Abstract
We analyzed cDNA libraries from developing endosperm of the B73 maize inbred line to evaluate the expression of storage protein genes. This study showed that zeins are by far the most highly expressed genes in the endosperm, but we found an inverse relationship between the number of zein genes and the relative amount of specific mRNAs. Although α-zeins are encoded by large multigene families, only a few of these genes are transcribed at high or detectable levels. In contrast, relatively small gene families encode the γ- and δ-zeins, and members of these gene families, especially the γ-zeins, are highly expressed. Knowledge of expressed storage protein genes allowed the development of DNA and antibody probes that distinguish between closely related gene family members. Using in situ hybridization, we found differences in the temporal and spatial expression of the α-, γ-, and δ-zein gene families, which provides evidence that γ-zeins are synthesized throughout the endosperm before α- and δ-zeins. This observation is consistent with earlier studies that suggested that γ-zeins play an important role in prolamin protein body assembly. Analysis of endosperm cDNAs also revealed several previously unidentified proteins, including a 50-kD γ-zein, an 18-kD α-globulin, and a legumin-related protein. Immunolocalization of the 50-kD γ-zein showed this protein to be located at the surface of prolamin-containing protein bodies, similar to other γ-zeins. The 18-kD α-globulin, however, is deposited in novel, vacuole-like organelles that were not described previously in maize endosperm.
INTRODUCTION
Genes encoding storage proteins are highly expressed in developing seed, and this has greatly facilitated their molecular cloning and characterization. As a result, we know much about the structural relationships between these proteins and the mechanisms by which storage proteins are synthesized (Herman and Larkins, 1999; Shewry and Casey, 1999). Traditionally, storage proteins were identified on the basis of their solubility in water or aqueous solvents containing salt, alcohol, and acid or alkali, and this led to their classification as albumins, globulins, prolamins, and glutelins, respectively. Proteins in these solubility classes occur in every seed, but major types predominate in certain plant families. For example, legume seed contain primarily storage globulins, whereas cereal seed contain primarily prolamins.
As the molecular structure of storage proteins was deduced from DNA sequences, our understanding of their evolutionary relationships improved immensely. However, this information created the challenge of developing a method that systematically identifies them in a context that fits both their structure and their solubility characteristics. Today, three major types of storage proteins are recognized: the family of 2S albumins, the 7S and 11S families of storage globulins, and the family of alcohol-soluble prolamins (Shewry and Casey, 1999). Other types of proteins (e.g., lectins, protease inhibitors, thionins, etc.) often are abundant in seed, but presumably they have functions other than amino acid storage, so they are not conventionally considered storage proteins (Shewry and Casey, 1999).
Although maize kernels contain albumins, globulins, prolamins, and glutelins (Landry and Moureaux, 1970), two types of storage proteins predominate in the seed: the embryo contains a 7S globulin, similar to that found in dicot embryos (Kriz, 1999), and the endosperm, the major site of storage protein accumulation, contains predominantly prolamins, the so-called zein fraction (Landry and Moureaux, 1970; Wilson, 1983). As is true of all prolamins, proteins in the zein fraction share the property of solubility in 70% ethanol, but they are structurally distinct. Classification of the various zein proteins was confounded by their differential solubility in aqueous solvents and by the ability of several proteins to form disulfide bonds (Wilson, 1983, 1985). Eventually, a widely accepted nomenclature system was developed that classified these proteins on the basis of their solubility and structural relationships as α-, β-, γ-, and δ-zeins (Esen, 1987; Coleman and Larkins, 1998).
Zeins accumulate as accretions, called protein bodies, that form within the lumen of the rough endoplasmic reticulum (RER) (Lending and Larkins, 1989). The smallest zein-containing protein bodies, which are observed in subaleurone cells, consist primarily of β- and γ-zeins. Larger protein bodies found in subaleurone and starchy endosperm cells contain, in addition to β- and γ-zeins, mainly α- and some δ-zeins. It appears that as storage protein synthesis progresses, the α- and δ-zein proteins penetrate the matrix of β- and γ-zeins, expanding the protein body into a spherical structure that reaches a diameter of 1 to 2 μm. Some evidence suggests the γ- and β-zein proteins play a role in α- and δ-zein retention in the RER (Coleman et al., 1996; Bagga et al., 1997), although the mechanisms by which this occurs have not been investigated.
α-Zeins were among the first storage protein genes to be described (Wienand et al., 1981; Pedersen et al., 1982). Early studies showed that α-zeins are encoded by a large multigene family, although estimates of its size and complexity varied significantly (Wilson and Larkins, 1984). On the basis of DNA and RNA renaturation kinetics, Viotti et al. (1979) concluded that there are at least 15 non-cross-hybridizing zein sequences and 110 to 130 genes. Additional evidence supporting the large size and complexity of the gene families encoding these proteins came from the heterogeneity observed among zeins after separation by isoelectric focusing and SDS-PAGE. In contrast, Pedersen et al. (1980) determined that there are only two to four distinct zein mRNAs expressed in maize endosperm, each containing some unique and some repetitive nucleotide sequences and encoded by one to five gene copies. DNA gel blot hybridization provided evidence for a large number of α-zein genes in the maize genome, with estimates ranging from 75 to 100 coding sequences (Hagen and Rubenstein, 1981; Wilson and Larkins, 1984). The subsequent isolation and characterization of cDNA sequences from endosperm libraries provided evidence for the expression of multiple α-zein genes, but not nearly as many as were implied by the number of coding sequences in the genome (Marks et al., 1985a, 1985b). Because of the conflicting results and conclusions of these early studies, a number of questions remain unresolved regarding the nature and expression of α-zein genes in maize endosperm.
In contrast to the α-zeins, genes encoding the 15-kD β-zein (Pedersen et al., 1986), the 27- and 16-kD γ-zeins (Prat et al., 1987), and the 10- and 18-kD δ-zeins (Kirihara et al., 1988; Chui and Falco, 1995) occur in only one or two copies in the genome. The evolutionary and physiological significance of the difference in the number of these genes, compared with α-zeins, is unknown. It has been demonstrated that a number of α-zein genes contain mutations that could affect their expression, suggesting that many of them are pseudogenes (Spena et al., 1983; Kridl et al., 1984). Indeed, based on nuclear run-on transcription assays, the γ-zein genes are much more highly expressed than are the α-zein genes, in spite of the greater quantity of the latter (Kodrzycki et al., 1989; Or et al., 1993).
Nuclear run-on transcription assays showed expression of the various types of zein genes to be coordinately regulated during endosperm development and to begin ∼10 days after pollination (DAP) (Kodrzycki et al., 1989). Using in situ hybridization techniques, Dolfini et al. (1992) showed evidence for striking spatial differences in the expression of 19- and 22-kD α-zein genes and the 27-kD γ-zein gene. The significance of these observations for gene regulation and protein body formation was not investigated further.
In recent years, the development of extensive maize cDNA libraries, along with computer software to systematically characterize them, has made it possible to analyze the genes expressed in developing maize endosperm more thoroughly. We used this information to identify storage protein gene transcripts and to reassess the complexity of zein gene families and their expression in maize endosperm. Here, we report an analysis of zein gene complexity in the B73 maize inbred line and the development of gene-specific and antibody-specific probes that allowed us to investigate the pattern of storage protein gene expression in developing endosperm. This analysis revealed that, whereas there are multiple α-zein coding sequences in the genome, a relatively small number of them are expressed in the endosperm. Using in situ hybridization, we demonstrated differences in the temporal and spatial expression among some zein gene families. The results extend previous observations describing the temporal accumulation of zeins in protein bodies and imply a functional role for γ-zeins in protein body initiation.
Our analysis of endosperm expressed sequence tag (EST) libraries identified three seed protein genes that were not reported previously. These include a novel 50-kD γ-zein protein, an α-globulin protein similar to one found in rice endosperm, and a protein with characteristics of a legume 11S globulin. The results of these studies provide a comprehensive assessment of the expression of storage protein genes in maize endosperm.
RESULTS
Abundance and Classification of Endosperm Storage Protein Gene cDNAs in EST Databases
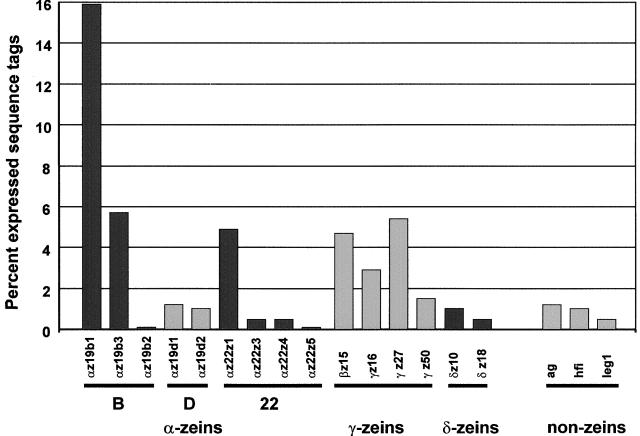
Cluster analysis of ESTs from cDNA libraries is a powerful tool to estimate the diversity and percentage of unique transcripts in mRNA populations. To estimate the abundance of individual RNA transcripts, a cluster analysis restricted to ESTs from nonnormalized libraries of the B73 inbred line was performed (see Methods). The longest clones, representing a unique cluster of endosperm-specific sequences, were obtained from these libraries and sequenced in their entirety. This identified 15 different, complete coding sequences that were grouped into the α-, γ-, and δ-zein gene subfamilies of seed proteins and three abundant, endosperm-specific, non-zein seed protein sequences (Figure 1). Most of these sequences have been described previously (Marks et al., 1985b; Coleman and Larkins, 1998), but the 50-kD γ-zein, the 18-kD α-globulin, and legumin 1 are novel gene products. The percentage of these unique endosperm sequences was estimated by tallying the numbers of clustered ESTs of the corresponding contig and comparing that number to the total number of ESTs in the B73 nonnormalized endosperm libraries. Although each of these cDNA sequences contributed >0.1% of the clones, their abundance varied widely. As expected (Shen et al., 1994), zein-specific cDNA sequences accounted for nearly 50% of the clones, with α-zeins (∼30%) and γ-zeins (∼15%) representing the most abundant transcripts.
Figure 1.
Storage Protein Transcript Levels in B73 Endosperm as Reflected by Their Percentage among Randomly Chosen Clones from 15- to 40-DAP Endosperm-Specific cDNA Libraries.
For abbreviations, see Table 1.
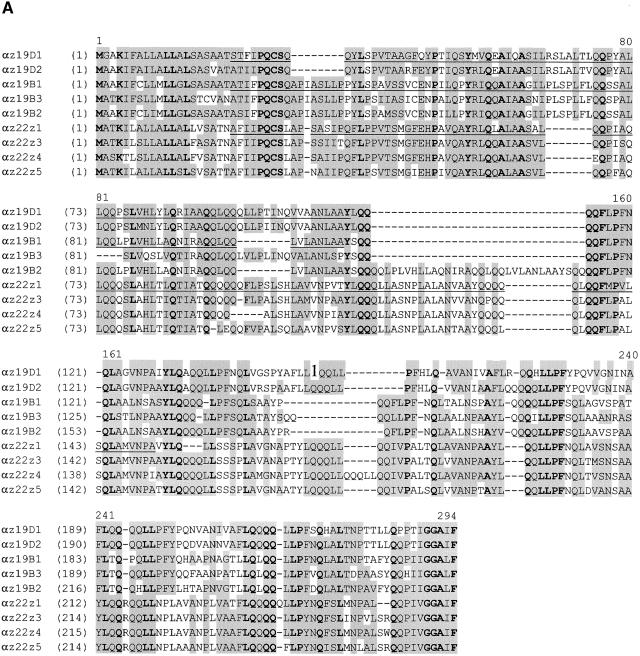
Of the five 19-kD α-zein transcripts detected, most ESTs were derived from only two genes, the 19-kD B1 and the 19-kD B3 α-zeins. Similarly, the 22-kD Z1 α-zein, a nonmutant allele of the floury2 gene (Coleman et al., 1997), accounted for most of the 22-kD α-zein transcripts. Figure 2 shows an alignment and dendrogram of deduced amino acid sequences of the nine expressed members of the α-zein gene family. Although it is common to divide α-zeins into two groups based on their SDS-PAGE migration (Coleman and Larkins, 1998), this sequence alignment clearly reveals a division into three subgroups: 22-kD α-zeins and the B and D subfamilies of 19-kD α-zeins. Family members within each protein subgroup share 75 to 95% amino acid identity, whereas between subgroups only 40 to 55% of the residues are conserved. These differences provided the basis to develop specific antibodies for proteins in each subgroup. The underlined residues in Figure 2 indicate partial peptide sequences that were produced in bacteria and used for this purpose.
