Abstract
To study the role of initiation codon context in chloroplast protein synthesis, we mutated the three nucleotides immediately upstream of the initiation codon (the −1 triplet) of two chloroplast genes in the alga Chlamydomonas reinhardtii. In prokaryotes, the −1 triplet has been proposed to base pair with either the 530 loop of 16S rRNA or the extended anticodon of fMet-tRNA. We found that in vivo, none of the chloroplast mutations affected mRNA stability. However, certain mutations did cause a temperature-sensitive decrease in translation and a more dramatic decrease at room temperature when combined with an AUU initiation codon. These mutations disrupt the proposed extended base pairing interaction with the fMet-tRNA anticodon loop, suggesting that this interaction may be important in vivo. Mutations that would still permit base pairing with the 530 loop of the 16S rRNA also had a negative effect on translation, suggesting that this interaction does not occur in vivo. Extended base pairing surrounding the initiation codon may be part of a mechanism to compensate for the lack of a classic Shine-Dalgarno rRNA interaction in the translation of some chloroplast mRNAs.
INTRODUCTION
Chloroplast gene expression frequently is regulated at the post-transcriptional level, especially at the steps of RNA processing, stability, and translation (reviewed by Monde et al., 2000; Zerges, 2000). Because of the endosymbiotic origin of the chloroplast, its protein synthesis machinery generally resembles that of prokaryotes (reviewed by Harris et al., 1994; Sugiura et al., 1998). Despite many similarities, however, there are some important differences between chloroplast and prokaryotic translation. Translation of chloroplast mRNAs requires nucleus-encoded translational activators (reviewed by Barkan and Goldschmidt-Clermont, 2000) and ribosomal proteins that have no Escherichia coli counterpart (Yamaguchi and Subramanian, 2000; Yamaguchi et al., 2000). Another striking difference is the lack of functional Shine-Dalgarno (SD) sequences in many chloroplast mRNAs. In E. coli, the SD sequence, located approximately seven nucleotides upstream of the initiation codon, base pairs with a complementary sequence at the 3′ end of the 16S rRNA to facilitate proper ribosome positioning and translation initiation (McCarthy and Brimacombe, 1994). However, only one-third of the higher plant chloroplast genes examined have a SD sequence in the expected position relative to the initiation codon (Bonham-Smith and Bourque, 1989; Sugiura et al., 1998). The remainder have SD-like sequences much farther upstream in the 5′ untranslated region (UTR) or appear to lack them completely. In the unicellular green alga Chlamydomonas reinhardtii, the sequences upstream of the initiation codons are extremely [A+U] rich, and SD-like sequences rarely are present near the initiation codon (Stern et al., 1997). Mutations in most putative SD sequences have little or no effect on chloroplast translation in vivo (Sakamoto et al., 1994; Fargo et al., 1998) or in vitro (Hirose and Sugiura, 1996). In the two cases in which putative SD mutations abolished translation (Mayfield et al., 1994; Hirose et al., 1998), there was no direct evidence that the altered sequences functioned as SD sequences by base pairing with 16S rRNA.
In the absence of a SD element, other cis sequences within chloroplast mRNAs must be required for ribosome positioning and efficient translation initiation. Sequences farther upstream in chloroplast 5′ UTRs are known to be required for translation initiation, probably by interacting with nucleus-encoded translational activators (Sakamoto et al., 1994; Hirose and Sugiura, 1996; Zerges et al., 1997; Higgs et al., 1999; Nickelsen et al., 1999; Ossenbuhl and Nickelsen, 2000). Moreover, sequences downstream of the initiation codon have been shown to affect translational efficiency (Kuroda and Maliga, 2001). However, the role of sequences immediately surrounding the initiation codon has remained enigmatic.
We previously used a molecular genetic approach to address the mechanism of initiation codon recognition in the chloroplast using Chlamydomonas as a model system. By mutating the AUG initiation codon of the petA and petD genes, we found that the site of initiation is fixed but that the initiation codon sequence strongly influences its efficiency (Chen et al., 1993, 1995). To demonstrate that initiation occurred from the usual start site in the petA initiation codon mutants, we mutated the three nucleotides immediately upstream of the initiation codon (the −1 triplet) from AUU to UAA. This mutation had no effect on translation from a wild-type AUG initiation codon but severely decreased translation with an AUU initiation codon (Chen et al., 1995). Here, we have built on this work by examining the effect of the context of the initiation codon on translation initiation.
