Abstract
Stamen and carpel identities are specified by the combinatorial activities of several floral homeotic genes, APETALA3, PISTILLATA, AGAMOUS (AG), SEPALLATA1 (SEP1), SEPALLATA2 (SEP2), and SEPALLATA3 (SEP3), all of which code for MADS domain DNA binding proteins. AG and the SEP genes also control floral determinacy. HUA1 and HUA2 were identified previously as regulators of stamen and carpel identities and floral determinacy because the recessive hua1-1 or hua2-1 allele affected these processes in plants with a lower dosage of functional AG (either homozygous for the weak ag-4 allele or heterozygous for the strong ag-1 allele). HUA2 was cloned previously and shown to code for a novel protein. We isolated the HUA1 gene using a map-based approach and show that it encodes a protein with six CCCH-type zinc finger motifs that is also found in yeast, Caenorhabditis elegans, Drosophila melanogaster, and mammalian proteins. Several such genes from invertebrates and mammals are known to play key regulatory roles in development. Therefore, HUA1 are another example of non–MADS domain proteins involved in organ identity specification. We demonstrated that HUA1 binds ribohomopolymers, preferentially poly rU and poly rG, but not double-stranded DNA in vitro. This finding suggests that HUA1, like several mammalian CCCH zinc finger proteins, is an RNA binding protein. Therefore, HUA1 likely participates in a new regulatory mechanism governing flower development.
INTRODUCTION
The identities of the four floral organ types in Arabidopsis (sepal, petal, stamen, and carpel) are specified by the combinatorial activities of several classes of homeotic genes (Coen and Meyerowitz, 1991; Meyerowitz et al., 1991; Pelaz et al., 2000). For example, stamen identity requires APETALA3 (AP3), PISTILLATA (PI), AGAMOUS (AG), and one of three SEPALLATA genes, SEPALLATA1, 2, or 3 (SEP1, SEP2, or SEP3). AG and the SEP genes determine carpel identity and, in addition, confer determinacy to the floral meristem. These floral homeotic genes code for MADS domain proteins (Yanofsky et al., 1990; Ma et al., 1991; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994; Mandel and Yanofsky, 1998). The combinatorial interaction among these genes may result from physical interaction among the floral homeotic proteins (Honma and Goto, 2001). Although it is likely that these MADS domain proteins act as transcription factors, it is unclear how these potential transcription factors specify organ identity at the molecular level. Little is known about other players in the homeotic pathways.
HUA1 and HUA2 were identified as regulators of stamen/carpel identities and floral determinacy in a sensitized screen in the ag-4 background (Chen and Meyerowitz, 1999). Although flowers of severe loss-of-function ag alleles (such as ag-1) show stamen-to-petal transformation in the third whorl (Bowman et al., 1989), flowers of the weak ag-4 allele contain stamens in the third whorl (Sieburth et al., 1995). Recessive hua1-1 and hua2-1 mutations alter the identity of the third whorl organs in ag-4 flowers. ag-4 hua1-1 or ag-4 hua2-1 flowers contain petaloid stamens in the third whorl. ag-4 hua1-1 hua2-1 flowers have petals in the third whorl. That HUA1 and HUA2 also play a role in carpel identity and floral determinacy is obvious in the ag-1/+ background: both carpel identity and floral determinacy are lost in ag-1/+ hua1-1 hua2-1 flowers but not in ag-1/+ flowers. HUA2 codes for a novel protein with a conserved domain found in mammalian growth factors and transcription coactivators (Chen and Meyerowitz, 1999).
To begin to understand the molecular function of HUA1, we isolated the HUA1 gene using a map-based approach. With the knowledge of the HUA1 sequence, we screened T-DNA insertion libraries and isolated another hua1 allele, hua1-2. We showed that hua1-2, like hua1-1, also caused stamen-to-petal transformation in ag-4 flowers, confirming the role of HUA1 in stamen identity specification. HUA1 belongs to a family of nine Arabidopsis genes containing multiple, tandem, CCCH-type zinc finger motifs. Proteins with similar zinc fingers are found in diverse eukaryotes from yeast to human. Several such proteins from Caenorhabditis elegans and mouse play regulatory roles in development. We demonstrated that HUA1 binds RNA and single- stranded DNA but not double-stranded DNA in vitro, suggesting that HUA1, like several mammalian CCCH zinc finger proteins, is an RNA binding protein.
RESULTS
Positional Cloning of HUA1
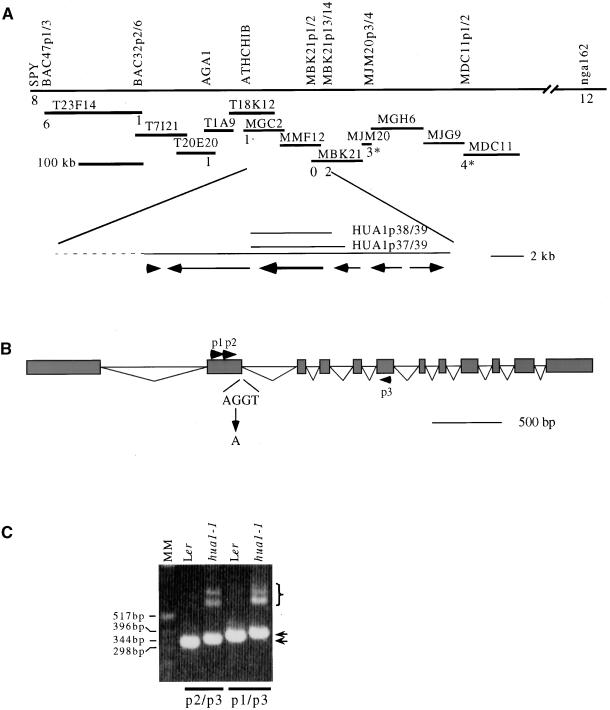
HUA1 was mapped in an 86-kb region between polymorphism markers ATHCHIB and MBK21p3/4 on chromosome III (see Methods; Figure 1A). Three candidate genes from this region were sequenced from Landsberg erecta (Ler) and hua1-1. A G-to-A mutation was found in one of the genes. The mutated nucleotide resides immediately next to a potential intron splice donor site (Figure 1B), changing the sequence next to the splice donor site from a preferred (AG) to a rare (AA) context in Arabidopsis genes (Brown et al., 1996). To determine whether this potential intron was in fact an intron and whether this mutation affected the splicing of this intron, we isolated cDNAs corresponding to this gene. The identity of the intron was confirmed by comparison of the cDNA and genomic sequences (Figure 1B). Reverse transcriptase–mediated polymerase chain reaction (RT-PCR) was performed using RNA from Ler and hua1-1 inflorescences with two sets of primers that flanked the intron (Figure 1B). Whereas a single PCR fragment of the expected size was obtained from Ler with each set of primers, several fragments of larger sizes were found in hua1-1 samples (Figure 1C). No products of the correct size were obtained from hua1-1. The p2/p3 fragment that was slightly larger than the corresponding Ler fragment was cloned and sequenced. A cryptic splice donor site 22 nucleotides downstream of the wild-type donor site apparently was used (data not shown). One of the much larger fragments also was cloned and sequenced and found to correspond to the inclusion of the entire intron (data not shown). These hua1-1 RNA species would result in premature translation termination in the polypeptide.