Figure 2.
Amino Acid Sequence Alignment of Members of the Maize α-Zein Protein Family.
(A) Polypeptides correspond to alleles of the 22-kD α-zeins and the B and D subfamilies of 19-kD α-zeins in the B73 inbred line (see Table 1 for GenBank accession numbers and abbreviations). Identical residues are shown in boldface, and similar amino acids and conservative changes are shaded. Underlined residues designate peptide sequences that were expressed in bacteria and used to develop subfamily-specific antibodies (see Methods).
(B) Dendrogram showing an alignment of α-zein polypeptides that correspond to W64A alleles (GenBank accession numbers M12143 [A1], M12146 [C1], M12144 [D1], and M12141 [22C]) and to B73 alleles (Table 1). The bootstrapping values of all internal branches exceeded 95% (data not shown), strongly supporting the correct topology of the dendrogram. The branches of the three α-zein subfamilies are indicated. The nomenclatures of Heidecker and Messing (1986) and Rubenstein and Geraghty (1986) are shown in parentheses. Family members within the three subgroups share 75 to 95% identical amino acid residues, and there is 40 to 55% identity between residues in subgroups.
The genetic map positions of several 19- and 22-kD α-zeins were reported (Soave et al., 1981; Soave and Salamini, 1984; Heidecker and Messing, 1986), but the map position of the D α-zein gene subfamily was unknown. Consequently, we mapped the two genes to the long arm of chromosome 1 by restriction fragment length polymorphism mapping (Table 1).
Table1.
Abundant Endosperm-Specific Protein cDNA (Inbred Line B73)
| Name (Abbreviation) | GenBank Accession Number | Map Loci (Pioneer Composite Map) | Percent EST in Developing B73 Endosperm cDNA Libraries | Mature Peptide (Amino Acid Residues) | Molecular Weight (Calculated) | Molecular Mass (SDS-PAGE) |
Antigenic Rabbit Polypeptide (Relative to CDS) |
|---|---|---|---|---|---|---|---|
| 27-kD γ-zein (gz27) | AF371261 | c7L, position 147, Bin 7.05a | 5.4 | 204 | 21,822 | 27 kD | 73–127,* be |
| 16-kD γ-zein (gz16) | AF371262 | c2L, position 128.1, Bin 2.07b | 2.9 | 163 | 17,663 | 16 kD | 38-58, sp |
| 50-kD γ-zein (gz50) | AF371263 | c7L, position 70.5, Bin 7.05b | 1.5 | 278 | 32,882 | 50 kD | 21-120, be |
| 15-kD β-zein (bz15) | AF371264 | c6S, position 28.9, Bin 6.01c | 4.7 | 160 | 17,458 | 15 kD | 53-77, sp |
| 18-kD δ-zein (dz18) | AF371265 | c6L, position 84, Bin 6.04d | 1.0 | 190 | 21,220 | 18 kD | 158-182, sp |
| 10-kD δ-zein (dz10) | AF371266 | c9L, position 67.6, Bin 9.03e | 0.5 | 129 | 14,431 | 10 kD | 98-121, sp |
| 19-kD α-zein D1 (az19D1)f | AF371267 | c1L, position 123.3, Bin 1.06b | 1.2 | 222 | 24,818 | 19 kD | 21-110, be |
| 19-kD α-zein D2 (az19D2) | AF371268 | c1L, position 122.4, Bin 1.06b | 1.0 | 220 | 24,706 | 19 kD | NA |
| 19-kD α-zein B1 (az19B1) | AF371269 | 4L, 7Sg | 15.9 | 213 | 23,359 | 19 kD | 21-110,** be |
| 19-kD α-zein B2 (az19B2) | AF371270 | 4L, 7Sg | 0.1 | 246 | 27,128 | 22 kD | NA |
| 19-kD α-zein B3 (az19B3) | AF371271 | c4L, c7S, c10Lh | 5.7 | 219 | 24,087 | 19 kD | NA |
| 19-kD α-zein B5 (az19B5) | AF371272 | 4L, 7Sg | 0.05 | Truncated cDNA | NA | NA | NA |
| 19-kD α-zein B4 (az19B4) | AF371273 | 4L, 7Sg | 0.06 | In-frame stop codon | NA | NA | NA |
| 22-kD α-zein Z1 (az22z1) | AF371274 | c4S, position 27.3, Bin 4.02i | 4.9 | 242 | 26,359 | 22 kD | 21-151, be |
| 22-kD α-zein Z3 (az22z3) | AF371275 | c4S, position 27.3, Bin 4.02i | 0.5 | 245 | 26,751 | 22 kD | NA |
| 22-kD α-zein Z4 (az22z4) | AF371276 | c4S, position 27.3, Bin 4.02i | 0.5 | 246 | 26,923 | 22 kD | NA |
| 22-kD α-zein Z5 (az22z5) | AF371277 | c4S, position 27.3, Bin 4.02i | 0.1 | 245 | 26,701 | 22 kD | NA |
| 18-kD α-globulin (ag) | AF371278 | c6L, position 99.5, Bin 6.05b | 1.2 | 184 | 20,299 | 18 kD | 46-206, be |
| 50-kD legumin 1 (leg1) | AF371279 | c6S, position 31.9, Bin 6.01b | 0.5 | 447 | 49,317 | 50 kD | 37-483, be |
| Hageman factor inhibitor (hfi) | AF371280 | NA | 1.0 | 127 | 13,578 | NA | NA |
Rabbit and chicken.
Rabbit and rat. NA, not applicable; be, bacterially expressed; sp, synthetic peptide.
a Burr et al., 1988; Lopes et al., 1995.
b This paper.
c Weerakoon et al., 1993.
d Swarup et al., 1995.
e Benner et al., 1989.
f Zein subfamily z1D/SF3 (Heidecker and Messing, 1986; Rubenstein and Geraghty, 1986).
g Zein subfamily z1B/SF2 (Heidecker and Messing, 1986; Rubenstein and Geraghty, 1986).
h Zein subfamily z1A/SF1 (Heidecker and Messing, 1986; Rubenstein and Geraghty, 1986).
i Zein subfamily z1C/SF4 (Llaca and Messing, 1998; Rubenstein and Geraghty, 1986).
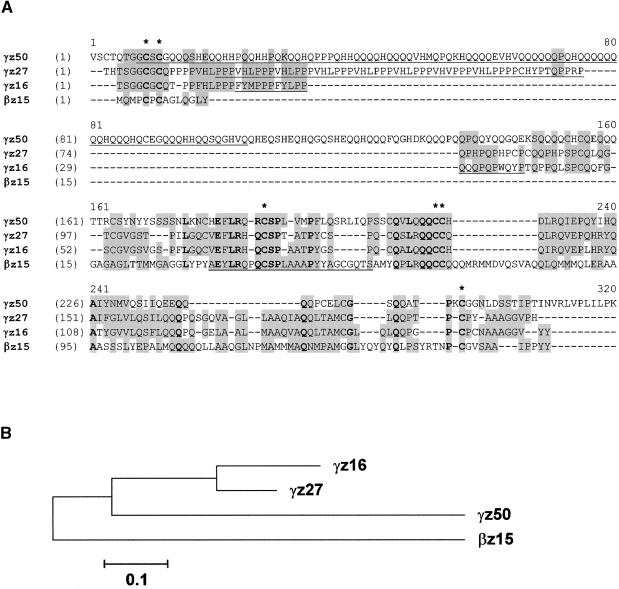
In contrast to the α-zeins, the γ-zein gene family is not complex and is represented by a relatively large number of ESTs. The 27-kD γ-zein and the 15-kD β-zein each contribute ∼5% of the endosperm transcripts, followed by the 16- and 50-kD γ-zeins, with ∼3 and 1.5%, respectively (Figure 1). These levels of expression were corroborated by in situ hybridization (see below). The amino acid alignment and dendrogram of the γ-zein protein family are shown in Figure 3. The 50-kD γ-zein was not described previously. Interestingly, it contains seven Lys residues (2.7% by weight), making it unique among zein proteins, which are devoid of this essential amino acid. We named the 50-kD γ-zein protein according to the convention to identify zeins based on their relative migration during SDS-PAGE (see below), but its predicted molecular mass is ∼33 kD (Table 1).
Figure 3.
Amino Acid Sequence Alignment of Members of the Maize γ-Zein Protein Family.
(A) The polypeptides correspond to alleles of the 16-, 27-, and 50-kD γ-zeins and the 15-kD β-zein in the B73 inbred line (for GenBank accession numbers and abbreviations, see Table 1). Conserved Cys residues are indicated with asterisks. Identical residues are shown in boldface, and similar amino acids and conservative changes are shaded. Underlined residues indicate peptide sequences that were either expressed in bacteria or synthesized chemically and used to develop monospecific antibodies (see Methods).
(B) Dendrogram of the alignment shown in (A). The bootstrapping values of all internal branches exceeded 95% (data not shown).
Alleles of the 50-kD γ-zein gene from different inbred lines are highly conserved. Compared with the cDNA from B73, only one polymorphism, a 3-bp deletion at positions 409 to 411 of the nucleotide sequence, was detected in the corresponding cDNA from the Mo17 inbred line. This deletion was used to map the 50-kD γ-zein gene by pyrosequencing (Nyren et al., 1993) of DNA of a Mo17 × B73 F2 population (Burr and Burr, 1991) to the long arm of chromosome 7 (Table 1). The map position of the 16-kD γ-zein was determined by restriction fragment length polymorphism mapping to be on the long arm of chromosome 2 (Table 1).
All γ-zeins share six highly conserved Cys residues as well as some other conserved polypeptide domains (Figure 3). These features led to the classification of the 15-kD β-zein as a member of the γ-zein family. Although its name does not accurately reflect this relationship, we retained the β-zein designation for historical reasons. Both the 27- and 50-kD γ-zeins contain a large block of multiple tandem repeats at the N terminus. This block is missing in the 15-kD β- and 16-kD γ-zeins, which consequently are of lower molecular weight. The block of repeats in the 27-kD γ-zein containing the hexapeptide PPPVHL is remarkably different from the much larger block of polyglutamine repeats in the 50-kD γ-zein. These differences in amino acid sequence allowed us to develop specific antibodies against these otherwise similar proteins. For this purpose, polypeptides (Figure 3, underlined) were either expressed in bacteria or synthesized chemically and used for immunization.
The sulfur amino acid–rich δ-zeins (Kirihara et al., 1988; Chui and Falco, 1995; Swarup et al., 1995) are encoded by only two genes and have the least abundant RNA transcripts among zeins (Figure 1). As shown by the alignment in Figure 4, the 18-kD δ-zein differs from the 10-kD δ-zein by a highly repetitive and Met-rich 53–amino acid insertion in the central part of the molecule. Otherwise, the amino acid sequences are very similar to each other at the N and C termini. Figure 4 also compares the amino acid sequences of the alleles of the 18-kD δ-zeins in Mo17 and B73, which differ markedly (10%) from each other. Specific antibodies were raised with synthetic polypeptides against the region least conserved between the 10- and 18-kD δ-zeins from the inbred line B73 (underlined in Figure 4). Interestingly, the B73 18-kD δ-zein antibody did not cross-react with the 18-kD δ-zein protein from Mo17, indicating very high specificity (data not shown).