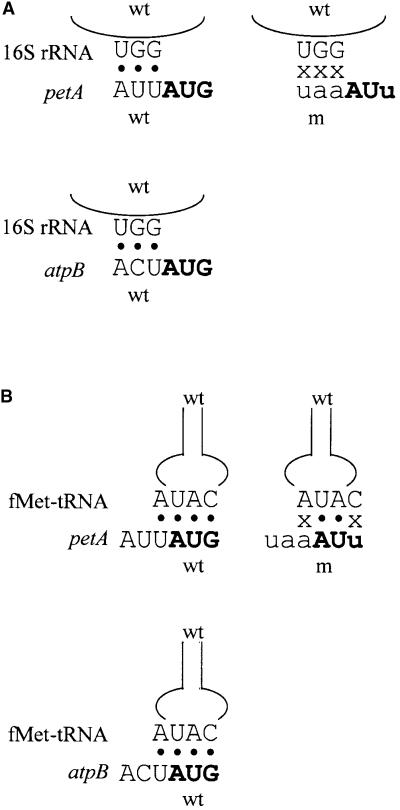
In E. coli, the sequence upstream of the initiation codon has been proposed to interact via base pairing with either of two components of the translational apparatus, and the AUU-to-UAA mutation in petA may have disrupted this interaction. One hypothesis is that the −1 triplet facilitates ribosome positioning by interacting with a universally conserved 5′-GGU-3′ sequence in the 530 loop of 16S rRNA, as shown in Figure 1A (Lagunez-Otero, 1993). This hypothesis is based on sequence bias in the −1 triplet of prokaryotic genes. An alternate idea is that the −1 nucleotide base pairs with the nucleotide downstream of the initiator fMet-tRNA anticodon, as shown in Figure 1B (Eckhardt and Luhrmann, 1981). Although this position is usually a modified adenine or guanine (Sprinzl et al., 1998), the initiator tRNAs of prokaryotes and chloroplasts are notable exceptions because they possess an unmodified adenine (Dube et al., 1968; Sprinzl et al., 1998), which would be available for Watson-Crick base pairing with a uridine upstream of the initiation codon. Evidence for such an extended codon–anticodon interaction comes from in vitro work examining initiation complex formation and dipeptide synthesis in response to short RNAs with an extended complementarity to the fMet-tRNA anticodon loop (Ganoza et al., 1978, 1982; Eckhardt and Luhrmann, 1981) as well as from analysis of bias in prokaryotic −1 sequences (Ganoza et al., 1985).
Figure 1.
Proposed Base Pairing Interactions between Wild-Type or Mutant petA or atpB mRNA and the 16S rRNA 530 Loop or fMet-tRNA Anticodon Loop.
(A) Diagram of the proposed −1 triplet–530 loop interaction. Wild-type (wt) and mutant (m) sequences are in uppercase and lowercase letters, respectively. The initiation codon is in boldface. Black circles indicate a base pair, and x indicates the absence of a base pair.
(B) Diagram of the proposed extended codon–anticodon interaction. Wild-type (wt) and mutant (m) sequences are in uppercase and lowercase letters, respectively. The initiation codon is in boldface. Black circles indicate a base pair, and x indicates the absence of a base pair.
Although in vitro work and sequence analysis have been used to support these two hypotheses, neither has been systematically tested in vivo, either in bacteria or in chloroplasts. Our strategy has been to create mutations in the −1 triplets of two Chlamydomonas chloroplast genes, petA and atpB, that encode cytochrome f and the ATP synthase β-subunit, respectively. These mutations allow varying numbers and positions of base pairings with the Chlamydomonas 530 loop sequence 5′-GGU-3′ (Dron et al., 1982) or the Chlamydomonas fMet-tRNA 5′-CAUA-3′ anticodon loop. Our results do not support the hypothesis of 530 loop pairing, but they do support the presence of an extended codon–anticodon interaction in chloroplast translation initiation. Because this interaction is particularly important in the presence of a mutant initiation codon or at increased temperature, it may have a particular advantage under environmentally stressful conditions.