Figure 1.
Map-Based Cloning of HUA1.
(A) The chromosomal region containing HUA1 is represented by the top line with the cleaved-amplified polymorphic sequence/(CAPS) and simple sequence length polymorphism (SSLP) markers written above. The TAMU BAC (those starting with the letter T) and P1 (those starting with the letter M) clones are represented by the shorter, overlapping lines. The numbers below the corresponding markers indicate the numbers of recombination break points between these markers and HUA1 in 1448 chromosomes or 1038 chromosomes (numbers with asterisks). HUA1 was eventually mapped between ATHCHIB and MBK21p3/4. The potential genes in this region are indicated by arrows. A mutation is found in one of the genes (the thicker arrow) in hua1-1. Two genomic clones, HUA1p38/39 and HUA1p37/39, are used for plant transformation.
(B) The genomic organization of this gene, with exons represented by boxes and introns represented by triangles. The G-to-A mutation is at the end of the second exon at the exon-intron boundary. p1, p2, and p3 are three primers used in RT-PCR shown in (C).
(C) RT-PCR with p2 and p3 or p1 and p3 from Ler and hua1-1 samples. Although only one band is found in Ler samples, at least three bands are present in hua1-1 samples, one of which (indicated by arrows) is slightly larger than the corresponding Ler band. The larger products are indicated by a bracket. MM, molecular mass.
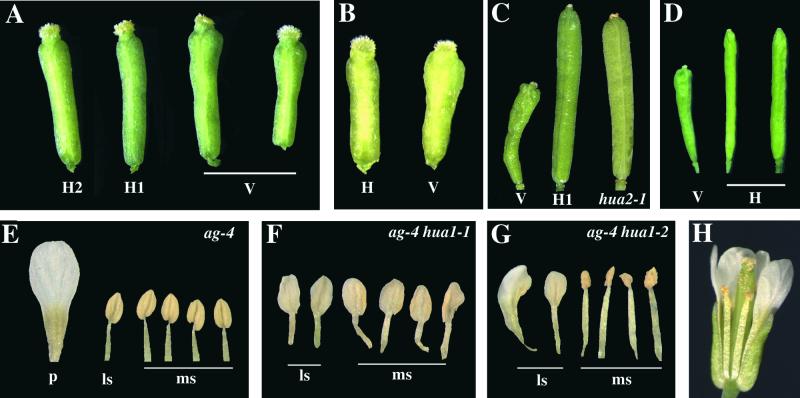
To confirm that this is the HUA1 gene, we transformed hua1-1 hua2-1 plants with two genomic clones that contained only this gene (Figure 1A). The two clones differed in the amount of the potential promoter region of the gene (1.3 kb versus 540 bp). In each case, ∼50% of the transgenic lines (more than 40 for each) showed phenotypes that indicated complete rescue, and the remaining 50% showed partial rescue or no rescue. None of the 30 transgenic plants containing the cloning vector alone showed any sign of rescue (Figures 2A and 2C). hua1-1 hua2-1 gynoecia are consistently (100%) enlarged in the top portion of the ovary (Chen and Meyerowitz, 1999) starting from stage 12 (stages according to Smyth et al. [1990]). Plants showing full rescue exhibited completely normal stage 12 gynoecia (Figure 2A), as found in wild-type plants. The phenotypic rescue was most evident in mature flowers. The rescued siliques were much longer than hua1-1 hua2-1 siliques at maturity and comparable in length to hua2-1 siliques (Figure 2C). Phenotypic rescue also was observed, although at a lower frequency, in hua1-1 hua2-1 plants transformed with 35S:: HUA1cDNA (data not shown). Therefore, we most likely cloned the HUA1 gene. However, it remains to be determined whether this gene rescues the floral homeotic phenotype of ag-4 hua1-1.
Figure 2.
Plant Phenotypes.
(A) to (D) Phenotypic rescue by HUA1 genomic or GFP-HUA1 clones in hua1-1 hua2-1.
(A) Stage 12 gynoecia of hua1-1 hua2-1 transgenic plants containing HUA1 genomic clones HUA1p38/39 (H2) or HUA1p37/39 (H1) (see Figure 1A) or the vector alone (V).
(B) Stage 12 gynoecia of hua1-1 hua2-1 transgenic plants containing 35S::GFP-HUA1 (H) or the vector alone (V).
(C) Mature siliques of hua1-1 hua2-1 transgenic plants containing vector alone (V) or HUA1p37/39 (H1) and a hua2-1 silique for comparison.
(D) Mature siliques of hua1-1 hua2-1 transgenic plants containing vector alone (V) or 35S::GFP-HUA1 (H).
(E) to (G) Dissected second and third whorl organs from ag-4, ag-4 hua1-1, and ag-4 hua1-2 flowers, respectively. p, petal; ls, lateral stamen; ms, medial stamen.
(H) A hua1-2 flower.
hua1-2
Previous genetic analyses of HUA1 were performed on the single hua1-1 allele. To test the conclusions from these analyses, we isolated another hua1 allele, hua1-2, from the Tom Jack T-DNA collection (Arabidopsis Stock Center, Ohio State University, Columbus) using a PCR-based approach. An ∼5-kb T-DNA was inserted in the second exon 24 bp upstream of the point mutation found in hua1-1 (data not shown).
We crossed hua1-2 to ag-4 to determine if hua1-2 affects organ identity in the third whorl of ag-4 flowers, as hua1-1 does. Indeed, ag-4 hua1-2 third whorl organs differed from those in ag-4 in that the lateral stamens were transformed to petals or petaloid stamens (cf. Figures 2G and 2E). The medial stamens appeared normal. This phenotype was consistent in all flowers of ag-4 hua1-2 plants. To rule out the possibility that this phenotype is attributable to differences in the genetic background of the two parental strains (hua1-2 in Columbia [Col] and ag-4 in Ler), we analyzed 55 ag-4 homozygous plants in the F2 population. Six were homozygous for hua1-2, whereas the others were hua1-2/+ or +/+. Only ag-4 hua1-2 plants exhibited the third whorl phenotype, suggesting that the homeotic transformation is caused by hua1-2 and that hua1-2 is recessive. hua1-2 seemed weaker than hua1-1, because all six stamens in ag-4 hua1-1 appeared petaloid (Figure 2F) (Chen and Meyerowitz, 1999). Nonetheless, this confirmed the role of HUA1 in stamen identity specification. hua1-2 hua2-1 double mutant flowers exhibited the same defects in the gynoecia as hua1-1 hua2-1 flowers (data not shown). Like hua1-1 flowers, single mutant hua1-2 flowers have no obvious phenotypes (Figure 2H).
