyzer (BAS2000; Fuji Photo Film, Tokyo, Japan).
Accession Number
The DDBJ/EMBL/GenBank accession number for the nucleotide sequence of the DAD1 gene is AB060156.
Acknowledgments
The authors thank Ken Matsuoka, Kenzo Nakamura, Katsunori Hatakeyama, Takeshi Takasaki, Kokichi Hinata, Kenji Matsui, Yasuo Kowyama, Daisuke Shibata, Fusao Motoyoshi, Ayumi Tanaka, Takashi Araki, Yoshibumi Komeda, Yoshiro Shimura, and all members of the Okada laboratory, especially Ryuji Tsugeki, Takuji Wada, and Tokitaka Oyama, for their helpful discussions. We also thank Rie Ishiguro for invaluable technical assistance. We are grateful to John G. Turner for coi1 seed, Yasuo Niwa for pTH-2 vector, and Hiroyuki Anzai for pARK5 vector and bialaphos. dde1/opr3 seed was provided by the Arabidopsis Biological Resource Center at Ohio State University. This work was supported in part by Grants-in-Aid for Special Research on Priority Areas (grant no. 07281101 to S.I. and grant no. 10182101 to K.O.) and for the Encouragement of Young Scientists (grant No. 12740437 to S.I.) and by Special Coordination Funds for Promoting Science and Technology (to S.I. and K.O.) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan. This work also was funded by grants from the Research Institute of Seed Production and from the Research for the Future Program of the Japan Society for the Promotion of Science to S.I. and from the Mitsubishi Foundation to K.O.
References
- Akama, K., Shiraishi, H., Ohta, S., Nakamura, K., Okada, K., and Shimura, Y. (1992). Efficient transformation of Arabidopsis thaliana: Comparison of the efficiencies with various organs, plant ecotypes and Agrobacterium strains. Plant Cell Rep. 12, 7–11. [DOI] [PubMed] [Google Scholar]
- Beals, T.P., and Goldberg, R.B. (1997). A novel cell ablation strategy blocks tobacco anther dehiscence. Plant Cell 9, 1527–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [Google Scholar]
- Bell, E., and Mullet, J.E. (1993). Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol. 103, 1133–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, E., Creelman, R.A., and Mullet, J.E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 92, 8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesgen, C., and Weiler, E.W. (1999). Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta 208, 155–165. [DOI] [PubMed] [Google Scholar]
- Block, M.A., Dorne, A.J., Joyard, J., and Douce, R. (1983). Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. J. Biol. Chem. 258, 13281–13286. [PubMed] [Google Scholar]
- Bonner, L.J., and Dickinson, H.G. (1990). Anther dehiscence in Lycopersicon esculentum. New Phytol. 115, 367–375. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brady, L., Brzozowski, A.M., Derewenda, Z.S., Dodson, E., Dodson, G., Tolley, S., Turkenburg, J.P., Christiansen, L., Huge-Jensen, B., Norskov, L., Thim, L., and Menge, U. (1990). A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature 343, 767–770. [DOI] [PubMed] [Google Scholar]
- Browse, J., and Somerville, C. (1991). Glycerolipid synthesis: Biochemistry and regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 467–506. [Google Scholar]
- Browse, J., Warwick, N., Somerville, C.R., and Slack, C.R. (1986). Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem. J. 235, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, J.L., Ploense, S.E., Snustad, D.P., and Silflow, C.D. (1992). Preferential expression of an alpha-tubulin gene of Arabidopsis in pollen. Plant Cell 4, 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Conconi, A., Miquel, M., Browse, J., and Ryan, C.A. (1996). Intracellular levels of free linolenic and linoleic acids increase in tomato leaves in response to wounding. Plant Physiol. 111, 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71, 791–801. [DOI] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene–mediated disease resistance in Arabidopsis, has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.F., Benedetti, C.E., Penford, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, W., Heinz, E., and Zeus, M. (1973). The suitability of lipase from Rhizopus arrhizus delemar for analysis of fatty acid distribution in dihexosyl diglycerides, phospholipids and plant sulfolipids. Hoppe-Seyler's Z. Physiol. Chem. 354, 1115–1123. [DOI] [PubMed] [Google Scholar]