RESULTS
Generation of petA −1 Triplet Mutants Using Chloroplast Transformation
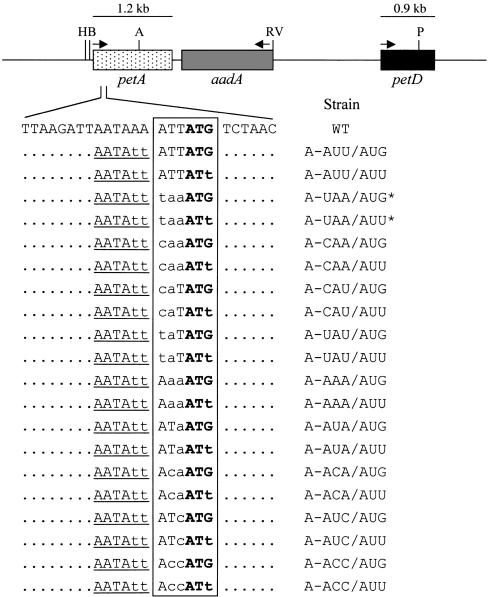
A map of the petA-petD region of the chloroplast genome is shown at the top of Figure 2. Sixteen Chlamydomonas strains carrying one of eight mutations in the −1 triplet of the petA gene and either a wild-type AUG or mutant AUU initiation codon were created by particle bombardment. To facilitate the selection and screening of chloroplast transformants, transforming plasmids included the aadA selectable marker inserted into the petA-petD intergenic region in the antisense orientation, and two A-to-T changes were introduced at positions −4 and −5 to create an SspI site tightly linked to the mutations of interest. Plasmids also were constructed carrying the SspI site but without any −1 triplet mutation (strains A-AUU/AUG and A-AUU/AUU; Figure 2). The antisense orientation was chosen to simplify RNA analysis by avoiding the accumulation of dicistronic petA-aadA transcripts.
Figure 2.
The petA-petD Region of the Chlamydomonas Chloroplast Genome and Initiation Region Mutant Strains Used in This Study.
The petA, aadA, and petD genes are indicated by stippled, gray, and black boxes, respectively. The nucleotides surrounding the petA initiation region are shown, with mutated nucleotides in lowercase letters. Unchanged nucleotides outside of the sequence of interest are shown as dots. The introduced SspI restriction site is underlined. The −1 triplet and initiation codon are boxed, with the initiation codon in boldface. The names of the strains are given at right. Asterisks indicate that strains A-UAA/AUG* and A-UAA/AUU* (Chen et al., 1995) have a spectinomycin resistance mutation in 16S rRNA and lack the aadA gene. Arrows indicate the direction of transcription, and the transcript size is shown above each gene. The petA 5′ UTR is ∼275 nucleotides long (Matsumoto et al., 1991). H, HincII; B, BglII; A, AccI; RV, EcoRV; P, PstI; WT, wild type.
Transformants were initially selected for resistance to spectinomycin and subsequently screened using polymerase chain reaction (PCR) to amplify the petA initiation region, which was then digested with SspI. Initially, transformants were heteroplasmic, containing both wild-type and mutant chloroplast genomes, leading to incomplete digestion of the PCR product. Transformants possessing the SspI site were subjected to single-colony cloning on spectinomycin-containing medium until digestion with SspI was complete, indicating that they were homoplasmic (data not shown). To verify the presence of the desired mutation at the initiation region, sequencing of the petA initiation region was performed after its PCR amplification from total Chlamydomonas DNA (data not shown). The resulting strains, listed on the right side of Figure 2, are designated (pet)A-[−1 triplet sequence]/[initiation codon sequence]. Among these, A-UAA/ AUG* and A-UAA/AUU* were described previously (Chen et al., 1995) and contain a point mutation in the 16S rRNA conferring resistance to spectinomycin (Harris et al., 1989) instead of the aadA cassette. The remainder did contain the aadA cassette and were used to test interactions between the −1 triplet and either the 530 loop sequence or the fMet-tRNA anticodon loop.