Figure 4.
Amino Acid Sequence Alignment of Members of the Maize δ-Zein Protein Family.
The polypeptides correspond to the 18- and 10-kD δ-zein alleles of the B73 inbred line (see Table 1 for GenBank accession numbers and abbreviations) and the δ-zein allele of the Mo17 inbred line (Swarup et al., 1995; GenBank accession number U31541). Identical residues are shown in boldface, and conservative changes are shaded. Underlined residues indicate peptide sequences that were synthesized chemically and used to develop specific antibodies (see Methods).
Only three endosperm-specific non-zein seed protein genes were identified in the analysis of endosperm cDNA libraries, and they represent 0.5 to 1.5% of the clones (Figure 1). The Hageman factor inhibitor is an endosperm-specific protease inhibitor, and the 18-kD α-globulin and legumin 1 are proteins with close homologs in other plants, including other cereals (see below). With the exception of eEF1-α (GenBank accession number AF136823), α-1 tubulin (X73980), and enolase (P26301), no single gene with a nonstorage function contributed >0.1% of the endosperm cDNAs.
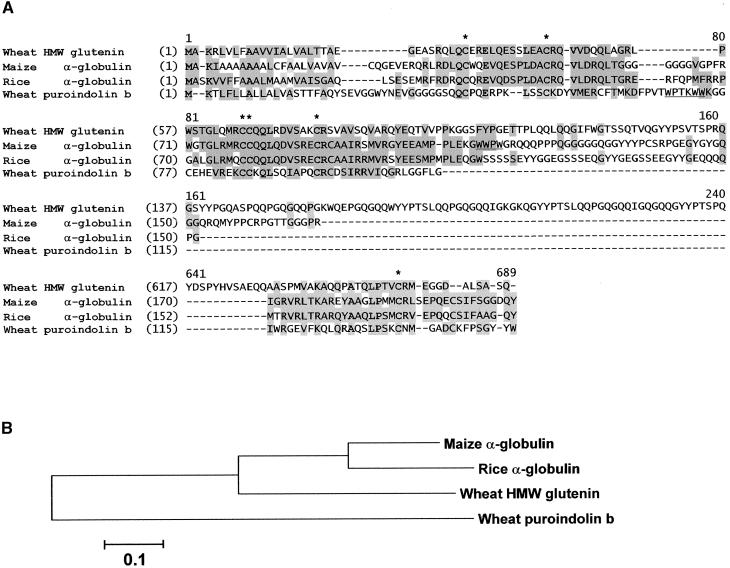
Based on the cDNA database analysis, the 18-kD α-globulin appears to be the only member of this protein family in maize. DNA gel blot analysis also identified only one gene (data not shown). The Mo17 and B73 alleles of the 18-kD α-globulin are polymorphic, and this enabled the gene to be mapped by pyrosequencing (Nyren et al., 1993) of DNA from a Mo17 × B73 F2 population to the long arm of chromosome 6 (Table 1). Diverse homologs of the 18-kD α-globulin can be recognized in other cereal species, as shown in the sequence alignment presented in Figure 5.
Figure 5.
Amino Acid Sequence Alignment of the 18-kD α-Globulin and Related Protein Sequences.
(A) Conserved Cys residues are indicated with asterisks. Identical residues are shown in boldface, and similar amino acids and conservative changes are shaded. The “Trp box” in wheat puroindolin and a similar Trp-rich sequence in the maize 18-kD α-globulin are underlined. The unique wheat HMW glutenin repeat domain between residues 217 and 616 was omitted for brevity. GenBank accession numbers are as follows: maize α-globulin, AF371278; wheat HMW glutenin, P08488; rice α-globulin, D50643; and wheat puroindolin b, X69912.
(B) Dendrogram of the alignment shown in (A). The bootstrapping values of all internal branches exceeded 95% (data not shown).
Identification of Maize Storage Proteins in Endosperm Extracts
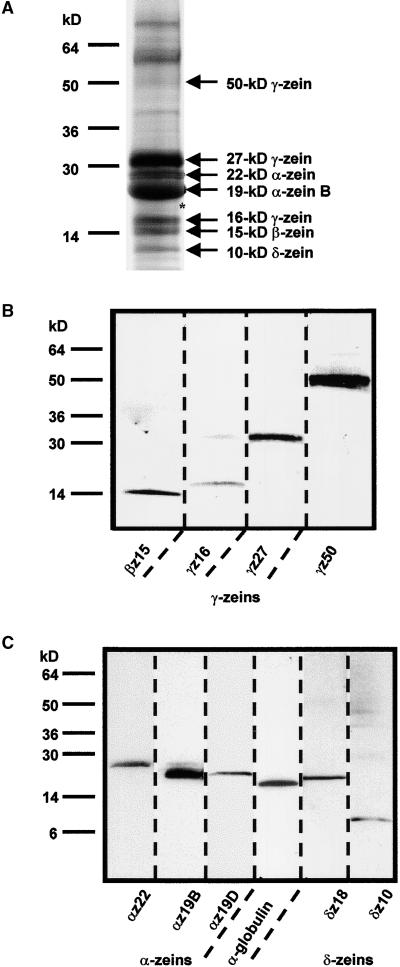
The analysis of endosperm storage protein genes made it possible to develop specific antibodies that could distinguish between members of each family of proteins (see above and Table 1) and, consequently, provided tools to identify them accurately in SDS-PAGE separations of endosperm proteins. Figure 6A shows a Coomassie blue–stained gel, and Figures 6B and 6C show the immunodetection of blotted polypeptide bands by monospecific antibodies. The α-zeins migrated as expected in two major bands at 19 and 22 kD and constituted ∼20 and 5%, respectively, of the SDS-extractable protein. The antibody against the 19-kD B α-zein subfamily binds, in addition to the 19-kD band, a faint band at 22 kD (Figure 6C, lane 2). The latter protein probably corresponds to the polypeptide encoded by the 19-kD B2 α-zein gene (Figure 2), which, in contrast to other 19kD α-zein polypeptides, contains a 33–amino acid insertion in the central part of its sequence. The 22-kD α-zein antibody labels a single protein band at 22 kD, and the antibody against the 19-kD α-zein D subfamily binds a protein that migrates slightly faster than does the B subfamily of α-zein polypeptides (Figure 6C, lanes 1 to 3).
Figure 6.
Immunodetection of Proteins in Maize Endosperm Extracts.
(A) Total protein from B73 endosperm was separated by 4 to 20% gradient SDS-PAGE and stained with Coomassie blue. Positions of molecular mass markers are indicated at left. Proteins that were clearly identified with monospecific polyclonal antibodies are specified at right. The asterisk indicates the approximate position of the 19-kD α-zein D, the 18-kD δ-zein, and the 18-kD α-globulin polypeptides, all of which migrated to a similar position.
(B) and (C) Immunoblots of the protein extract in (A) separated by 4 to 20% gradient SDS-PAGE (B) and by 10 to 20% SDS-PAGE (C). Identical lanes were cut from the blots and incubated with the designated monospecific antibody (for abbreviations, see Table 1).
Among the γ-zeins, the strongest Coomassie blue–stained band is the 27-kD γ-zein, which represents an estimated 15 to 20% of the SDS-extractable endosperm protein. The next most abundant proteins are the 15-kD β- and 16-kD γ-zeins. Least abundant, but clearly discernible, is the 50-kD γ-zein (Figures 6A and 6B). The relative mobility of this protein differs markedly from its calculated molecular mass of ∼33 kD (Table 1). The 10-kD δ-zein is a moderately abundant polypeptide that is clearly detectable after Coomassie blue staining. The 18-kD δ-zein and the 18-kD α-globulin both migrate slightly ahead of the 19-kD α-zeins (Figure 6C, lanes 4 and 5). However, both are minor polypeptides and are possibly obscured by the α-zeins. They are not clearly discernible in SDS-PAGE gels after Coomassie blue staining (Figure 6A, asterisk).
Storage Protein Genes Have Different Spatial and Temporal Patterns of Expression during Endosperm Development
We examined the spatial and temporal expression of the 50-kD γ-zein and 18-kD α-globulin genes relative to previously described classes of zein genes (Dolfini et al., 1992). Longitudinal sections of W64A kernels were prepared at various stages of development and hybridized with antisense RNA probes. To minimize variation between kernels used for these experiments, two or three comparable serial sections of kernels at the same developmental stage were used for hybridization with sense and antisense probes for each of the genes examined. The sense probes revealed no background hybridization and therefore are not illustrated, because the embryo and pericarp in each section provided an internal negative control. The alkaline phosphatase (AP) reaction was allowed to proceed to the point of nonspecific color reaction in the pericarp and embryo.
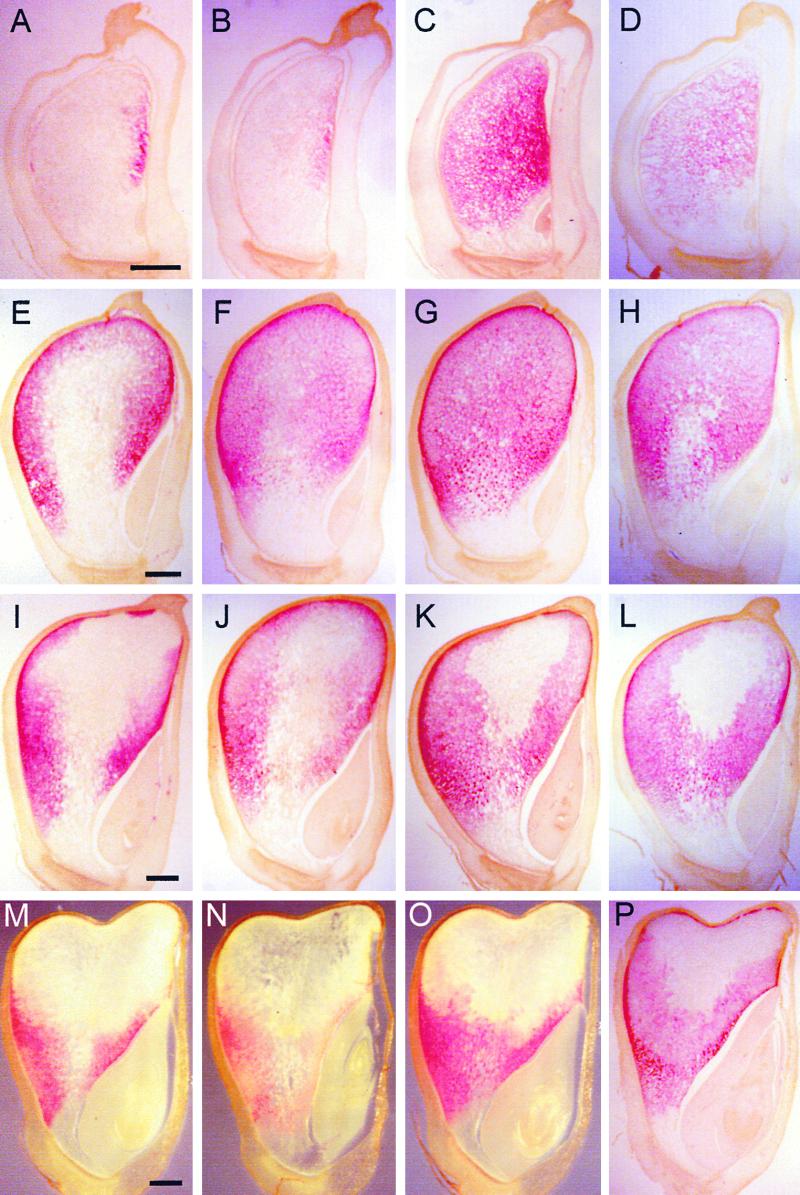
Figures 7 and 8 show typical images of in situ hybridization for eight different seed protein genes at four developmental stages. We examined a D subfamily (19-kD α-zein D1) and two B subfamily (19-kD α-zein B1 and B3) genes, the latter of which correspond to the W64A A/B1/B2 and C1 sequences, respectively (Marks et al., 1985a). Because the spatial and temporal expression of these genes were identical but the level of the 19-kD B1 α-zein transcript was greater than that of B3 and D1, only the results of the 19-kD B1 α-zein are presented. At 10 DAP, transcripts encoding the 22-kD α-zeins appeared in a narrow vertical stripe on the adgerminal side of the endosperm. The pattern of 19-kD B1 α-zein transcript distribution is nearly identical to that of the 22-kD α-zeins at this stage, suggesting that this pattern generally is characteristic of α-zein gene expression (Figures 7A and 7B). By 15 DAP, 22-kD α-zein transcripts were detected in most areas except the central and basal regions of the endosperm. The intensity of the signal was greater in the peripheral than in the central region of the endosperm (Figure 7E). At this stage, expression of the 19-kD B1 α-zein was greatest at the periphery of the endosperm, although it was detected in the central, starchy endosperm cells as well. At 20 DAP, the transcripts of 22-kD α-zeins were confined mainly to the peripheral regions of the endosperm, whereas they were restricted to the peripheral regions of the lower half of the endosperm at 25 DAP (Figures 7I and 7M). The 19-kD B1 α-zein transcripts were found throughout most regions of the 20-DAP endosperm, but their spatial distribution pattern was similar to that of 22-kD α-zein at 25 DAP (Figures 7J and 7N).