- Froehlich, J.E., Itoh, A., and Howe, G.A. (2001). Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 125, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatt, S., and Barenholz, Y. (1969). Phospholipase A1 from rat brain, specific for a′ position of lecithin. Methods Enzymol. 14, 197–203. [Google Scholar]
- Goldberg, R.B., Beals, T.P., and Sanders, P.M. (1993). Anther development: Basic principles and practical applications. Plant Cell 5, 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz, T., Bergey, D.R., and Ryan, C.A. (1997). A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant Physiol. 114, 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y., Wang, T.W., Hudak, K.A., Schade, F., Froese, C.D., and Thompson, J.E. (2000). An ethylene-induced cDNA encoding a lipase expressed at the onset of senescence. Proc. Natl. Acad. Sci. USA 97, 8717–8722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard, J., Block, M.A., Douce, R., Browse, J., Warwick, N., Somerville, C.R., and Slack, C.R. (1991). Molecular aspects of plastid envelope biochemistry. Eur. J. Biochem. 199, 489–509. [DOI] [PubMed] [Google Scholar]
- Kates, M., and Sastry, P.S. (1969). Phospholipase D. Methods Enzymol. 14, 197–203. [Google Scholar]
- Keijzer, C.J. (1987). The processes of anther dehiscence and pollen dispersal. I. The opening mechanism of longitudinally dehiscing anthers. New Phytol. 105, 487–498. [DOI] [PubMed] [Google Scholar]
- Keijzer, C.J., Hoek, I.H.S., and Willemse, T.M. (1987). The processes of anther dehiscence and pollen dispersal. III. The dehydration of the filament tip and the anther in three monocotyledonous species. New Phytol. 106, 281–287. [Google Scholar]
- Koda, Y., and Kikuta, Y. (1992). Wound-induced accumulation of jasmonic acid in tissues of potato tubers. Plant Cell Physiol. 35, 751–756. [Google Scholar]
- Laudert, D., Pfannschmidt, U., Lottspeich, F., Hollander-Czytko, H., and Weiler, E.W. (1996). Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol. Biol. 31, 323–335. [DOI] [PubMed] [Google Scholar]
- Lee, S., Suh, S., Kim, S., Crain, R.C., Kwak, J.M., Nam, H.-G., and Lee, Y. (1997). Systemic elevation of phosphatidic acid and lysophospholipid levels in wounded plants. Plant J. 12, 547–556. [Google Scholar]
- Matsui, K., Kurishita, S., Hisamitsu, A., and Kajiwara, T. (2000). A lipid-hydrolysing activity involved in hexenal formation. Biochem. Soc. Trans. 28, 857–860. [PubMed] [Google Scholar]
- Matsuoka, K., and Nakamura, K. (1991). Propeptide of a precursor to a plant vacuolar protein required for vacuolar targeting. Proc. Natl. Acad. Sci. USA 88, 834–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maucher, H., Hause, B., Feussner, I., Ziegler, J., and Wasternack, C. (2000). Allene oxide synthases of barley (Hordeum vulgare cv. Salome): Tissue specific regulation in seedling development. Plant J. 21, 199–213. [DOI] [PubMed] [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M., Creelman, R.A., Bell, E., Mullet, J.E., and Browse, J. (1997). Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 5473–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, M.J. (1997). Enzymes involved in jasmonic acid biosynthesis. Physiol. Plant. 100, 653–663. [Google Scholar]