Temperature-Sensitive Accumulation of Cytochrome f in Certain petA Mutant Strains with an AUG Initiation Codon
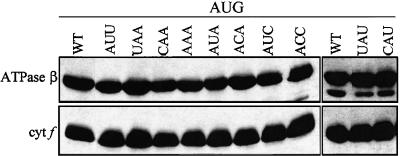
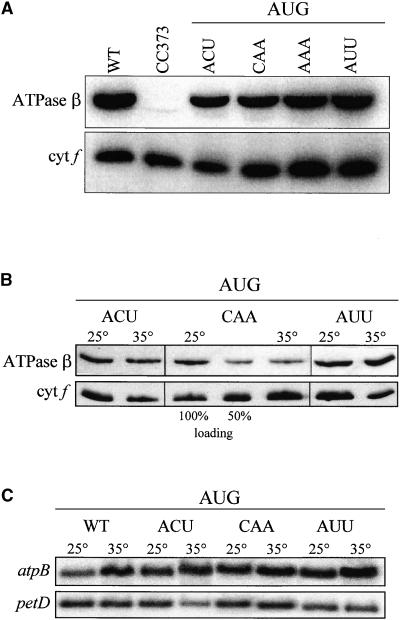
We wished to determine the effect of −1 triplet mutations in the presence of an AUG initiation codon. Because these mutations do not alter the petA coding region, any effect on cytochrome f accumulation can be presumed to result from a change in RNA stability or translation initiation rather than protein stability. To test the first possibility, RNA gel blot analyses were performed, and no decrease in petA RNA accumulation was detected in any of the mutant strains (data not shown). To test the second possibility, immunoblot analyses were performed. Because cytochrome f accumulation invariably correlates with its rate of synthesis (Kuras and Wollman, 1994; Chen et al., 1995), decreased accumulation results from decreased translation initiation. Previous work has shown that the A-UAA/AUG* mutation (with a 16S rRNA mutation instead of aadA as a selectable marker) had no effect on cytochrome f accumulation under normal growth conditions (Chen et al., 1995). Thus, it was not surprising that none of the eight additional mutations had any effect on cytochrome f accumulation with an AUG initiation codon under normal growth conditions (25°C, with Tris-acetate-phosphate [TAP] medium containing acetate as a carbon source), as shown in Figure 3. The −4 and −5 mutations creating the SspI site also had no effect on their own, as shown by the A-AUU/AUG strain (lane AUU).
Figure 3.
Cytochrome f Accumulation in petA −1 Triplet Mutants with an AUG Initiation Codon.
Cells were grown in TAP liquid medium at 25°C, and whole-cell proteins were separated on an SDS–polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and reacted with the antibodies indicated at left. The chloroplast ATPase β-subunit served as a loading control. cyt f, cytochrome f; WT, wild type.
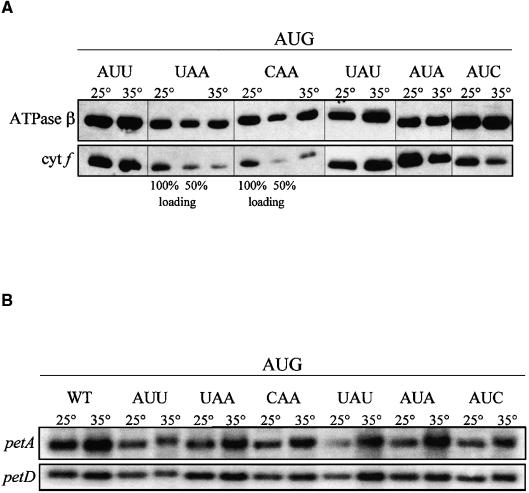
It was of interest to determine if there were other conditions under which an effect of the −1 triplet mutations would become apparent. First, the mutants were grown photoautotrophically in minimal medium lacking acetate. Cytochrome f accumulation was not altered in any of the −1 triplet mutants (data not shown). Next, selected mutants were grown at an increased temperature (35°C) that might destabilize a base pairing interaction in which these nucleotides were involved. Figure 4A shows that growth at 35°C did have an effect on some of the −1 triplet mutants with an AUG initiation codon. There was no decrease in cytochrome f at 35°C with the wild-type AUU −1 triplet or with the mutant UAU −1 triplet. However, cytochrome f accumulation was reduced by ∼50% in the mutants with UAA, CAA, AUA, and AUC −1 triplets, the latter two differing from the wild-type sequence at only the −1 position. Thus, a U at the −1 position appeared to be necessary for a wild-type level of translation at increased temperature, supporting the idea that this nucleotide may base pair with the A downstream of the initiator tRNA anticodon. On the other hand, base pairing with the 530 loop is not supported, because a −1 C could pair with the G in the putative complementary sequence. To ensure that this effect was not caused by a change in RNA stability, RNA gel blot analysis was performed. As shown in Figure 4B, no decrease in petA RNA accumulation, relative to the control petD transcript, was detected in any of the mutant strains at increased temperature.
Figure 4.
Cytochrome f and petA mRNA Accumulation in petA −1 Triplet Mutants with an AUG Initiation Codon Grown at 35°C.