The HUA1 Protein
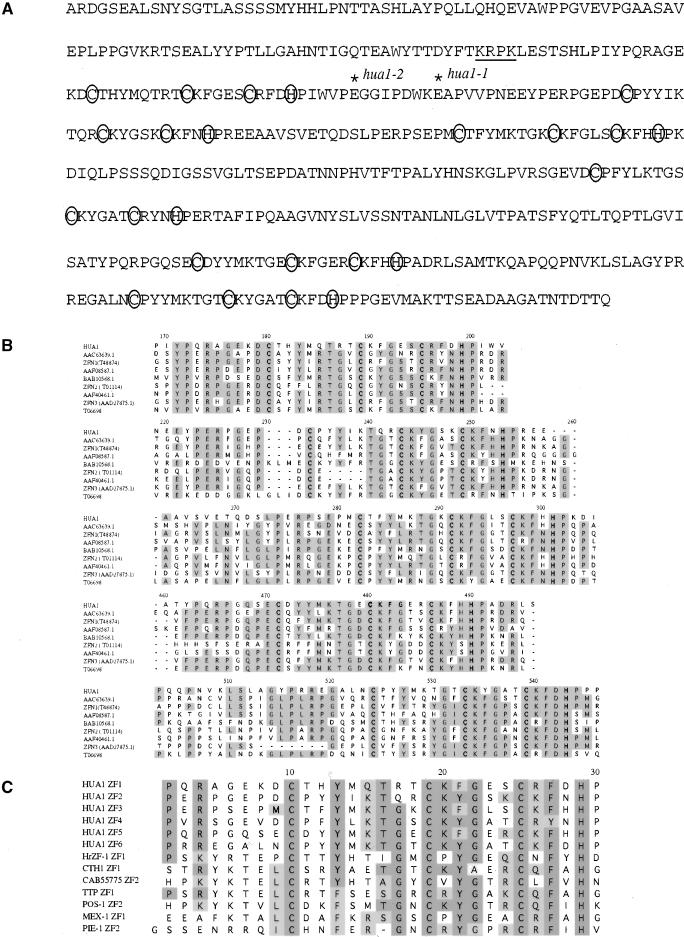
The most prominent feature of the 524–amino acid HUA1 protein is the presence of six tandem CCCH-type zinc finger motifs that span the C-terminal two-thirds of the protein (Figure 3A). A potential nuclear localization signal (KRPK) is found in the N-terminal one-third of the protein (Figure 3A). The hua1-1 mutation that results in aberrant splicing of the transcript presumably would cause translation termination after the first zinc finger motif.
Figure 3.
The HUA1 Protein.
(A) The HUA1 amino acid sequence with the critical CCCH zinc finger residues circled. The putative nuclear localization signal is underlined. The positions of the hua1-1 and hua1-2 mutations are indicated.
(B) A Clustal W alignment of the HUA1 family members at the corresponding zinc finger regions. GenBank accession numbers for the HUA1 paralogs are indicated. The cysteine and histidine residues of the zinc fingers are in boldface letters. Shaded letters represent identical residues.
(C) A Clustal W alignment of the six HUA1 zinc fingers with those from animal proteins. Identical and similar residues are shaded with darker and lighter gray, respectively. HrZF-1 ZF1, zinc finger 1 of HrZF1 (BAA81905) from Halocynthia roretzi; CTH1 ZF1, zinc finger 1 of CTH1 (CAA71245) from Cyprinus carpio; CAB55775 ZF2, zinc finger 2 of a zebrafish protein; TTP ZF1, POS-1 ZF2, MEX-1 ZF1, and PIE-1 ZF2 are the indicated zinc fingers from the mouse TTP (S04743) and the C. elegans POS-1 (T37246), MEX-1 (U81043), and PIE-1 (AAB17868) proteins, respectively.
HUA1 belongs to a family of nine Arabidopsis genes with highly similar, tandem, CCCH-type zinc finger motifs. The other members of the family include three previously cloned genes of unknown function, ZFN1, ZFN2, and ZFN3, and five genes predicted from the genome sequence (Figure 3B). These proteins all contain five tandem zinc fingers, whereas HUA1 has six. When these protein sequences are aligned, similarity is found only in the zinc finger regions, with HUA1 zinc fingers 1, 2, 3, 5, and 6 aligning with zinc fingers 1, 2, 3, 4, and 5 of the other proteins, respectively. The similarity is not confined to the cysteine and histidine residues of the zinc fingers but instead persists at least over a block of 30 amino acids. Interestingly, the six HUA1 zinc finger regions are less similar among themselves than they are to their counterparts in the paralogous proteins (cf. Figures 3B and 3C). Genes with zinc fingers of the same type and organization are found in other plant species such as rice and garden pea. There exist other Arabidopsis CCCH-type zinc finger proteins that do not belong to this family. One example is ZFWD1 (Terol et al., 2000), which contains a single CCCH-type zinc finger distantly related to the HUA1 zinc fingers (data not shown).
Besides plant proteins, BLAST searches revealed that many animal proteins contain similar zinc fingers, although the similarity between the HUA1 zinc fingers and the zinc fingers from these animal proteins is not as extensive as that among the HUA1 family members (Figure 3C). These proteins are found in diverse organisms such as Halocynthia roretzi, C. elegans, zebrafish, mouse, and human.
HUA1 RNA Is Found in All Major Plant Organs
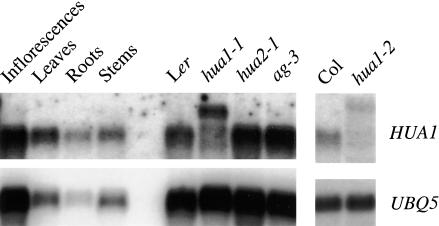
Among the known floral homeotic genes, those encoding MADS domain proteins are expressed primarily in flowers (Yanofsky et al., 1990; Ma et al., 1991; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994; Mandel and Yanofsky, 1998), whereas AP2 and HUA2 are expressed in nonfloral as well as floral tissue (Jofuku et al., 1994; Chen and Meyerowitz, 1999). We examined the expression of HUA1 with RNA filter hybridization. HUA1 RNA was found throughout the plant, in inflorescences, stems, leaves, and roots (Figure 4).