Figure 7.
Detection of mRNAs in Maize Endosperm by in situ Hybridization of Antisense Probes.
Longitudinal median sections of maize kernels at 10 ([A] to [D]), 15 ([E] to [H]), 20 ([I] to [L]), and 25 DAP ([M] to [P]) were hybridized with antisense RNA probes of 22-kD α-zein ([A], [E], [I], and [M]), 19-kD B1 α-zein ([B], [F], [J], and [N]), 27-kD γ-zein ([C], [G], [K], and [O]), and 16-kD γ-zein ([D], [H], [L], and [P]). Most sections for a particular developmental stage were derived serially from one or two kernels. Embryos are found at the lower right corners of the sections. Bars = 1 mm.
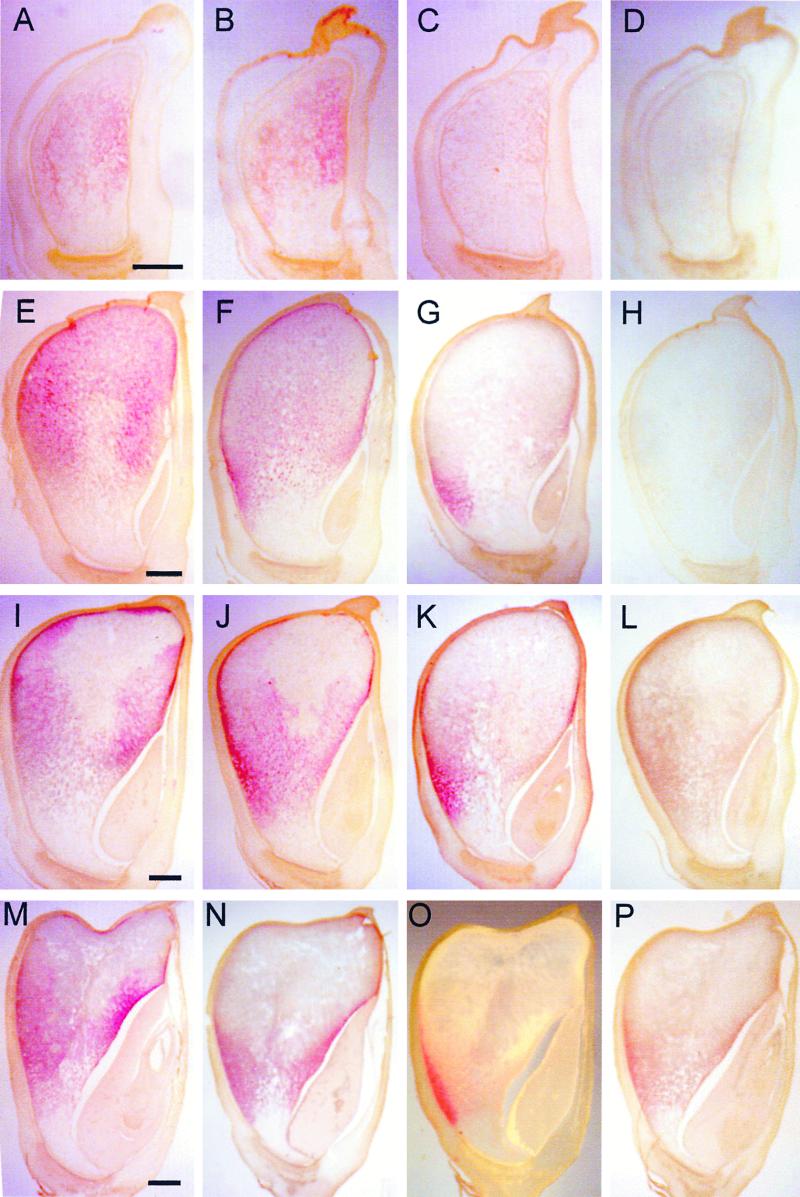
Figure 8.
Detection of mRNAs in Maize Endosperm by in situ Hybridization of Antisense Probes.
Developing endosperm and tissue sections were prepared as in Figure 7. The kernel sections were hybridized with antisense RNA probes of 50-kD γ-zein ([A], [E], [I], and [M]), 15-kD β-zein ([B], [F], [J], and [N]), 10-kD δ-zein ([C], [G], [K], and [O]), and 18-kD α-globulin ([D], [H], [L], and [P]). Bars = 1 mm.
In contrast to α-zein genes, the 27-kD γ-zein was highly expressed throughout the endosperm by 10 and 15 DAP (Figures 7C and 7G). At these stages, the 27-kD γ-zein RNA showed the highest AP signal intensity with the shortest incubation time, indicating that it is likely to be the most abundant zein transcript during early stages of endosperm development. Like the 22-kD α-zein, expression of the 27-kD γ-zein was not detected in some cells of the central starchy endosperm by 20 DAP (Figure 7K). However, the 27-kD γ-zein transcripts occurred more extensively than did the α-zeins in the central starchy endosperm at 20 and 25 DAP. Instead of a gradual decrease in signal intensity, as observed in Figure 7I, the very central starchy endosperm cells showed no evidence of 27-kD γ-zein RNA at 20 DAP, causing a clear boundary between cells in this region. By 25 DAP, expression of the 27-kD γ-zein was restricted to the lower half of the endosperm (Figure 7O).
The spatial expression pattern of the 16-kD γ-zein was similar to that of the 27-kD γ-zein at all developmental stages examined (Figures 7D, 7H, 7L, and 7P). However, the intensity of 16-kD γ-zein signal was consistently less than that of 27-kD γ-zein, even though the AP color reaction was performed for a few hours longer. A relatively high signal for the 16-kD γ-zein in the subaleurone cells of the crown region of 25-DAP endosperm was characteristic and distinguished it from the spatial pattern of the 27-kD γ-zein gene (Figure 7P).
Although the spatial expression of the 50-kD γ-zein was similar to that of the 16- and 27-kD γ-zeins at all developmental stages, its RNA transcripts were significantly less abundant than those of either of the other two γ-zeins. At 10 DAP, the signal for the 50-kD γ-zein RNAs was detected only weakly throughout the endosperm, but it increased by 15 DAP (cf. Figures 8A and 8E). In 20- and 25-DAP endosperms, relatively high levels of 50-kD γ-zein RNAs were found in the crown and the adgerminal regions (Figures 8I and 8M).
The spatial expression of the 15-kD β-zein was very similar to that of the 27-kD γ-zein during endosperm development; however, the level of RNAs was more similar to that of the 50-kD γ-zein at the stages examined (Figures 8B, 8F, 8J, and 8N). A higher level of 15-kD β-zein RNA in the peripheral region of abgerminal endosperm at 20 DAP was one characteristic that distinguished it from the expression patterns of the 16- and 50-kD γ-zeins (Figure 8J).
There was little evidence of 10-kD δ-zein RNA transcripts in 10-DAP endosperm (Figure 8C). We also observed expression of the 10-kD δ-zein in only two small regions of the adgerminal and abgerminal endosperm at 15 and 20 DAP (Figures 8G and 8K). At 25 DAP, the signal was restricted mainly to the abgerminal endosperm (Figure 8O).
The temporal and spatial expression of the 18-kD α-globulin had to be analyzed at a higher level of magnification because of its extremely low level of expression. RNA transcripts, which were detectable only by 20 DAP (Figure 8L), were confined primarily to the subaleurone layer and two or three of the outermost starchy endosperm cell layers. Although the apparent level of RNAs for all of the other genes examined decreased after 15 DAP, the 18-kD α-globulin showed a noticeable increase in transcript level at 25 DAP (Figure 8P).
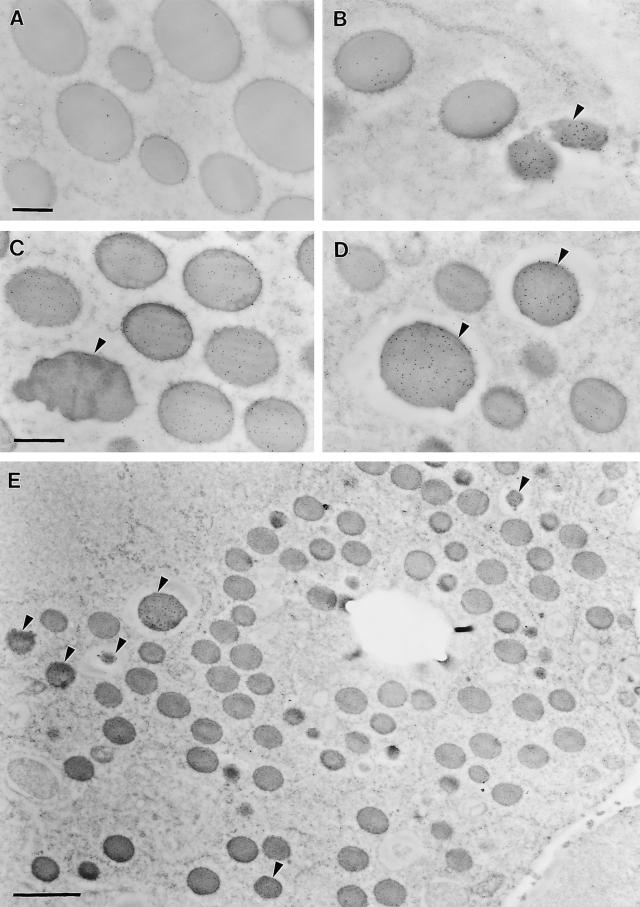
Localization of the 50-kD γ-Zein and 18-kD α-Globulin Proteins in Endosperm Cells
Despite having an apparently lower level of RNA transcripts than the 16- and 27-kD γ-zein genes, the 50-kD γ-zein is widely expressed throughout the endosperm with a comparable pattern of spatial and temporal expression. The distribution of the 50-kD γ-zein protein in protein bodies was examined by immunolocalization with a rabbit polyclonal antibody and gold-conjugated secondary antibodies. The 10-nm gold particles identifying the 50-kD γ-zein protein were found mostly at the periphery of protein bodies, which are surrounded by RER membranes (Figures 9A and 9C). In contrast, 5-nm gold particles labeling a 19-kD α-zein were distributed primarily in the central region of the protein bodies (Figure 9C). Single immunolabeling of the 50-kD γ-zein (Figure 9A) resulted in a slightly greater number of gold particles per protein body than with double labeling (Figure 9C). It was observed consistently that the density of the 50-kD γ-zein labeling was greater in the small protein bodies of subaleurone cells than in the larger protein bodies of starchy endosperm cells (data not shown). This feature of 50-kD γ-zein localization in protein bodies is similar to that of the 27-kD γ-zein and the 15-kD β-zein, as described previously by Lending and Larkins (1989).