(A) Cells were grown in TAP liquid medium at 35°C. Accumulation of cytochrome f (cyt f) in each strain was compared with accumulation in the same strain grown at 25°C. The ATPase β-subunit served as a loading control. Two dilutions (100 and 50%) of protein from A-UAA/AUG* and A-CAA/AUG grown at 25°C were used to estimate accumulation.
(B) Cells were grown in TAP liquid medium at 35°C, and total RNA was separated on a 1.2% agarose/2.2 M formaldehyde gel, transferred to a nylon membrane, and hybridized with the probes indicated at left. Accumulation of petA in each strain was compared with accumulation in the same strain grown at 25°C. The chloroplast petD transcript served as a loading control. WT, wild type.
Cytochrome f Accumulation Responds to the −1 Triplet in the Context of an AUU Initiation Codon
Our previous work has shown that an AUU initiation codon reduced the synthesis and accumulation of cytochrome f to ∼20% of the wild-type level (Chen et al., 1995). RNA gel blot analysis showed that the accumulation of petA mRNA was not decreased in this strain or in any of the newly constructed mutant strains with the AUU initiation codon (Figure 2 and data not shown). To test the effects of the newly constructed mutations on translation, the accumulation of cytochrome f was determined with the strains grown at 25°C under heterotrophic conditions.
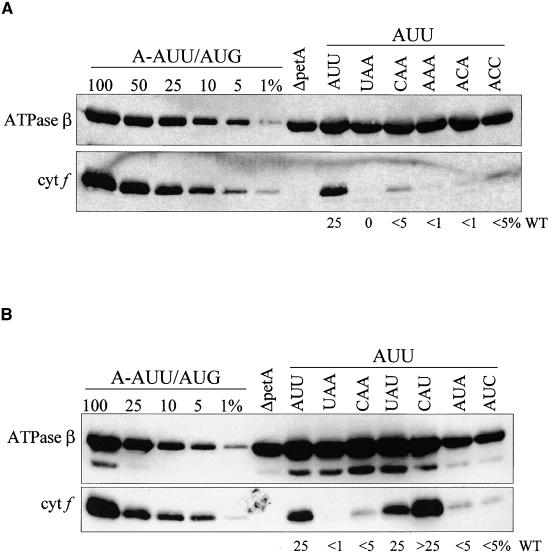
Figure 5A shows an immunoblot with a series of strains designed to test the hypothesis of pairing with the 530 loop sequence 5′-GGU-3′. The mutant petA −1 triplets could theoretically make zero (UAA, CAA), one (AAA), two (ACA), or three (ACC) base pairs (the hypothetically paired positions are underlined). If the 530 loop hypothesis were correct, protein accumulation would increase in accordance with the number of base pairs. However, each mutation reduced the level of cytochrome f from ∼25% of wild type in A-AUU/AUU to <5% (see Methods for quantification information), suggesting that any interaction with the 530 loop does not occur simply by base pairing. Furthermore, strains such as CAA, which could not base pair, had equivalent protein accumulation to ACC, which could make three base pairs. Thus, there is little if any support for the 530 loop hypothesis based on these data.
Figure 5.
Cytochrome f Accumulation in petA −1 Triplet Mutants with an AUU Initiation Codon.
(A) −1 triplet mutants with an AUU initiation codon designed to test 530 loop base pairing were grown in TAP medium at 25°C. Proteins were separated on an SDS–polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and reacted with the antibodies specified at left. The ATPase β-subunit served as a loading control. ΔpetA, a strain lacking the petA gene, served as a negative control. Estimates of cytochrome f (cyt f) accumulation, given at bottom as percentage of wild-type (WT), are based on comparison with a dilution series of proteins from A-AUU/AUG (a wild-type strain) from multiple blots.
(B) −1 triplet mutants with an AUU initiation codon designed to test initiator tRNA base pairing were grown in TAP medium at 25°C, and proteins were analyzed as described for (A).
Figure 5B shows an immunoblot with a series of strains designed to test the hypothesis that the A downstream of the initiator tRNA anticodon base pairs with the −1 position relative to the initiation codon. As for the 530 loop hypothesis, strains UAA and CAA do not have base pairing potential at this position, nor do strains AUC and AUA. However, strains CAU and UAU could pair at the −1 position (the hypothetically paired position is underlined). The results show that in strains UAA, CAA, AUC, and AUA, the cytochrome f level decreases from ∼25% in A-AUU/AUU to <5%. In striking contrast, strains CAU and UAU, which can make the −1 base pair, have an equal (UAU) or even somewhat increased (CAU) level of cytochrome f with respect to A-AUU/AUU. Together, these results support an interaction with the initiator tRNA extended anticodon because it would permit the formation of four contiguous base pairs in the mRNA-tRNA duplex.