Figure 4.
HUA1 RNA Accumulation in Various Organs and in Inflorescences of Various Genotypes as Indicated.
UBIQUITIN5 (UBQ5) RNA was used to monitor the total amount of RNA in the samples. Ler is the wild-type control for hua1-1, hua2-1, and ag-3. Col is the wild-type control for hua1-2.
To determine if HUA1 expression is regulated by AG or HUA2, two other genes with similar floral homeotic functions, we examined HUA1 RNA abundance in wild-type, ag-3, and hua2-1 inflorescences. Comparable levels of HUA1 RNA were found in these genotypes (Figure 4), suggesting that HUA1 is not regulated at the transcript level by either AG or HUA2.
In hua1-1, the major RNA species was ∼400 nucleotides longer than the wild-type HUA1 RNA (Figure 4), suggesting that the second intron was retained in most hua1-1 RNA. This major hua1-1 transcript was detected at 12% of the abundance of wild-type HUA1 RNA. Other hua1-1 RNA species of smaller sizes and lower abundance also were detected (Figure 4). The prevalence of the RT-PCR products 22 bp longer than the wild-type products in Figure 1C likely was caused by preferential amplification by PCR. In hua1-2, a major transcript larger than the wild-type transcript was detected at 22% of the wild-type abundance (Figure 4).
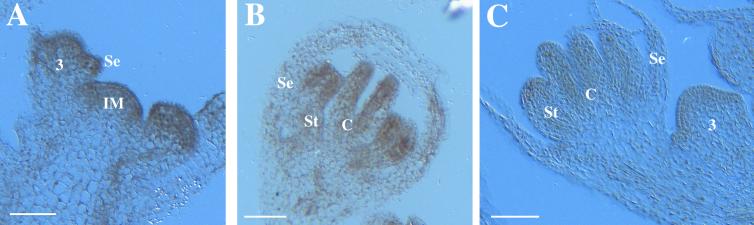
We also examined the localization of HUA1 transcript in wild-type inflorescences by in situ hybridization. Like HUA2 RNA, HUA1 RNA was detected with radioactive probes in all cells in the inflorescence meristem, the inflorescence stem, and flowers of all stages (data not shown). With nonradioactive probes that are less sensitive, HUA1 RNA was detected throughout the inflorescence meristem and young floral primordia (Figures 5A and 5C). In stage 7 and older flowers, HUA1 RNA was more concentrated in petals (data not shown), stamens, and carpels (Figures 5B and 5C).
Figure 5.
HUA1 RNA Localization in Wild-Type Inflorescences as Determined by in Situ Hybridization.
(A) and (B) Longitudinal sections of an inflorescence and a stage 8 to 9 flower, respectively, reacted with a HUA1 antisense probe.
(C) A longitudinal section of a stage 3 and a stage 7 flower reacted with a HUA1 sense probe.
Se, sepal; St, stamen; C, carpel; IM, inflorescence meristem; 3, a stage 3 flower. Bars = 50 μm.
HUA1 Is a Nuclear Protein
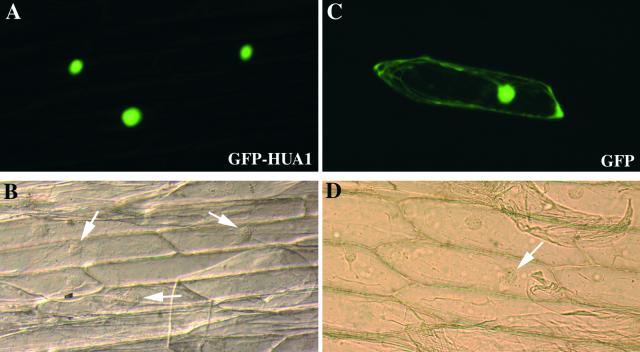
Because HUA1 has a poor potential nuclear localization signal and several C. elegans proteins with similar zinc fingers are found in the cytoplasm, we examined the subcellular localization of HUA1 by fusing the full-length cDNA to the green fluorescent protein (GFP) gene. The fusion protein was functional because it was able to rescue the hua1-1 hua2-1 mutant phenotype (Figures 2B and 2D). In onion epidermal cell transient expression assays, GFP-HUA1 was found to be in the nucleus, whereas GFP alone was present throughout the cell (Figure 6). The N-terminal one-third of the protein without the zinc fingers (HUA1NT) and the C-terminal two-thirds containing the zinc fingers (HUA1CT) also were fused separately to GFP. The GFP-HUA1NT fusion protein was found exclusively in the nucleus, and the GFP-HUA1CT fusion protein was present throughout the cell (data not shown), suggesting that the N-terminal one-third of the protein contains signals for nuclear import or protein degradation in the cytoplasm. GFP-HUA1 localization also was examined in the roots of transgenic Arabidopsis plants, where GFP fluorescence was detected only in the nuclei (data not shown).
Figure 6.
Onion Epidermal Cell Transient Expression Assay for HUA1 Subcellular Localization.
(A) and (B) Fluorescent and light field images, respectively, of cells expressing GFP-HUA1. Arrows indicate nuclei.
(C) and (D) Fluorescent and light field images, respectively, of cells expressing GFP. Arrows indicate nuclei.
Nucleic Acid Binding Properties of HUA1
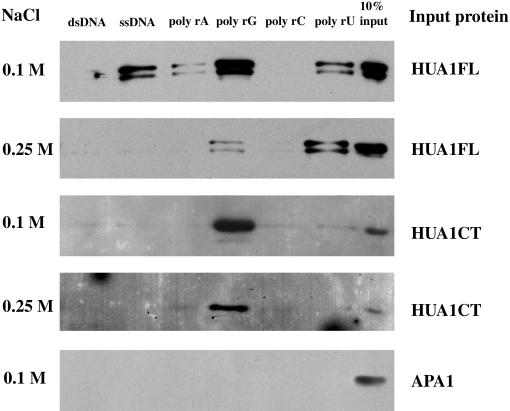
Many CCCH-type zinc finger proteins from animals have been shown to bind RNA and/or to be involved in post-transcriptional regulation (see Discussion). To begin to understand the molecular functions of HUA1, we examined the nucleic acid binding properties of HUA1 using recombinant HUA1 proteins purified from Escherichia coli. Under moderate salt concentrations (0.1 and 0.25 M) that resemble the in vivo situation, HUA1 was found to bind ribohomopolymers but not double-stranded DNA (Figure 7). Specifically, at 0.1 M NaCl, the full-length protein bound poly rG, poly rU, poly rA, and single-stranded DNA but not double-stranded DNA. At 0.25 M NaCl, the full-length protein bound only poly rU and poly rG. The binding to poly rU can be detected at salt concentrations as high as 0.5 M (data not shown). An N-terminally truncated HUA1 protein with only the zinc fingers (HUA1CT) bound only poly rG under the two salt concentrations. An unrelated protein (APA1; X. Chen and E.M. Meyerowitz, unpublished results) did not bind any nucleic acid under the same conditions. This finding suggests that HUA1 is an RNA binding protein and that the N-terminal portion of the protein affects the HUA1 RNA binding properties.