Figure 9.
Immunodetection of the 50-kD γ-Zein and the 18-kD α-Globulin in Protein Bodies of 20-DAP Endosperm.
Protein bodies were labeled singly with 50-kD γ-zein antibodies (A) or 18-kD α-globulin antibodies (B) and double labeled with 50-kD γ-zein antibodies (10-nm gold beads) and 19-kD α-zein antibodies (5-nm gold beads) (C) or 18-kD α-globulin antibodies (10-nm gold beads) and 19-kD α-zein antibodies (5-nm gold beads) (D). (E) shows a representative transmission electron micrograph showing a region of the third to fifth starchy endosperm cell layers labeled with 18-kD α-globulin antibodies (10-nm gold beads) and 19-kD α-zein antibodies (5-nm gold beads). The large empty hole is the former location of a starch grain. α-Globulin–containing protein bodies are indicated by arrowheads in (B), (C), (D), and (E). Bars = 0.4 μm in (A) and (C) and 2 μm in (E).
Immunogold labeling with antibodies against the 18-kD α-globulin identified a novel type of protein body in maize endosperm cells (arrowheads in Figures 9B, 9D, and 9E). This protein was found mainly in protein accretions enclosed in smooth, vacuole-like membranes, distinct from the RER-surrounded, zein-containing protein bodies (cf. Figures 9A and 9C). A few gold particles labeling the 18-kD α-globulin were found decorating the RER-surrounded zein protein bodies, and these were more obvious in single immunolabeled images (Figure 9B). Considering the low level of background labeling, as exemplified in the double labeled images by the total absence of 19-kD α-zein label from the vacuole-like protein bodies, this result suggests the possibility of a small amount of 18-kD α-globulin in zein-containing protein bodies (Figures 9B and 9D).
The18-kD α-globulin–containing protein bodies were distinguished readily from zein-containing protein bodies based on the empty space between the protein accretion and the surrounding membrane, which is not decorated by ribosomes (Figures 9B, 9D, and 9E). In the absence of immunostaining for the 18-kD α-globulin, these protein bodies were hardly distinguishable from zein-containing protein bodies, especially when protein completely filled the vacuole-like structure. However, there are two additional physical features that distinguish these vacuole-like protein bodies from zein-containing protein bodies in the RER: (1) they have a dark appearance resulting from higher, more homogenous heavy metal staining throughout the protein body, and (2) they generally have an irregular shape, particularly those in subaleurone and the first and second starchy endosperm cell layers. In the subaleurone layer, 18-kD α-globulin–containing protein bodies are small and irregular in shape, and they occupy only a portion of the vacuole-like structure. In cells of the third starchy endosperm layer, these protein accretions are more spherical and fill the entire vacuole-like structure, so empty space between the protein body and the membrane was not apparent. This may indicate that these protein bodies begin to form in a vacuole-like structure of more or less fixed size and then gradually enlarge to fill the entire vacuole as the cell matures. By the eighth inner starchy endosperm cell layer, these vacuole-like protein bodies are up to two to three times the size of zein protein bodies and become more spherical (data not shown). In addition, the immunolabeling density of the 18-kD α-globulin decreased notably (data not shown), suggesting the deposition of other kinds of proteins. In these cells, the frequency of vacuole-like protein bodies was estimated at ∼3 to 5% of the total number of protein bodies, based on transmission electron micrographs like that shown in Figure 9E.
DISCUSSION
EST Databases Reveal Novel Information Regarding Patterns of Gene Expression
Despite the fact that maize endosperm storage protein genes have been studied for many years and were among the first plant genes characterized in detail, many questions regarding their sequence relationships and expression levels have not been resolved. The development of tools for genome-wide studies of gene families and for large-scale gene expression analysis make a comprehensive analysis of storage protein gene expression in maize endosperm possible. We obtained qualitative and quantitative information on maize endosperm genes by screening the DuPont-Pioneer genome database of ∼480,000 ESTs. Approximately one-tenth of the ESTs in this collection originated either from dissected endosperm or from whole kernel cDNA libraries and, based on computational tools (cluster analysis using the PHRAP algorithm), this subset contains ∼12,000 distinct sequences. It must be emphasized that these ESTs are the result of single-pass DNA sequencing reactions; consequently, they contain many errors and ambiguities. These types of errors tend to contribute to an inflated count of distinct or unique sequences. The number of ESTs in clusters can be used to estimate tissue-specific transcript levels. These numbers are potentially biased, because certain RNA transcripts might be poor substrates for cDNA synthesis and certain cDNAs might be affected selectively during cloning. Although software algorithms (e.g., assignment of PHRED scores [Ewing et al., 1998]) have become more and more sophisticated in addressing some of these issues, most of these problems are inherent in EST-based analysis and cannot be resolved without additional experimentation. A satisfactory solution to some questions requires the complete genome sequence. For instance, based on EST data, the number of distinct human genes was estimated to exceed 100,000 (Liang et al., 2000). However, based on the initial analysis of the human genome sequence (Lander et al., 2001; Venter et al., 2001), the number of genes is surprisingly only approximately one-third of that estimate.
For this study, we applied a number of procedures to increase the accuracy of the data and improve the validity of our conclusions. First, throughout most of the analyses, we considered ESTs from only one maize inbred line (B73). This reduced the EST pool size to ∼250,000, which was important to accurately estimate gene expression, which could differ between inbred lines. But even more important, this increases the accuracy of distinguishing alleles among gene paralogs. Extensive sequence polymorphisms exist between alleles of maize endosperm storage protein cDNA sequences in public and proprietary databases (data not shown), reflecting the diversity of the maize germplasm. This allelic polymorphism often exceeds sequence differences between paralogs (i.e., between different but phylogenetically closely related genes). For example, the 22-kD α-zein genomic locus of the inbred line BSSS53 (Llaca and Messing, 1998) has been sequenced completely, and it contains 23 different 22-kD α-zein paralogs. However, we were able to identify with certainty only one 22-kD α-zein gene in B73 with a corresponding BSSS53 allele.
The estimates of gene expression levels presented in this study were based exclusively on ESTs of randomly selected clones (i.e., from libraries with the smallest biases). Although the corresponding EST set from endosperm comprised only 6732 sequences, a relatively small number compared with the whole data set, it appeared large enough to identify medium level and abundantly expressed transcripts (represented at >0.1%). Our analysis specifically targeted seed protein–related gene sequences (i.e., transcripts that would fall into this category). The ESTs were BLAST searched against known seed protein sequences from maize and other species, and all relevant, putatively distinct full-length or conspicuous sequences were verified by full-insert DNA sequencing. In this context, ∼50% of the ESTs were recognized as seed protein–related sequences, mostly zeins, in accordance with earlier studies (Shen et al., 1994), and full-insert sequences were obtained for >300 clones. Noteworthy, and illustrating the precautions to be taken when using solely computer-based analysis of EST data sets, the ESTs of most clones selected for sequencing fell into the category of singletons based on the PHRED-PHRAP algorithm. Nevertheless, after sequencing, the majority of these “singletons” turned out to be either cDNA cloning artifacts (fusions between different transcripts, large deletions, etc.) or matched contig sequences with >99% identity. In addition to ambiguities among ESTs that are the result of sequencing errors, real mutations can be introduced during cDNA cloning. Full-insert sequencing does not necessarily reveal these errors as cloning artifacts. To avoid overreporting of such sequences as independent gene derivatives, we considered sequences to represent a distinct gene only when at least two perfectly matched sequences were found among cDNAs from independently generated libraries. Applying this criterion, one 19-kD α-zein gene (GenBank accession number AF371273) was found to encode a truncated polypeptide. All four independent cDNA clones of this gene contained the same in-frame stop codon. Genomic sequences of zein genes that encode truncated polypeptides have been described previously (Wandelt and Feix, 1989; Llaca and Messing, 1998). However, it is interesting to find transcriptional activity associated with such a gene.
After the database search and reiterative cluster analysis and sequencing, we identified 20 unique endosperm-specific cDNAs. In addition to the ESTs from randomly chosen endosperm clones, a second EST data set of ∼60,000 sequences, derived from normalized and subtracted endosperm and kernel libraries, was searched for additional unique endosperm expressed protein genes. However, none were found. Therefore, we are confident that the sequences reported here constitute all of the highly expressed seed protein genes in the inbred line B73. Our data generally corroborate the results from earlier studies using RNA hybridization–based approaches (Marks et al., 1985a; Kodrzycki et al., 1989; Liu and Rubenstein, 1993; Or et al., 1993). However, aside from cloning biases (see above), the EST tallying approach allowed direct comparison of expression levels among different genes, and as long as contigs were not overly clustered, it also allowed differentiation between closely related genes. These types of data cannot be obtained easily by hybridization methods, including hybridization-based microarray technologies.
A Relatively Small Number of α-Zein Genes Are Highly Expressed in Maize Endosperm
The α-zeins in B73 are encoded by a large multigene family containing ∼75 members. Three almost equally diverged subfamilies (19-kD α-zein B, 19-kD α-zein D, and 22-kD α-zein) can be recognized (Figure 2). The nucleotide sequence identities between and within them are between 56 and 67% and between 80 and 99%, respectively. Consequently, it is possible to differentiate between members of these subfamilies (but not within) by hybridization with labeled RNA or DNA probes. The inbred line W64A expresses genes corresponding to all three α-zein gene subfamilies (Marks et al., 1985a, 1985b), and this relationship permitted us to use B73 cDNAs to create RNA probes for in situ hybridization experiments with W64A endosperm to assess the temporal and spatial expression of these genes. The B73 19-kD B1 α-zein would cross-hybridize to the 19A, 19B1, and 19B2 sequences of W64A, and the B73 19-kD B3 sequence would cross-hybridize to the 19C1 and 19C2 sequences of W64A (Marks et al., 1985b). The 19A and 19B genes are two of the most highly expressed α-zeins in W64A (Marks et al., 1985a); there is near sequence identity between the 22-kD α-zeins in both inbred lines. However, W64A expresses the 10-kD δ-zein but not the 18-kD δ-zein.
Approximately 200 different α-zein–related DNA sequences have been submitted to GenBank during the last 20 years (ESTs not included). These submissions by different authors relate to sequences from a variety of maize inbred lines. There were attempts to classify these genes (Heidecker and Messing, 1986; Rubenstein and Geraghty, 1986), but unfortunately, conventions for naming α-zeins are inconsistent and confusing. However, a comprehensive phylogenetic analysis of α-zeins, in conjunction with the assignment of coherent names, should await complete sequencing of genomic loci. In this regard, a genomic sequence containing the 22-kD α-zein gene subfamily on chromosome 4S of the BSSS53 inbred line was reported recently (Llaca and Messing, 1998).
It is striking that in B73 endosperm only three 22-kD α-zein genes were found to be highly expressed, one at a very high level (Figure 1), indicating that the majority of the genes in this subfamily are either inactive or expressed at very low levels. A similar pattern of gene expression for 22-kD α-zeins in other maize inbred lines has been observed as well (J. Messing, personal communication). The 19-kD α-zein gene subfamilies also have been estimated to be large, consisting of ∼55 members (Wilson and Larkins, 1984). However, like the expression pattern of the 22-kD α-zeins, we detected only three cDNAs of 19-kD α-zein B genes and two 19-kD α-zein D genes in B73 endosperm. The expression of the gene encoding the 19-kD α-zein B1 is remarkably high; it contributes almost 16% of the RNA transcripts. Consequently, these data suggest large differences in promoter strength or mRNA stability among the α-zein genes. These differences could be an important consideration for the selection of potentially strong promoters for use in transgenic plants. However, most α-zein genes are the result of recent gene duplications, and their promoters show considerable sequence similarity. The elements that direct large differences in promoter strength, therefore, are not obvious; thus, the identification of a strong promoter might not be a simple task.