Mutations in the atpB −1 Triplet Support an Extended Interaction with the Anticodon Loop and Parallel Those for petA
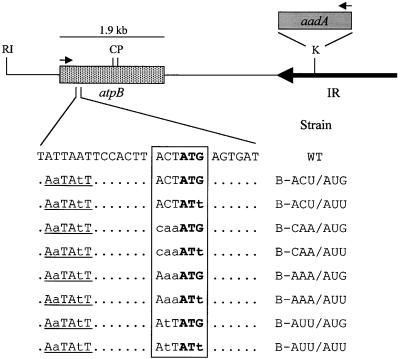
To determine whether the importance of the −1 triplet for translation initiation was specific to petA or if it might be a more general feature of chloroplast translation, a limited set of analogous mutations was made in another chloroplast gene, atpB. The wild-type −1 triplet of atpB is ACU, which could form three base pairs with the 530 loop or support the −1 interaction with the initiator tRNA anticodon loop. As shown in Figure 6, six Chlamydomonas strains carrying one of three mutations in the −1 triplet, along with a wild-type AUG initiation codon or a mutant AUU initiation codon, were created by particle bombardment. As for petA, the aadA selectable marker cassette was inserted downstream of atpB in the antisense orientation (to avoid accumulating dicistronic atpB-aadA transcripts), and an SspI restriction fragment length polymorphism (RFLP) was introduced through mutations at −12 and −15. Control strains carrying the RFLP alone (B-ACU/AUG) or with only an AUU initiation codon mutation (B-ACU/AUU) were described previously (Rimbault et al., 2000).
Figure 6.
The atpB Region of the Chlamydomonas Chloroplast Genome and Initiation Region Mutant Strains Used in This Study.
The atpB and aadA genes are indicated by stippled and gray boxes, respectively. The large inverted repeat (IR) of the chloroplast genome is represented by a horizontal arrow. The nucleotides surrounding the atpB initiation region are shown, with mutated nucleotides in lowercase letters. Unchanged nucleotides outside of the sequence of interest are shown as dots. The introduced SspI restriction site is underlined. The −1 triplet and initiation codon are boxed, with the initiation codon in boldface. The names of the strains are given at right. Arrows indicate the direction of transcription, and the transcript size is shown above each gene. The atpB 5′ UTR is ∼350 nucleotides long (Blowers et al., 1990). RI, EcoRI; C, ClaI; P, PstI; K, KpnI; WT, wild type.
After transformation, colonies were selected for resistance to spectinomycin and subsequently screened for the presence of the SspI site. To verify the presence of the desired mutation in the initiation region, sequencing of PCR-amplified Chlamydomonas DNA was performed. The resultant strains are listed at the bottom of Figure 6 and designated (atp)B-[−1 triplet sequence]/[initiation codon sequence].
To determine if the mutations in the atpB −1 triplet affected transcript stability, RNA gel blot analysis was performed. As for the petA mutants, there was no significant change in mRNA accumulation (data not shown). Subsequently, immunoblot analysis was performed to determine if the −1 triplet mutations affected translation. Figure 7A shows that ATPase β-subunit accumulation in strain B-ACU/AUG (lane ACU) was slightly reduced compared with the wild-type control, indicating that one or both of the point mutations at −12 and −15 and/or the presence of the aadA gene reduced translation. However, no further reduction relative to B-ACU/AUG was seen with strains containing −1 triplet mutations with an AUG initiation codon when grown heterotrophically at 25°C. This agrees with results obtained for petA. The atpB strains shown in Figure 7A were tested subsequently at 35°C, as shown in Figure 7B. No decrease in ATPase β-subunit accumulation was seen at 35°C with the wild-type ACU −1 triplet or the mutant AUU −1 triplet. However, its accumulation with the CAA −1 triplet was reduced by at least 50% at 35°C. To ensure that the differences in protein level were not attributable to changes in RNA stability, RNA gel blot analysis was performed, as shown in Figure 7C. No decrease in atpB mRNA was observed at high temperature relative to the control petD transcript. Thus, these results parallel the petA results and support the hypothesis that a U at −1 is important for efficient translation initiation.