Figure 7.
In Vitro Nucleic Acid Binding by HUA1.
The full-length HUA1 protein (HUA1FL), an N-terminally truncated version that contains all six zinc fingers (HUA1CT), and an unrelated protein (APA1) were expressed as His-tagged proteins, purified, and incubated with various nucleic acids attached to agarose or cellulose beads. HUA1FL binds single-stranded DNA (ssDNA), poly rA, poly rG, and poly rU at 0.1 M NaCl and poly rG and poly rU at 0.25 M NaCl. HUA1CT binds poly rG under both conditions. APA1 does not bind any nucleic acid under these conditions. Ten percent of the input proteins is shown in the right lane. dsDNA, double-stranded DNA.
Expression of Floral Homeotic Genes in hua1-1 Flowers
The RNA localization patterns of AP1 and AG in hua1-1 hua2-1 flowers were reported previously (Chen and Meyerowitz, 1999). Those of the two B function genes, AP3 and PI, are similar in hua1-1 hua2-1 and wild-type flowers (X. Chen and E.M. Meyerowitz, unpublished results). We examined the RNA accumulation patterns of the SEP1, SEP2, and SEP3 genes, which were recognized recently as regulators of floral organ identities and potential partners of AG (Pelaz et al., 2000; Honma and Goto, 2001). The expression patterns of all three SEP genes in hua1-1 hua2-1 were similar to those in wild-type controls and to those reported previously (data not shown; Ma et al., 1991; Mandel and Yanofsky, 1998), suggesting that the HUA genes do not regulate the expression domains of the SEP genes.
Because HUA1 is a nuclear RNA binding protein, it is conceivable that HUA1 regulates AG at the level of RNA splicing. We performed RT-PCR to determine if partially spliced AG RNAs could be detected in hua1-1 but not wild-type inflorescences. No difference was found between the hua1-1 and wild-type samples (data not shown).
DISCUSSION
Biological Role of HUA1
HUA1 was identified initially as a gene involved in conferring reproductive organ identities as well as floral determinacy in Arabidopsis. Although these proposed roles were based on analyses of the single hua1-1 allele, the fact that hua1-2 and hua1-1 show similar effects in the ag-4 and hua2-1 backgrounds suggests that the previously analyzed hua1-1 defects likely reflect the true biological functions of HUA1.
In the flower, available genetic evidence suggests that HUA1 acts only in the third and fourth whorls. However, HUA1 RNA is clearly detected in all four floral whorls in early flowers and is concentrated in the inner three whorls in later flowers. Translational or post-translational processes theoretically can account for the whorl-specific activities of HUA1. Alternatively, HUA1 also plays a role in the perianth organs, a role not recognized because of the presence of one or more genes with overlapping functions. It is worth noting that the HUA1 expression patterns in the flower resemble those of SEP1, 2, or 3, although the functional significance of this, if any, remains to be determined.
HUA1 also regulates plant size (Chen and Meyerowitz, 1999). Although hua1-1 or hua2-1 single mutants show no obvious phenotypes, hua1-1 hua2-1 plants are shorter and smaller than are wild-type plants. Further backcrosses are necessary to determine if hua1-2 hua2-1 plants exhibit similar phenotypes, because the two alleles are from two ecotypes that differ in size (Ler and Col). The fact that HUA1 is expressed in stems and leaves is consistent with its proposed role in vegetative development. Both HUA1 and HUA2 (Chen and Meyerowitz, 1999) are expressed in the root. No obvious root defects have been observed with either the hua1-1 or hua2-1 single mutant or the double mutant on sucrose-containing plates (T. Western and X. Chen, unpublished results).
HUA1 Belongs to a Gene Family
HUA1 belongs to a family of nine genes in Arabidopsis that contain tandem CCCH-type zinc fingers. Genes of this family also are found in other higher plants. Not all Arabidopsis genes containing CCCH-type zinc fingers belong to this family. This family of genes is unique in that the CCCH-type zinc fingers are found in tandem and the corresponding zinc fingers among genes are more similar than the zinc fingers in any one gene. HUA1 is the only gene of this family with a known biological function. Proteins with similar zinc fingers are found in invertebrates and vertebrates. However, the degree of similarity is much less than that among HUA1 family members, and most of these animal proteins contain only two tandem zinc fingers.
Despite the established function of HUA1 in conferring reproductive organ identity and floral determinacy in flower development, neither hua1-1 nor hua1-2 single mutant flowers exhibit any floral defects. It is likely that neither hua1 nor hua2 is a null allele (transcript found in both) and that the lack of floral phenotypes is attributable to the residual activities of the mutant proteins. Alternatively, the lack of floral phenotypes in the hua1 single mutants can be attributable mainly to the presence of other genes with overlapping functions. Given that examples of structurally related genes with redundant or similar functions exist (e.g., SEP1, SEP2, and SEP3 [Pelaz et al., 2000]; SCARECROW and SHORT ROOT [Di Laurenzio et al., 1996; Helariutta et al., 2000]), it is tempting to postulate that some of the HUA1 family members share overlapping roles with HUA1 in flower development. This hypothesis can be tested by the loss-of-function phenotypes of these genes.