The detection of a small number of highly expressed α-zein genes in B73 endosperm is consistent with the results of Pedersen et al. (1980) and Marks et al. (1985a) in studies of W64A. Because only a few distinct mRNAs constitute the zein cDNA population, with α-zeins containing short, repeated nucleotide sequences (Marks et al., 1985b), we would predict complex Rot hybridization kinetics and a Cot value that is two to five times the single-copy rate. Other members of these gene families could be expressed at very low levels; hence, their transcripts would not be detected among the ESTs. However, we believe it is more likely that these genes are repressed by one mechanism or another (Lund et al., 1995).
The expansion of multigene families by gene duplication is thought to provide evolutionary advantages by providing a mechanism to develop new or enhanced gene functions (Lynch and Conery, 2000). For instance, under certain conditions, increased expression caused by a duplicated gene could provide a selective benefit. In this regard, the expansion of the α-zein gene family could be viewed as an adaptation to meet the need to massively synthesize α-zein proteins as a nitrogen reserve in seed. However, this “gene dosage” hypothesis does not appear to be valid in view of the surprising finding that very few α-zein genes, possibly <10% of the gene family members, are actively expressed. α-Zeins are small proteins that contain multiple peptide repeats, and the respective coding regions create repetitive nucleotide sequences (Coleman and Larkins, 1998). Repeated DNA sequences are especially prone to unequal crossing over during recombination, which can lead to high rates of gene mutation. The expansion of the α-zein gene family as an adaptation to counteract such mutations has been proposed by Messing and co-workers (Heidecker and Messing, 1986; Das et al., 1990; Heidecker et al., 1991). Although this is a plausible explanation, other highly expressed storage protein genes (e.g., the γ-zeins) also contain regions of repetitive nucleotide sequences, but the ancestors of these genes did not undergo extensive gene duplications and expand the gene family.
γ- and δ-Zein Genes Are Highly Expressed Relative to Their Copy Number
In addition to α-zeins, Figure 1 shows transcript levels of eight other endosperm protein genes, including γ- and δ-zeins and several non-zein proteins, that are highly expressed in maize endosperm. The genetic map positions of several of these genes were known (Benner et al., 1989; Weerakoon et al., 1993; Lopes et al., 1995; Swarup et al., 1995; Llaca and Messing, 1998), and we determined those of several other genes in the course of this study (Table 1). DNA gel blotting (data not shown) indicated one copy of each gene at these loci. Consequently, in contrast to the α-zeins, we found no evidence for inactive genes or pseudogenes.
The γ-zein gene family in B73 is composed of four members. Two of the encoded polypeptides, the 15-kD β-zein and the 16-kD γ-zein, are relatively low molecular weight polypeptides and perhaps similar to the γ-zein ancestral gene. It is possible that the two other members of this gene family, the 27- and 50-kD γ-zeins, arose independently from duplicated copies of the ancestral gene as a result of the insertion of tandem repeat domains. The low complexity Pro-rich domains of the 27-kD γ-zein and the Gln-rich peptide domains of the 50-kD γ-zein have the potential to be involved in protein–protein interactions (Chen et al., 1997) and the oligomerization of proteins (Hughes and Olson, 2001). These domains could play a role in the initiation of protein body formation, a function consistent with previous reports (Bagga et al., 1997; Coleman et al., 1996; see below).
The 50-kD γ-zein is a novel protein. By creating monospecific antibodies (Figure 5), we were able to identify the corresponding protein after SDS-PAGE with an apparent molecular mass of 50 kD. This electrophoretic mobility is surprising given the calculated molecular mass of 33 kD for the mature protein (Table 1). In previous studies, the 50-kD γ-zein was misidentified as a dimer of the 27-kD γ-zein (Lopes and Larkins, 1991) because it cross-reacts with antibodies developed against the complete 27-kD γ-zein. Because of this result and similar observations, we decided to develop monospecific antisera against each of the zein proteins based on predicted amino acid sequences using either bacterially expressed or chemically synthesized peptides as antigens (Table 1). In contrast to a method that relies on protein purification from seed, this approach entirely avoids possible cross-contamination with other endosperm proteins and thus allows a more rigorous interpretation of the immunodetection data. As a result, we obtained antibodies that are selectively able to distinguish between each of the four γ-zein proteins, each of the two δ-zein proteins, and the three subfamilies of α-zein proteins. With these antisera, we were able to clarify several questions regarding the identity of polypeptide bands separated by SDS-PAGE (Figure 5). As mentioned above for the 50-kD γ-zein, a remarkable property of many maize endosperm storage proteins is a large disparity between the observed electrophoretic mobility in SDS-PAGE and the predicted mobility based on calculated molecular weights (summarized in Table 1). All α- and δ-zein polypeptides, as well as the two low molecular weight γ-zeins, migrate ∼20 to 25% faster in SDS-PAGE than predicted. The 27- and 50-kD γ-zein proteins, in contrast, show 25 and 50% retardation, respectively, in electrophoretic mobility. We observed similar migration discrepancies with their bacterially expressed glutathione S-transferase fusion proteins. The unusual electrophoretic behavior of zeins, therefore, likely is associated with the secondary structure of the tandem peptide repeat regions of these proteins.
Among maize prolamins, δ-zeins are encoded by the smallest number of transcripts; they appear to be expressed at about the same level as the 18-kD α-globulin and legumin 1 (see below). The sequence polymorphism between the 18-kD δ-zeins of the B73 and Mo17 inbred lines is remarkable (Figure 4). The polypeptides are 90% identical, but the B73 allele encodes five fewer Met residues than the Mo17 allele. Alleles from other maize inbred lines show a similar degree of polymorphism but of different haplotypes (data not shown). This variability suggests low evolutionary pressure on the 18-kD δ-zein genes to maintain a primary protein structure and also indicates little or no selection for alleles during the domestication of maize. This supports earlier findings of Swarup et al. (1995) indicating that the high Met trait in wild and exotic germplasm may have escaped selection during the early cultivation of maize.
Seed Protein Genes Have Distinct Patterns of Temporal and Spatial Expression
Dolfini et al. (1992) described the localization of RNAs encoding a 19- and a 22-kD α-zein and the 27-kD γ-zein based on in situ hybridization in W64A endosperm. They found that transcript levels peaked at 17 DAP, and the spatial pattern of expression was distinctly different for 19- and 22-kD α-zein genes. Furthermore, expression of the 27-kD γ-zein was not uniform during the early stages of endosperm development. In contrast, we found similar patterns of transcript distribution for different α-zein subfamilies and clear temporal and spatial differences in the distribution of other storage protein RNA transcripts. Although RNAs of most genes were detected by 10 DAP, at this stage, α-zein transcripts are not highly abundant and are restricted to a small area on the adgerminal side of the endosperm. In contrast, members of the γ-zein family, in particular the 27- and 16-kD γ-zein and the 15-kD β-zein, are highly and uniformly expressed throughout the starchy endosperm. This finding suggests that the earliest forms of protein bodies would contain primarily γ-zeins, consistent with the immunolocalization studies of Lending and Larkins (1989). These data support the hypothesis of a functional role for γ-zeins in protein body assembly, which has been suggested by several other studies (Bagga et al., 1997; Coleman et al., 1996).
RNA transcripts of α- and γ-zeins were detected throughout most regions of the starchy endosperm by 15 DAP, except the aleurone and basal transfer cells. However, expression of the 10-kD δ-zein and the 18-kD α-globulin, which occurred at a reduced level compared with the other seed protein genes, was restricted primarily to a few cells of the abgerminal side of the endosperm. The physiological significance of the localized expression of these genes is unclear, but this observation implies that the composition of protein bodies varies in different regions of the endosperm. By 20 DAP, we did not detect significant amounts of zein RNAs near the crown and center of the starchy endosperm. Our inability to detect zein RNA transcripts, particularly the γ-zeins, in these regions could be attributable to high concentrations of starch in these cells. However, it has been shown that cell death commences in these areas between 16 and 20 DAP (Young et al., 1997), and the sharp boundary we observed between the cells that contained RNA transcripts and those that did not is consistent with this explanation. After 20 DAP, the most active area of storage protein synthesis appeared to be restricted to the peripheral and basal regions of the endosperm.
Novel Globulin Proteins in Maize Endosperm
Although zeins are by far the most abundant seed proteins in maize endosperm, our results demonstrate that in the B73 inbred line, at least, there are two novel types of globulin proteins. The maize legumin 1 (GenBank accession number AF371279) differs from other members of the 11S globulin protein family by the lack of an almost canonical asparaginyl endopeptidase cleavage site (R. Jung, unpublished data). The second novel globulin from endosperm, the 18-kD α-globulin, shares significant sequence homology with a previously described rice α-globulin gene (Nakase et al., 1996) and a wheat high molecular weight glutenin (GenBank accession number D50643). Despite contrasting solubility characteristics and, consequently, the classification of these sequences as globulins and prolamins (glutenins), all members of the α-globulin family share conserved Cys residues and homologous N-terminal as well as C-terminal peptide domains. The 18-kD α-globulin contains a Trp-rich cluster (Figure 5, underlined sequence). A similar Trp-rich sequence (Figure 5, underlined) was found in puroindolin b (Gautier et al., 1994), a protein implicated in the endosperm hardness of wheat grain. The closest related sequence to the 18-kD α-globulin found in the GenBank database is a rice α-globulin (accession number D50643; Figure 5), and because the maize polypeptide is soluble in saline solutions (data not shown), that is, it is a globulin, we derived its name in reference to the rice protein. This protein appears to be deposited primarily in vacuole-like structures in endosperm cells, although it was detected in zein-containing protein bodies as well when immunolabeling was done with 18-kD α-globulin antiserum alone. We could not confirm the identity of these structures as protein storage vacuoles, because they could not be immunolabeled with an α-TIP antiserum from bean (data not shown); however, the membranes showed no evidence of ribosomes, in clear contrast to the zein-containing protein bodies. Because the density of immunogold labeling decreased as these protein bodies enlarged, we believe that they may contain other types of proteins, such as the 11S globulin–related protein. Unfortunately, the antiserum we produced against this protein reacted in immunoblots but failed to detect the legumin 1 protein in thin sections of developing endosperm. Experiments are in progress to characterize the synthesis and subcellular trafficking of the 18-kD α-globulin and legumin 1 proteins in developing maize endosperm.
METHODS
Construction of Endosperm cDNAs and Expressed Sequence Tag Databases
RNAs were isolated using TriZol reagent (Gibco BRL, Gaithersburg, MD) from dissected frozen maize (Zea mays) endosperm (inbred line B73) harvested from greenhouse-grown plants at 11, 13, 15, 18, 21, 24, 27, 29, 30, 35, and 40 days after pollination (DAP). cDNA synthesis was performed using a SuperScript II kit, and cloning was into NotI-SalI sites of the pSPORT1 vector (Gibco BRL). Inserts of ∼600 randomly chosen clones from each library were sequenced by Human Genome Systems (Rockville, MD) from the 5′ end to obtain expressed sequence tags (ESTs). The sequence information of 6732 EST accessions pertaining to these cDNA libraries is maintained in the central DuPont-Pioneer genomics database and is accessible via database interface software. We also analyzed ∼480,000 maize ESTs representing 215 cDNA libraries, including 25 endosperm and whole kernel cDNA libraries from other inbred lines, and identified >10,000 different accessions that either resemble previously described maize storage protein gene family members or show homology with seed storage protein genes from other species.