Figure 7.
ATPase β-Subunit and atpB mRNA Accumulation in −1 Triplet Mutants with an AUG Initiation Codon Grown at 25 or 35°C.
(A) −1 triplet mutants with AUG initiation codons were grown in TAP liquid medium at 25°C, and whole-cell proteins were separated on an SDS–polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, reacted with the indicated antibodies and 125I-protein A, and visualized with a PhosphorImager. Cytochrome f (cyt f) served as a loading control. WT, wild type.
(B) Cells were grown in TAP liquid medium at 35°C. Accumulation of ATPase β-subunit in each strain was compared with accumulation in the same strain grown at 25°C. Cytochrome f (cyt f) served as a loading control. Dilutions (100 and 50%) of protein from B-CAA/AUG grown at 25°C were used to estimate accumulation.
(C) Cells were grown in TAP liquid medium at 35°C, and total RNA was separated on a 1.2% agarose/2.2 M formaldehyde gel, transferred to a nylon membrane, and hybridized with the probes indicated at left. Accumulation of petA in each strain was compared with accumulation in the same strain grown at 25°C. The chloroplast petD transcript served as a loading control. WT, wild type.
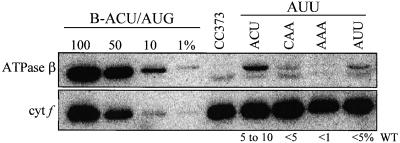
Figure 8 shows the accumulation of the ATPase β-subunit in the −1 triplet mutants possessing an AUU initiation codon. Two immunoreactive proteins were seen in each sample, the faster migrating of which also was present in the sample from strain CC373, in which the majority of the atpB coding region is deleted. Although the precise end points of the deletion are not known, it is highly unlikely that this species represents a genuine atpB-encoded species, especially because the other strains possess a full-length coding region. Therefore, this lower band was discounted. In terms of the upper band, presumed to be bona fide β-subunit, the level in strain B-ACU/AUU (ACU triplet) was 5 to 10% of the wild-type level, somewhat less than the level in the analogous petA and petD mutants. Among the remaining strains, each had less β-subunit than B-ACU/AUU. The greatest amount was in the strain with an AUU triplet, with additional reductions in strains CAA and AAA. The apparent preference for U in the −1 position is in agreement with results for petA, although the overall low accumulation made precise quantification problematic.
Figure 8.
ATPase β-Subunit Accumulation in atpB −1 Triplet Mutants with an AUU Initiation Codon.
−1 triplet mutants with AUU initiation codons were grown in TAP medium at 25°C, and proteins were analyzed as described for Figure 7A. Estimates of protein accumulation given at bottom are based on quantification of 125I-protein A signal from several blots using a PhosphorImager. cyt f, cytochrome f; WT, wild type.
DISCUSSION
We have shown that mutations in the sequence immediately upstream of the initiation codons of two chloroplast mRNAs, petA and atpB, can have remarkable effects on translation initiation under stress conditions or when the initiation codon itself has been weakened by mutation. Two alternative hypotheses were tested in this study: an interaction between the −1 triplet and the 530 loop of 16S rRNA, and an extended codon–anticodon interaction between the mRNA and the initiator tRNA. Both scenarios had been evoked previously for E. coli translation (Eckhardt and Luhrmann, 1981; Lagunez-Otero, 1993); however, neither had been tested by mutation of the rRNA or tRNA to restore complementarity to mutated mRNAs. Our unpublished results, using a strategy that parallels the strategy originally used to define interactions such as that between the SD element and the 3′ end of 16S rRNA (Hui and de Boer, 1987), also favor an extended interaction with the initiator tRNA. Together, our results best support an extended codon–anticodon interaction that facilitates selection of the correct initiation codon, at least for these two genes in the Chlamydomonas chloroplast.