Potential Molecular Functions of HUA1
The presence of the CCCH-type zinc fingers in HUA1 provides some hints to the potential molecular functions of HUA1. Although no conclusive evidence exists for CCCH-type zinc fingers being involved in DNA binding, many proteins with this type of zinc finger have been shown to bind RNA or to be associated with RNA metabolism. The murine TTP protein (Figure 3C) regulates the expression of tumor necrosis factor α (TNF-α) at the post-transcriptional level. TTP binds to an AU-rich sequence in the 3′ untranslated region of the TNF-α mRNA and destabilizes it (Carballo et al., 1998). Both zinc fingers in TTP are required for this RNA–protein interaction (Lai et al., 1999). Three other mammalian proteins and one Xenopus laevis protein with double zinc fingers also were shown to bind to TNF-α RNA (Lai et al., 2000). In C. elegans, three genes encoding proteins with double zinc fingers of the CCCH type (Figure 3C), Pie-1, Mex-1, and Pos-1, specify the identity of germline blastomeres in early embryonic development with distinct mechanisms. Pie-1 is a bifunctional protein that acts in the nucleus to repress germline transcription and in the cytoplasm to regulate the expression of nanos-2 RNA at the post-transcriptional level (Tenenhaus et al., 2001). The two zinc fingers in the protein are not required for transcription repression, but the second zinc finger is required for the cytoplasmic function of PIE-1 (Tenenhaus et al., 2001). Mex-1 acts to restrict the localization of PIE-1 (Guedes and Priess, 1997). Pos-1 appears to regulate the translation of maternal RNAs in the germline blastomeres (Tabara et al., 1999). All three proteins are known to be associated with large, germline-specific RNA–protein complexes known as the P granules in the cytoplasm (Mello et al., 1996; Guedes and Priess, 1997; Tabara et al., 1999). In addition, several yeast, Drosophila melanogaster, and mammalian proteins with CCCH fingers have been demonstrated to function in splicing, polyadenylation, or RNA cleavage (Zhang et al., 1992; Bai and Tolias, 1996; Barabino et al., 1997).
The fact that HUA1 binds poly rG, poly rU, and single- stranded DNA in vitro suggests that HUA1 is an RNA binding protein. This and the nuclear localization of HUA1 point to a potential role in nuclear RNA metabolism, such as RNA processing, RNA stability, or RNA export.
Molecular Function of HUA1 in the AG Pathway
The previously isolated floral homeotic genes all seem to code for potential transcription factors. AP1, AP3, PI, AG, and the SEP proteins contain the MADS domain known to bind to DNA (Huang et al., 1993; Shiraishi et al., 1993). AP2 contains another DNA binding domain, the AP2 domain (Weigel, 1995). HUA2 does not have any known DNA binding domain, but it has been shown to possess transcription activation activity in yeast (X. Chen, unpublished results). Two other genes that play a role in carpel development, CRABS CLAW (CRC) and SPATULA (SPT), both encode proteins with canonical DNA binding motifs (Bowman and Smyth, 1999; Heisler et al., 2001). Although we cannot exclude the possibility that HUA1 also functions in transcriptional regulation, HUA1 likely functions in post-transcriptional processes at the RNA level.
How would HUA1, a nuclear, potential RNA binding protein, act in the floral homeotic AG pathway? One hypothesis is that HUA1 promotes the expression of AG at the post-transcriptional level, such as through RNA processing (capping, splicing, and polyadenylation), RNA stability, or RNA export from the nucleus. The hua1-1 mutation does not affect the spatial arrangement of AG RNA in flowers (Chen and Meyerowitz, 1999). Protein gel blot experiments with dissected inflorescences containing stage 6 and younger flowers showed that hua1-1 does not affect AG protein levels in these young flowers (Chen and Meyerowitz, 1999). However, it is possible that HUA1 acts to maintain AG expression in later floral stages. Although no aberrantly spliced forms of AG RNA can be detected in hua1-1 flowers, it is possible that these forms are unstable and rapidly degraded. Therefore, it is still possible that HUA1 regulates AG expression at the level of RNA splicing. A second hypothesis is that HUA1 acts together with AG in regulating the expression of downstream targets. It is well documented that transcription and RNA processing are closely coordinated (Ladomery, 1997; Proudfoot, 2000). It is possible that HUA1 acts as an RNA processing factor that works on AG target gene transcripts.
In addition, many genes known to play a role in the development of the gynoecium may be regulated by HUA1 or serve as regulators of HUA1. Examples are CRC (Alvarez and Smyth, 1999; Bowman and Smyth, 1999), SPT (Alvarez and Smyth, 1999; Heisler et al., 2001), ETTIN (Sessions and Zambryski, 1995), SHATTERPROOF1 and -2 (Liljegren et al., 2000), FRUITFUL (Férrandiz et al., 2000), LEUNIG (Liu et al., 2000), and AINTEGUMENTA (Liu et al., 2000). The relationships between HUA1 and these genes are unknown at present.
METHODS
Strains and Transgenic Lines
The mutant strains used in this study, ag-3 (Bowman et al., 1991), ag-4 (Sieburth et al., 1995), hua1-1 (Chen and Meyerowitz, 1999), hua2-1 (Chen and Meyerowitz, 1999), and hua1-1 hua2-1, are in the Landsberg erecta (Ler) background of Arabidopsis thaliana. hua1-2, isolated in this study, is in the Columbia (Col) background. Plants were grown at 23°C in Pro-mix BX (a peat-based growing medium from Premier Horticulture Inc., Red Hill, PA) under 16-hr-light/8-hr-dark cycles.
hua1-2 was isolated from the Tom Jack T-DNA lines using a polymerase chain reaction (PCR) screening approach. A HUA1-specific primer (HUA1p24; 5′-aaggcatcaaactttttgagtcctccttt-3′) was used in combination with a T-DNA border primer (JL-202; 5′-cattttataataacgctgcggacatctac-3′) to amplify from pooled DNA obtained from the Arabidopsis Stock Center (Ohio State University). The PCR product was confirmed by DNA filter hybridization and sequencing. An individual homozygous line was identified from one of the subpools.
hua1-1 hua2-1 plants were transformed with HUA1 genomic DNA, 35S::HUA1cDNA, and 35S::GFP-HUA1 fusion constructs by the vacuum infiltration method (Bechtold et al., 1993). Transgenic seedlings were selected on AT medium (Haughn and Somerville, 1986) containing 50 μg/mL kanamycin and transferred to soil. Floral phenotypes were recorded with a Fuji (Tokyo, Japan) HC300-Z digital camera.
Positional Cloning of HUA1
The mapping population was derived from the cross ag-4/+ hua1-1 hua2-1 × Col. In the F2 population of this cross, ag-4/+ hua1-1 hua2-1 and ag-4 hua1-1 hua2-1 plants were chosen based on their floral phenotypes. The HUA1 mapping population consisted of 724 such plants, among which 529 were used previously for the positional cloning of HUA2.