- Mueller, M.J., Brodschelm, W., Spannagl, E., and Zenk, M.H. (1993). Signaling in the elicitation process is mediated through the octadecanoid pathway leading to jasmonic acid. Proc. Natl. Acad. Sci. USA 90, 7490–7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschietti, J., Dircks, L., Vancanneyt, G., and McCormick, S. (1994). LAT52 protein is essential for tomato pollen development: Pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 6, 321–338. [DOI] [PubMed] [Google Scholar]
- Nagai, Y., Aoki, J., Sato, T., Amano, K., Matsuda, Y., Arai, H., and Inoue, K. (1999). An alternative splicing form of phosphatidylserine-specific phospholipase A1 that exhibits lysophosphatidylserine-specific lysophospholipase activity in humans. J. Biol. Chem. 274, 11053–11059. [DOI] [PubMed] [Google Scholar]
- Nárvaez-Vásquez, J., Florin-Christensen, J., and Ryan, C.A. (1999). Positional specificity of a phospholipase A activity induced by wounding, systemin, and oligosaccharide elicitors in tomato leaves. Plant Cell 11, 2249–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, S., Mita, S., Hattori, T., and Nakamura, K. (1990). Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol. 31, 805–813. [Google Scholar]
- Sanders, P.M., Bui, A.Q., Weterings, K., McIntire, K.N., Hsu, Y.-C., Lee, P.Y., Truong, M.T., Beals, T.P., and Goldberg, R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322. [Google Scholar]
- Sanders, P.M., Lee, P.Y., Biesgen, C., Boone, J.D., Beals, T.P., Weiler, E.W., and Goldberg, R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12, 1041–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., Aoki, J., Nagai, Y., Dohmae, N., Takio, K., Doi, T., Arai, H., and Inoue, K. (1997). Serine phospholipid-specific phospholipase A that is secreted from activated platelets: A new member of the lipase family. J. Biol. Chem. 272, 2192–2198. [DOI] [PubMed] [Google Scholar]
- Schaller, F. (2001). Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J. Exp. Bot. 52, 11–23. [PubMed] [Google Scholar]
- Soldatova, L., Kochoumian, L., and King, T.P. (1993). Sequence similarity of a hornet (D. maculata) venom allergen phospholipase A1 with mammalian lipases. FEBS Lett. 320, 145–149. [DOI] [PubMed] [Google Scholar]
- Song, W.C., Funk, C.D., and Brash, A.R. (1993). Molecular cloning of an allene oxide synthase: A cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proc. Natl. Acad. Sci. USA 90, 8519–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, R., Truernit, E., Gahrtz, M., and Sauer, N. (1999). The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J. 19, 269–278. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97, 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J., Taylor, C., and Wang, T.-W. (2000). Altered membrane lipase expression delays leaf senescence. Biochem. Soc. Trans. 28, 775–777. [PubMed] [Google Scholar]
- Ueda, J., and Miyamoto, K. (1994). Separation of a new type of plant growth regulator, jasmonates, by chromatographic procedures. J. Chromatogr. A 658, 129–142. [Google Scholar]
- Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. [DOI] [PubMed] [Google Scholar]
- Wang, C., Zien, C.A., Afitlhile, M., Welti, R., Hildebrand, D.F., and Wang, X. (2000). Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 12, 2237–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, F.K., D'Arcy, A., and Hunziker, W. (1990). Structure of human pancreatic lipase. Nature 343, 771–774. [DOI] [PubMed] [Google Scholar]
- Woolley, P., and Petersen, S.B. (1994). Lipases, Their Structure, Biochemistry and Application. (Cambridge, UK: Cambridge University Press).
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Ziegler, J., Stenzel, I., Hause, B., Maucher, H., Hamberg, M., Grimm, R., Ganal, M., and Wasternack, C. (2000). Molecular cloning of allene oxide cyclase: The enzyme establishing the stereochemistry of octadecanoids and jasmonates. J. Biol. Chem. 275, 19132–19138. [DOI] [PubMed] [Google Scholar]