Clustering, Identification, and Analysis of Abundant Endosperm and/or Storage Protein ESTs and cDNA Sequences
EST sequences from B73 endosperm libraries were first evaluated using PHRED-assigned quality scores (Ewing et al., 1998) and, after removal of short, low-complexity, and low-quality sequences, the similarity relationship (clustering) between ESTs was established using the BLAST algorithm (Altschul et al., 1990). EST clusters were subjected subsequently to a sequence assembly process using the PHRAP algorithm (http://www.phrap.org/phrap.docs/phrap.html). The resulting database of contigs and singletons was annotated systematically by searching (BLAST) against publicly available DNA and protein sequences. Sequences with homology with known seed storage proteins and ESTs that clustered into contigs of at least 50 sequences (termed abundant cDNAs) were tallied into a separate database. As a result, ∼3500 EST accessions formed an initial group of ∼200 mostly zein-related sequences, singletons, and contigs. These sequences were inspected thoroughly for excess and deficient clustering by visual and pairwise sequence alignments (BLAST, ClustalW, Gap), which resulted in ∼50 putatively unique sequences. The longest clone representing each of these sequences was obtained from the libraries, and both strands of each insert were sequenced by either primer walking or sequencing of nested sets of deletion subclones. The identification and clustering of unique storage protein or endosperm-abundant sequences were further refined by repeated pairwise BLAST searches and by searching the entire B73 endosperm EST subdatabase again using both the full-length insert nucleotide sequences and their deduced amino acid sequences. Finally, after several rounds of reiterative analysis and sequencing, 19 complete nucleotide sequences of unique full-length endosperm cDNAs (abundant or storage proteins) were identified (summarized in Figure 1 and Table 1). The percentage of each unique endosperm sequence in the database was estimated by tallying the number of clustered EST contigs. Phylogenetic analysis of selected sequences, including bootstrap tests of the resulting dendrograms (neighbor-joining method with 500 iterations), was performed using MEGA 2b3 software (Kumar et al., 1994; http://www.megasoftware.net).
In Vitro Transcription and Bacterial Expression of Seed Protein–Specific RNA and Polypeptides
The nucleotide and encoded polypeptide sequences of endosperm protein–specific cDNAs were analyzed for similarity (ClustalW, Gap). Regions of low similarity (nucleotide level, <70% global identity and <15 identical consecutive nucleotides; polypeptide level, <50% global identity and <6 identical consecutive amino acid residues) were selected for subcloning into bacterial vectors suitable for in vitro transcription or bacterial production of polypeptides. An overview of the subcloned DNA sequences is given in Table 1.
Two types of bacterial expression products were produced for each protein subsequence, a His tag fusion and a glutathione S-transferase (GST) fusion from subclones in pET28 (Novagen, Madison, WI) and from subclones in pGEX3 (Pharmacia, Piscataway, NJ), respectively. His tag and GST fusions were purified by affinity chromatography on Talon metal affinity resins (Clontech, Palo Alto, CA) and on glutathione–Sepharose 4B (Pharmacia), respectively. Protein expression and purification followed established procedures (Sambrook and Russel, 2001) and manufacturer-provided protocols.
Production of Monospecific Polyclonal Antibodies
Antibodies were produced in New Zealand White rabbits at HTI Bioproducts (Ramona, CA). Rabbits were sustained on a maize-free diet and prescreened for the absence of endosperm protein cross-reacting antibodies before immunization. Affinity-purified His tag fusions of protein sequences or, alternatively, synthetic oligopeptides coupled to keyhole limpet hemocyanin (Research Genetics, Huntsville, AL) were denatured in 5 M urea for 5 min at 100°C and used as antigens (Table 1). The resulting polyclonal antibodies were affinity purified with the respective bacterially expressed GST fusion proteins bound to Affigel 15 (Bio-Rad, Hercules, CA) according to standard procedures (Harlow and Lane, 1988) or, for peptide antibodies, with the immobilized synthetic oligopeptide antigens (Research Genetics). Selected antigen-generated antibodies also were produced in chickens and rats (Table 1).
Protein Extraction, Electrophoresis, and Immunodetection
Total protein was extracted from B73 endosperm in a 20-fold (w/v) excess of 2% SDS, 100 mM DTT, and 50 mM Tris-HCl, pH 6.8. The slurry was incubated immediately for 5 min at 100°C, vortexed vigorously for 1 min, and cleared by microcentrifugation for 5 min at ∼18,000g. Zein and non-zein protein fractions were extracted according to Wallace et al. (1990). Protein concentrations were estimated according to Bradford (1976) (Bio-Rad Protein Assay) with BSA (Pierce, Rockford, IL) as a standard. Protein samples were adjusted to 5 mg/mL in SDS-PAGE sample buffer (Laemmli, 1970). Proteins were separated electrophoretically using precast SDS-PAGE gradient gels and visualized by Coomassie Brilliant Blue R250 staining (Harlow and Lane, 1988) in the presence of 30% trichloroacetic acid to fix the proteins in the gel matrix. This procedure markedly reduced the loss of alcohol-soluble and low molecular mass polypeptides (e.g., 10-kD δ-zein) from the gel matrix. For immunodetection, electrophoretically separated polypeptides were transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore, Bedford, MA) using a semidry electroblotter (SemiPhor TE70; Hoefer, San Francisco, CA) as described (Matsudaira, 1987). Prestained molecular weight protein standards (SeeBlue; Novex, San Diego, CA) were used to monitor the electrophoresis and transfer. The detection of antigens on polyvinylidene difluoride blots was performed according to standard procedures (Harlow and Lane, 1988) using the ECL kit (Pharmacia) or the Western Blue reagent (Promega, Madison, WI) according to the manufacturer's instructions.
RNA in Situ Hybridization
The maize inbred line W64A was grown in a greenhouse at the University of Arizona research farm in October 1999. Kernels were taken from the middle of ears harvested at 5, 10, 15, 20, and 25 DAP. After trimming both sides of a kernel, 2- to 3-mm-thick longitudinal median sections containing embryos were obtained and immediately fixed in 3.7% paraformaldehyde and 0.2% picric acid in 50 mM potassium phosphate and 5 mM EGTA buffer, pH 6.8, which was prepared in diethyl pyrocarbonate–treated deionized, distilled water (DEPC-ddH2O). After fixation for 3 to 4 hr at room temperature, the thick sections were washed in the same buffer for 1 to 2 hr with three changes to fresh buffer and then processed through a standard paraffin embedding procedure that included dehydration in gradient ethanol, substitution with xylene, and gradual infiltration of paraffin. Paraffin-embedded tissues were cut into 15-μm-thick sections that were attached to Histogrip-coated (Zymed Laboratories, South San Francisco, CA) glass microscope slides. Fixation after rehydration and various treatments, such as with glycine, proteinase K, and acetic anhydride, and blocking with hybridization solution lacking the RNA probe were omitted because they were tested systematically and found to be unnecessary. However, before hybridization with RNA probes, the tissue sections were treated with 0.25% DEPC in PBS for 30 min to inactivate endogenous enzymes, including RNase and alkaline phosphatase (AP). Hybridization was at 63°C overnight.
The hybridization solution contained 400 ng/mL digoxigenin (DIG)-labeled RNA probe of each gene, 50% deionized formamide, 5% dextran sulfate (average molecular weight, 500,000), 1% Blocking Reagent (Roche Molecular Biochemicals, Indianapolis, IN), 20 μg/mL tRNA, 500 μg/mL poly(A), 40 μg/mL salmon sperm DNA, 300 mM NaCl, 10 mM Tris, pH 7.5, and 1 mM EDTA in DEPC-ddH2O. DIG-labeled sense and antisense RNA probes were synthesized in vitro by transcription of linearized pBluescript II KS+ plasmids containing the cDNA of interest with T3 or T7 RNA polymerase (Roche Molecular Biochemicals). After hybridization, slides were washed twice for 1 hr at 63°C with 0.2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) containing 0.05% SDS. Between these stringent washes, the slides were treated with 10 μg/mL RNase A for 30 min at 37°C. DIG-labeled probes were detected with anti-DIG antibody conjugated with AP and an AP color reaction using the Fast Red TR/Naphthol AS-MX system (Sigma, St. Louis, MO), which produces a red precipitate. These hybridization conditions were shown to minimize cross-hybridization among members of the same gene family, as indicated by consistent differences in spatial patterns of gene expression at particular developmental stages. The optimum incubation times for AP color reaction varied depending on the abundance of the transcript as follows: overnight, 19- and 22-kD α-zeins, 15-kD β-zein, 16-, 27-, and 50-kD γ-zeins; 2 days, 10-kD δ-zein and 18-kD α-globulin.
Immunogold Labeling of Endosperm Proteins
Approximately 0.5-mm-thick slices were excised from 20-DAP kernels midway between the crown and base; sections were prepared from the peripheral endosperm region, which has the highest density of protein bodies. The sections were fixed immediately in freshly prepared 4% paraformaldehyde and 1% glutaraldehyde in 50 mM potassium phosphate and 5 mM EGTA buffer, pH 6.8, and incubated overnight at 4°C. Before dehydration in graded ethanol concentrations, the slices were fixed in 2% osmium tetroxide for 2 hr at 4°C. The slices were infiltrated with LR White resin, cured under UV light at −10°C for 2 days, and further cured under UV light overnight at room temperature. Plastic tissue blocks were cut into 80- to 120-nm-thick sections and collected on formvar-coated nickel grids. The grids were incubated for 30 min in blocking solution, pH 8.2, containing 0.2% BSA and 0.06% Tween 20 in 20 mM Tris-HCl and 500 mM NaCl. The grids were labeled with primary antibodies overnight at 4°C, whereas labeling with secondary antibodies conjugated with gold particles was performed for 1 hr at room temperature. For simultaneous detection of 19-kD α-zein with 50-kD γ-zein and 19-kD α-zein with 18-kD α-globulin, the grids were incubated with 19-kD α-zein B1 rat antibodies and 50-kD α-zein or 18-kD α-globulin rabbit antibodies. The primary antibodies were detected with anti-rat or anti-rabbit antibodies conjugated with 5- and 10-nm gold particles, respectively. The grids were stained with 2.5% uranyl acetate for 15 min followed by three rinses in ddH2O. The grids were air dried and examined with a JEOL 100CX2 transmission electron microscope (JEOL USA, Peabody, MA).
Acknowledgments
We thank Dr. Choel Soo Kim for constructing zein transcription vectors for RNA in situ hybridization, and we are grateful to our colleagues who made critical suggestions to improve the manuscript, in particular Dr. Paul Anderson and Dr. Larry Beach at Pioneer Hi-Bred, who encouraged this work. We are indebted to Virginia Dress and Jan Barnikow for technical support. We further thank colleagues in the Pioneer Hi-Bred Analytical Biochemistry Department for their expert support and the Dupont-Pioneer genomics and bioinformatics groups for creating a comprehensive and searchable EST database of maize. This research was supported by grants from the Department of Energy (DE-FG03-95ER20183) and Pioneer Hi-Bred International to B.A.L.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 21, 5403–5410. [DOI] [PubMed] [Google Scholar]
- Bagga, S., Adams, H.P., Rodriguez, F.D., Kemp, J.D., and Sengupta- Gopalan, C. (1997). Coexpression of the maize δ-zein and β-zein genes results in stable accumulation of δ-zein in endoplasmic reticulum–derived protein bodies formed by β-zein. Plant Cell 9, 1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner, M., Phillips, R.L., Kirihara, J.A., and Messing, J. (1989). Genetic analysis of methionine-rich storage protein accumulation in maize. Theor. Appl. Genet. 78, 761–767. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Burr, B., and Burr, F.A. (1991). Recombinant inbreds for molecular mapping in maize: Theoretical and practical considerations. Trends Genet. 7, 55–60. [DOI] [PubMed] [Google Scholar]
- Burr, B., Burr, F.A., Thompson, K.H., Albertson, M.C., and Stuber, C.W. (1988). Gene mapping with recombinant inbreds in maize. Genetics 118, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.I., Einbond, A., Kwak, S.J., Linn, H., Koepf, E., Peterson, S., Kelly, J.W., and Sudol, M. (1997). Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J. Biol. Chem. 272, 17070–17077. [DOI] [PubMed] [Google Scholar]
- Chui, C.-F., and Falco, S.C. (1995). A new methionine-rich seed storage protein from maize. Plant Physiol. 107, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, C.E., Herman, E.H., Takasaki, K., and Larkins, B.A. (1996). The maize α-zeins and stabilizes its accumulation in protein bodies of transgenic tobacco endosperm. Plant Cell 8, 2335–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, C.E., Clove, A.M., Ranch, J.P., HIggins, R., Lopes, M.A., and Larkins, B.A. (1997). Expression of a mutant α-zein creates the floury2 phenotype in transgenic maize. Proc. Natl. Acad. Sci. USA 94, 7094–7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, C.E., and Larkins, B.A. (1998). The prolamins of maize. In Seed Proteins, P.R. Shewry and R. Casey, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 109–139.