DNA was isolated from each F2 plant either by the hexadecyl-trimethyl-ammonium bromide method (Reiter et al., 1992) or by a simpler preparation method (Edwards et al., 1991). HUA1 was first mapped to the top arm of chromosome III close to simple sequence length polymorphism (SSLP) markers nga162 and ATHCHIB and to a cleaved-amplified polymorphic sequence (CAPS) marker in a gene named AGA1 (X. Chen and E.M. Meyerowitz, unpublished results). AGA1 was used as a probe to screen Texas A&M University (TAMU) bacterial artificial chromosome (BAC) filters. One of the clones that hybridized to AGA1, T20E20, was sequenced from the ends. The end sequence was used to screen for overlapping BACs until eventually a contig of five TAMU BACs that overlapped with the Kazusa P1 contig in this region (see Figure 1) was established. CAPS markers were generated by sequencing the BAC or P1 ends in Ler and Col (in cases in which the Col sequence was not yet available from the Arabidopsis Genome Initiative) backgrounds. Eventually, HUA1 was mapped between markers ATHCHIB and MBK21p3/4. Information on the CAPS markers shown in Figure 1 as well as additional markers developed in this study can be viewed at the Chen laboratory World Wide Web site (http://waksman.rutgers.edu/~xuemei).
HUA1 genomic DNA for mutant rescue was amplified with HUA1p39 (5′ BglII-aaggcatcaaactttttgagtcctccttt 3′) in combination with either HUA1p37 (5′ BglII-cacaaaagcttctctgagt 3′) or HUA1p38 (5′ BglII-ttgctgactccatttttggtc 3′). The resulting 5.7- and 5-kb fragments, respectively, were cloned separately in the BamHI site of the plant transformation vector pPZP211 (Hajdukiewicz et al., 1994). The full-length HUA1 cDNA (see below) was amplified with HUA1p34 (5′ XbaI-gtcaaatcaagttttatggatc 3′) and HUA1p6 (5′ XbaI-tcattgagtagtgtcggtgttg 3′), cloned into pCGN1547 (McBride and Summerfelt, 1990) containing the 35S promoter of the Cauliflower mosaic virus and the nopaline synthase 3′ end, and sequenced.
HUA1 cDNA
The Weigel cDNA library was screened with a HUA1 genomic DNA probe. Six cDNA clones were obtained from 500,000 plaques screened. One cDNA clone, HUA1cDNA6, was full-length but contained a one-nucleotide deletion in the coding region. 5′ rapid amplification of cDNA ends (Gibco BRL) was performed according to the manufacturer's instructions to obtain a 650-bp fragment corresponding to the 5′ one-third of the HUA1 RNA. This fragment was cloned into pCR-Blunt (Invitrogen, Carlsbad, CA), and the resulting plasmid, pCR-MBKp46/50, was sequenced. A 350-bp AgeI-AccI fragment from pCR-MBKp46/50 was used to replace the corresponding fragment in HUA1cDNA6 to repair the one-nucleotide-deletion defect.
RNA Filter and in Situ Hybridization
Total RNA from various organs and genotypes was isolated using the Tri-reagent (Molecular Research Center, Cincinnati, OH). Poly(A+) RNA was isolated using a Rapid mRNA Purification Kit (Amresco, Solon, OH). Approximately 2 μg of poly(A+) RNA was separated on a 1.2% formaldehyde/agarose gel, transferred to a nylon membrane, and probed with 32P-labeled PCR fragments corresponding to either the full-length or the 3′ half of the HUA1 cDNA. Hybridization signals were quantified with a phosphorimager. The UBIQUITIN5 gene was used as a control for the amount of RNA used.
In situ hybridization with radioactive probes was performed as described (Chen and Meyerowitz, 1999). In situ hybridization with digoxigenin-labeled probes was performed according to www.wisc.edu/genetics/CATG/barton/protocols.html, except that UTP and digoxigenin-UTP were used at a 3:7 ratio in probe synthesis. A 1.55-kb fragment was amplified from HUA1 cDNA using primers HUA1p19 (5′ BglII-atggcacatcgtcaattgt 3′) and HUA1p6 and cloned into pCR-Blunt (Invitrogen). Two plasmids, pHUA1p19/6A and pHUA1p19/6S, with different orientations of the insert, were obtained. These two plasmids were digested with SpeI and transcribed with T7 RNA polymerase to generate the antisense and the sense probe for HUA1, respectively.
HUA1 Protein Localization
The full-length HUA1 coding region was amplified with Pwo polymerase (Roche Molecular Biochemicals, Indianapolis, IN) using primers HUA1p19 and HUA1p6 and cloned into pAVA321 (von Arnim et al., 1998), resulting in a fusion of green fluorescent protein (GFP) to the N terminus of HUA1 (pAVA321-HUA1). The 35S::GFP-HUA1 cassette was excised with SacI and KpnI and cloned into pPZP211 for plant transformation.
Transient expression assays were performed as described (von Arnim and Deng, 1994). Either pAVA321 or pAVA321-HUA1 DNA was delivered into onion epidermal cells by bombardment with the Bio-Rad Biolistic PDS-1000/He system. After bombardment, the epidermal peels were cultured at 22°C in darkness for 16 hr and observed under a Zeiss (Jena, Germany) Axioplan Universal transmitted light microscope. Images were captured with a digital 1X HRD100-N/K camera from Diagnostic Instruments (Sterling Heights, MI).
In Vitro Nucleic Acid Binding Assay
The full-length version and an N-terminally truncated version of the HUA1 gene were cloned into the pRSETA protein expression vector (Invitrogen). An unrelated gene, APA1, also was cloned into the same vector (X. Chen and E.M. Meyerowitz, unpublished results). The 6His-tagged proteins were purified from Escherichia coli using QIAexpressionist (Qiagen, Valencia, CA).
In vitro nucleic acid binding assays were performed as described (Yang et al., 1998). Five hundred nanograms of purified 6His-tagged HUA1 full-length protein, an N-terminally truncated HUA1 protein, and an unrelated protein (APA1) were incubated with 20 μL of poly rA, poly rG, poly rC, and poly rU attached to agarose beads and double-stranded and single-stranded calf thymus DNA attached to cellulose beads (Sigma) in 500 μL of RHPA binding buffer (10 mM Tris, pH 7.4, 2.5 mM MgCl2, 0.5% Triton X-100, and NaCl at various concentrations) with 1 mg/mL heparin. The concentrations of the nucleic acids were 0.35 to 0.7 mg/mL (poly rA), 0.25 to 1 mg/mL (poly rC), 1.5 to 4.5 mg/mL (poly rG), 0.1 to 1 mg/mL (poly rU), 0.75 to 1.25 mg/mL (single-stranded DNA), and 0.75 to 2 mg/mL (double-stranded DNA). After incubation at 4°C for 10 min, the beads were washed five times in RHPA buffer and then boiled in SDS loading buffer. The proteins were separated by SDS-PAGE and detected with a monoclonal anti-His antibody (Pharmacia).
GenBank Accession Number
The HUA1 cDNA sequence is in GenBank under accession number AY024357.
Acknowledgments
We are grateful to Dr. Elliot Meyerowitz for his support and advice on the initial efforts toward the map-based cloning of HUA1. We thank Dr. Zhongchi Liu for providing cloning vectors, Drs. Zhongchi Liu and Doris Wagner for technical advice, Dr. Tamara Western for discovering the one-nucleotide deletion in one of the HUA1 cDNAs, Dr. Pal Maliga for providing the gene gun, and Dr. Dan Klessig for helpful discussions. We thank Dr. Hugo Dooner, Dr. Tamara Western, Dr. Jun Liu, and Yulan Cheng for their comments on the manuscript. The isolation of hua1-2 was facilitated by the distribution of pooled DNA and seed stocks from the Arabidopsis Stock Center. This work was supported by National Institutes of Health Grant GM61146-02 to X.C.
References
- Alvarez, J., and Smyth, D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386. [DOI] [PubMed] [Google Scholar]
- Bai, C., and Tolias, P.P. (1996). Cleavage of RNA hairpins mediated by a developmentally regulated CCCH zinc finger protein. Mol. Cell. Biol. 16, 6661–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino, S.M.L., Hübner, W., Jenny, A., Minvielle-Sebastia, L., and Keller, W. (1997). The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 11, 1703–1716. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [Google Scholar]
- Bowman, J.L., and Smyth, D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Brown, J.W., Smith, P., and Simpson, C.G. (1996). Arabidopsis consensus intron sequences. Plant Mol. Biol. 32, 531–535. [DOI] [PubMed] [Google Scholar]
- Carballo, E., Lai, W.S., and Blackshear, P.J. (1998). Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Chen, X., and Meyerowitz, E.M. (1999). HUA1 and HUA2 are two members of the floral homeotic AGAMOUS pathway. Mol. Cell 3, 349–360. [DOI] [PubMed] [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio, L., Wysocka-Diller, J., Malamy, J.E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M.G., Feldmann, K.A., and Benfey, P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Férrandiz, C., Liljegren, S.J., and Yanofsky, M.F. (2000). FRUITFULL negatively regulates the SHATTERPROOF genes during Arabidopsis development. Science 289, 436–438. [DOI] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Guedes, S., and Priess, J.R. (1997). The C. elegans MEX-1 protein is present in germline blastomeres and is a P granule component. Development 124, 731–739. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Haughn, G., and Somerville, C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204, 430–434. [Google Scholar]
- Heisler, M.G., Atkinson, A., Bylstra, Y.H., Walsh, R., and Smyth, D.R. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Helariutta, Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., Hauser, M.T., and Benfey, P.N. (2000). The SHORT ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. [DOI] [PubMed] [Google Scholar]
- Huang, H., Mizukami, Y., Hu, Y., and Ma, H. (1993). Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 21, 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D., den Boer, B.G.W., Montagu, M.V., and Okamuro, J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladomery, M. (1997). Multifunctional proteins suggest connections between transcriptional and post-transcriptional processes. Bioessays 19, 903–909. [DOI] [PubMed] [Google Scholar]
- Lai, W.S., Carballo, E., Strum, J.R., Kennington, E.A., Phillips, R.S., and Blackshear, P.J. (1999). Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19, 4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, W.S., Carballo, E., Thorn, J.M., Kennington, E.A., and Blackshear, P.J. (2000). Interactions of CCCH zinc finger proteins with mRNA. J. Biol. Chem. 275, 17827–17837. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Franks, R.G., and Klink, V.P. (2000). Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell 12, 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H., Yanofsky, M.F., and Meyerowitz, E.M. (1991). AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 5, 484–495. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1998). The Arabidopsis AGL9 MADS-box gene is expressed in young flower primordia. Sex. Plant Reprod. 11, 22–28. [Google Scholar]
- Mandel, M.A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. [DOI] [PubMed] [Google Scholar]
- McBride, K.E., and Summerfelt, K.R. (1990). Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 14, 269–276. [DOI] [PubMed] [Google Scholar]
- Mello, C.C., Schubert, C., Draper, B., Zhang, W., Lobel, R., and Priess, J.R. (1996). The PIE-1 protein and germline specification in C. elegans embryos. Nature 382, 710–712. [DOI] [PubMed] [Google Scholar]
- Meyerowitz, E.M., Bowman, J.L., Brockman, L.L., Drews, G.N., Jack, T., Sieburth, L.E., and Weigel, D. (1991). A genetic and molecular model for flower development in Arabidopsis thaliana. Development S1, 157–167. [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N. (2000). Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 25, 290–293. [DOI] [PubMed] [Google Scholar]
- Reiter, R.S., Young, R.M., and Scolnik, P.A. (1992). Genetic linkage of the Arabidopsis genome: Methods for mapping with recombinant inbreds and random amplified polymorphic DNAs (RAPDs). In Methods in Arabidopsis Research, C. Koncz, N.-H. Chua, and J. Schell, eds (Singapore: World Scientific Publishing), pp. 177–178.
- Sessions, R.A., and Zambryski, P.C. (1995). Arabidopsis gynoecium structure in the wild-type and in ettin mutants. Development 121, 1519–1532. [DOI] [PubMed] [Google Scholar]
- Shiraishi, H., Okada, K., and Shimura, Y. (1993). Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 4, 385–398. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., Running, M.P., and Meyerowitz, E.M. (1995). Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell 7, 1249–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara, H., Hill, R.J., Mello, C.C., Priess, J.R., and Kohara, Y. (1999). pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126, 1–11. [DOI] [PubMed] [Google Scholar]
- Tenenhaus, C., Subramaniam, K., Dunn, M.A., and Seydoux, G. (2001). PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev. 15, 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terol, J., Bargues, M., and Pérez-Alonso, M. (2000). ZFWD: A novel subfamily of plant proteins containing a C3H zinc finger and seven WD40 repeats. Gene 260, 45–53. [DOI] [PubMed] [Google Scholar]
- von Arnim, A.G., and Deng, X.-W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045. [DOI] [PubMed] [Google Scholar]
- von Arnim, A.G., Deng, X.-W., and Stacey, M.G. (1998). Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43. [DOI] [PubMed] [Google Scholar]
- Weigel, D. (1995). The APETALA2 domain is related to a novel type of DNA-binding domain. Plant Cell 7, 388–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.Y., Yin, G.L., and Darnell, R.B. (1998). The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc. Natl. Acad. Sci. USA 95, 13254–13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]
- Zhang, M., Zamore, P.D., Carmo-Fonesca, M., Lamond, A.I., and Green, M.R. (1992). Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc. Natl. Acad. Sci. USA 89, 8769–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]