- Das, O.P., Levi-Minzi, S., Koury, M., Benner, M., and Messing, J. (1990). A somatic gene rearrangement contributing to genetic diversity in maize. Proc. Natl. Acad. Sci. USA 87, 7809–7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini, S.F., Landoni, M., Tonelli, C., Bernard, L., and Viotti, A. (1992). Spatial regulation in the expression of structural and regulatory storage protein genes in Zea mays endosperm. Dev. Genet. 13, 264–276. [Google Scholar]
- Esen, A. (1987). Proposed nomenclature for the alcohol-soluble proteins (zeins) of maize (Zea mays L.). J. Cereal Sci. 5, 117–128. [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M., and Green, P. (1998). Basecalling of automated sequencer traces using PHRED. I. Accuracy assessment. Genome Res. 8, 175–185. [DOI] [PubMed] [Google Scholar]
- Gautier, M.F., Aleman, M.E., Guirao, A., Marion, D., and Joudrier, P. (1994). Triticum aestivum puroindolines, two basic cystine-rich seed proteins: cDNA sequence analysis and developmental gene expression. Plant Mol. Biol. 25, 43–57. [DOI] [PubMed] [Google Scholar]
- Hagen, G., and Rubenstein, I. (1981). Complex organization of zein genes in maize. Gene 13, 239–249. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Heidecker, G., and Messing, J. (1986). Structural analysis of plant genes. Annu. Rev. Plant Physiol. 37, 439–466. [Google Scholar]
- Heidecker, G., Chaudhuri, S., and Messing, J. (1991). Highly clustered zein gene sequences reveal evolutionary history of the multigene family. Genomics 10, 719–732. [DOI] [PubMed] [Google Scholar]
- Herman, E.M., and Larkins, B.A. (1999). Protein storage bodies and vacuoles. Plant Cell 11, 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, R.E., and Olson, J.M. (2001). Therapeutic opportunities in polyglutamine disease. Nat. Med. 7, 419–423. [DOI] [PubMed] [Google Scholar]
- Kirihara, J.A., Petri, J.B., and Messing, J.W. (1988). Isolation and sequence of a gene encoding a methionine-rich 10-kDa zein protein from maize. Gene 71, 359–370. [DOI] [PubMed] [Google Scholar]
- Kodrzycki, R., Boston, R.S., and Larkins, B.A. (1989). The opaque2 mutation of maize differentially reduces zein gene transcription. Plant Cell 1, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridl, J.C., Vieira, J., Rubenstein, I., and Messing, J. (1984). Nucleotide sequence analysis of a zein genomic clone with a short open reading frame. Gene 28, 113–118. [DOI] [PubMed] [Google Scholar]
- Kriz, A. (1999). 7S globulins of cereals. In Seed Proteins, P.R. Shewry and R. Casey, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 477–498.
- Kumar, S., Tamura, K., and Nei, M. (1994). MEGA: Molecular Evolutionary Genetic Analysis software for microcomputers. Comput. Appl. Biosci. 10, 189–191. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Landry, J., and Moureaux, T. (1970). Heterogeneite des glutelines du grain de mais: Extraction selective et composition en acides amines des trios fractions isolees. Bull. Soc. Chim. Biol. 52, 1021–1037. [PubMed] [Google Scholar]
- Lending, C.R., and Larkins, B.A. (1989). Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell 1, 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F., Holt, I., Pertea, G., Karamycheva, S., Salzberg, S.L., and Quackenbush, J. (2000). Gene index analysis of the human genome estimates approximately 120,000 genes. Nat. Genet. 25, 239–240. [DOI] [PubMed] [Google Scholar]
- Liu, C.N., and Rubenstein, I. (1993). Transcriptional characterization of an α-zein gene cluster in maize. Plant Mol. Biol. 22, 323–336. [DOI] [PubMed] [Google Scholar]
- Llaca, V., and Messing, J. (1998). Amplicons of maize zein genes are conserved within genic but expanded and constricted in intergenic regions. Plant J. 15, 211–220. [DOI] [PubMed] [Google Scholar]
- Lopes, M.A., and Larkins, B.A. (1991). γ-Zein content is related to endosperm modification in quality protein maize. Crop Sci. 31, 1655–1662. [Google Scholar]
- Lopes, M.A., Takasaki, K., Bostwick, D.E., Helentjaris, T., and Larkins, B.A. (1995). Identification of two opaque2 modifier loci in quality protein maize. Mol. Gen. Genet. 247, 603–613. [DOI] [PubMed] [Google Scholar]
- Lund, G., Ciceri, P., and Viotti, A. (1995). Maternal-specific demethylation and expression of specific alleles of zein genes in the endosperm of Zea mays L. Plant J. 8, 571–581. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Conery, J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Marks, M.D., Lindell, J.S., and Larkins, B.A. (1985. a). Quantitative analysis of the accumulation of zein mRNA during maize endosperm development. J. Biol. Chem. 260, 16445–16450. [PubMed] [Google Scholar]
- Marks, M.D., Lindell, J.S., and Larkins, B.A. (1985. b). Nucleotide sequence analysis of zein mRNAs from maize endosperm. J. Biol. Chem. 260, 16451–16459. [PubMed] [Google Scholar]
- Matsudaira, P. (1987). Sequence from picomole quantities of protein electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262, 10035–10038. [PubMed] [Google Scholar]
- Nakase, M., Hotta, H., Adachi, T., Aoki, N., Nakamura, R., Masumura, T., Tanaka, K., and Matsuda, T. (1996). Cloning of the rice seed α-globulin-encoding gene: Sequence similarity of the 5′-flanking region to those of the genes encoding wheat high-molecular-weight glutenin and barley D hordein. Gene. 170, 223–226. [DOI] [PubMed] [Google Scholar]
- Nyren, P., Pettersson, B., and Uhlen, M. (1993). Solid phase DNA minisequencing by an enzymatic luminometric inorganic pyrophosphate detection assay. Anal. Biochem. 208, 171–175. [DOI] [PubMed] [Google Scholar]
- Or, E., Boyer, S.K., and Larkins, B.A. (1993). opaque2 modifiers act post-transcriptionally and in a polar manner on γ-zein gene expression in maize endosperm. Plant Cell 5, 1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, K., Bloom, K.S., Anderson, J.N., and Larkins, B.A. (1980). Analysis of the complexity and frequency of zein genes in the maize genome. Biochemistry 19, 1644–1650. [DOI] [PubMed] [Google Scholar]
- Pedersen, K., Devereux, J., Wilson, D.R., Sheldon, E., and Larkins, B.A. (1982). Cloning and sequence analysis reveal structural variation among related zein genes in maize. Cell 29, 1016–1026. [DOI] [PubMed] [Google Scholar]
- Pedersen, K., Argos, P., Naravanna, S.V.L., and Larkins, B.A. (1986). Sequence analysis and characterization of a maize gene encoding a high-sulfur zein protein of Mr 15,000. J. Biol. Chem. 261, 6279–6284. [PubMed] [Google Scholar]
- Prat, S., Perez-Grau, L., and Puigdomenech, P. (1987). Multiple variability in the sequence of a family of maize endosperm proteins. Gene 52, 41–49. [DOI] [PubMed] [Google Scholar]
- Rubenstein, I., and Geraghty, D.E. (1986). The genetic organization of zein. In Advances in Cereal Science and Technology, Y. Pomeranz, ed (St. Paul, MN: American Association of Cereal Chemists), pp. 286–315.
- Sambrook, J., and Russel, D. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).s
- Shen, B., Carneiro, N., Torres-Jerez, I., Stevenson, B., McCreery, T., Helentjaris, T., Baysdorfer, C., Almira, E., Ferl, R.J., Habben, J.E., and Larkins, B. (1994). Partial sequencing and mapping of clones from two maize cDNA libraries. Plant Mol. Biol. 26, 1085–1101. [DOI] [PubMed] [Google Scholar]
- Shewry, P.R., and Casey, R. (1999). Seed proteins. In Seed Proteins, P.R. Shewry and R. Casey, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–10.
- Soave, C., and Salamini, F. (1984). Organization and regulation of zein genes in maize endosperm. Philos. Trans. R. Soc. Lond. 304, 341–347. [Google Scholar]
- Soave, C., Reggiani, R., Di Fonzo, N., and Salamini, F. (1981). Clustering of genes for 20 kd zein subunits in the short arm of maize chromosome 7. Genetics 97, 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spena, A., Viotti, A., and Pirrotta, V. (1983). Two adjacent genomic zein sequences: Structure, organization and tissue-specific restriction pattern. J. Mol. Biol. 169, 799–811. [DOI] [PubMed] [Google Scholar]
- Swarup, S., Timmermans, M.C., Chaudhuri, S., and Messing, J. (1995). Determinants of the high-methionine trait in wild and exotic germplasm may have escaped selection during early cultivation of maize. Plant J. 8, 359–368. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., et al. (2001). The sequence of the human genome. Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- Viotti, A., Sala, E., Marotta, R., Alberi, P., Balducci, C., and Soave, C. (1979). Genes and mRNAs coding for zein polypeptides in Zea mays. Eur. J. Biochem. 102, 211–222. [DOI] [PubMed] [Google Scholar]
- Wallace, J.C., Lopes, M.A., Paiva, E., and Larkins, B.A. (1990). New methods for extraction and quantitation of zeins reveal a high content of γ-zein in modified opaque-2 maize. Plant Physiol. 92, 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandelt, C., and Feix, G. (1989). Sequence of a 21 kd zein gene from maize containing an in-frame stop codon. Nucleic Acids Res. 17, 2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerakoon, L.W., Weber, D.F., and Schneerman, M.C. (1993). Estimating genetic relatedness of maize to 23 plant species by RFLP analysis. Maydica 38, 231–237. [Google Scholar]
- Wienand, U., Langridge, P.L., and Feix, G. (1981). Isolation and characterization of a genomic sequence of maize coding for a zein gene. Mol. Gen. Genet. 182, 440–444. [Google Scholar]
- Wilson, C.M. (1983). Seed protein fractions of maize, sorghum and related cereals. In Seed Proteins: Biochemistry, Genetics, Nutritive Value, W. Gottschalk and H.P. Müller, eds (The Hague, The Netherlands: Martinus Nijhoof/Dr. W. Junk Publishers), pp. 271–307.
- Wilson, C.M. (1985). A nomenclature for zein polypeptides based on isoelectric focusing and sodium dodecyl sulfate gel electrophoresis. Cereal Chem. 62, 361–365. [Google Scholar]
- Wilson, D.R., and Larkins, B.A. (1984). Zein gene organization in maize and related grasses. J. Mol. Evol. 20, 330–340. [DOI] [PubMed] [Google Scholar]
- Young, T.E., Gallie, D.R., and DeMason, D.A. (1997). Ethylene-mediated programmed cell death during maize endosperm development of wild-type and shrunken2 genotypes. Plant Physiol. 115, 737–751. [DOI] [PMC free article] [PubMed] [Google Scholar]