In our study, a sequence with U at the −1 position was the favored sequence, which we suggest permits base pairing with an unmodified A downstream of the anticodon. Sequencing of this species from Chlamydomonas, spinach, and bean chloroplasts revealed an unmodified A (Sprinzl et al., 1998), in contrast to the N6-threonylcarbamoyladenosine in the bean cytoplasmic species (Sprinzl et al., 1998), which could not form a Watson-Crick base pair. Our observations support previous in vitro measurements of E. coli initiation complex formation, from which it was reported that recruitment of fMet-tRNA and dipeptide synthesis was most favorable in response to the tetranucleotide UAUG (Ganoza et al., 1978, 1982; Eckhardt and Luhrmann, 1981). Expression studies of reporter constructs or foreign genes in E. coli also have shown that a U at the −1 position has a favorable effect on translation (Hui et al., 1984; Gross et al., 1990; Morelle et al., 1991). Moreover, sequence analysis has shown that E. coli AUG initiation codons are more frequently preceded by a U than internal AUG codons (Ganoza et al., 1985). Furthermore, of the chloroplast genes known to be expressed in Chlamydomonas, 78% possess a U at the −1 position. The lack of a −1 U in the other chloroplast genes might have a regulatory role or might be compensated by another mechanism, such as a classic SD interaction, as in the case of psbA (Mayfield et al., 1994).
Although our results are most consistent with an extended codon–anticodon interaction, this does not account for any effects of −2 or −3 mutations. For example, translation in the petA and atpB CAA mutants was consistently higher than in the AAA mutants at 25°C with an AUU initiation codon. Moreover, cytochrome f expression in A-CAU/AUU was higher than in A-UAU/AUU, although both possess a U at −1. Thus, C at −3 appears to affect petA translation positively, a result that is consistent with mutagenesis of the −1 triplet in E. coli (Hui et al., 1984). Our results also parallel in vitro initiation complex formation measured by binding of fMet-tRNA in response to the RNA hexamers CAU/AUG, AAA/AUG, and UAA/AUG (Ganoza et al., 1978). Initiator tRNA recruitment was half as much with AAA/AUG than with CAU/AUG, whereas initiator tRNA recruitment with UAA/AUG was only 20% of that with CAU/AUG.
The −2 nucleotide affected initiation for atpB, exemplified by the slightly better translation of B-ACU/AUU compared with that of B-AUU/AUU. It was surprising that the AUU −1 triplet reduced translation, because that is the wild-type −1 triplet sequence for petA. Thus, although our data suggest that the −1 triplet is likely to be generally important in translation initiation, the optimal sequence may be context dependent. Because Hui et al. (1984) found a role for the −2 and −3 nucleotides as well as for the −1 U, they concluded that an extended codon–anticodon interaction was unlikely to have a major effect on translation and proposed that the −1 triplet interacts with another component of the translational apparatus, such as a ribosomal protein. Although our results best support the extended codon–anticodon interaction as the major role of this sequence, we cannot rule out the possibility that the −1 triplet interacts concurrently with a ribosomal protein or another region of the rRNA or also functions during another step of initiation complex formation.
Clearly, each chloroplast 5′ UTR has a unique set of cis elements that together determine the efficiency of translation initiation. In this study, the RFLP mutation at positions −12 and −15 in the atpB 5′ UTR appears to reduce translation somewhat, indicating that sequence is important for initiation, although we cannot rule out the aadA cassette as the cause of reduced translation. Moreover, the varying effects of AUU initiation codon mutations on the translational efficiency of different mRNAs are most likely the result of a specific combination of cis elements. Translation of mRNAs bearing an AUU initiation codon in Chlamydomonas ranges from a high of 25 to 45% for clpP (Majeran et al., 2000) to an undetectable level for psbD (Nickelsen et al., 1999). It is tempting to propose that the higher levels for clpP, petD (Chen et al., 1993), petA (Chen et al., 1995), and atpB (this study) are caused by the presence of a U at the −1 position, whereas the undetectable level observed in psbD AUU (Nickelsen et al., 1999) is caused by the presence of an A at this position. However, the presence of other cis elements and the requirement for nucleus-encoded translation factors suggest that such a simple explanation is unlikely.
METHODS
Construction of petA Transforming Plasmids
The plasmid pHcRV1.6 contains the entire petA gene within a 1.6-kb HincII-EcoRV subfragment of BamHI fragment 7 of the Chlamydomonas reinhardtii chloroplast genome (Harris, 1989) inserted into pBluescript SK+. Site-directed mutagenesis (Kunkel, 1985) was performed on pHcRV1.6 to produce mutations at the initiation region of petA. In each case, an SspI site was introduced upstream of the petA initiation codon by mutating the nucleotides at −4 and −5. After mutagenesis, larger plasmids containing these mutations were obtained by replacing the BglII-AccI fragment of the pQMAD plasmid containing the petA initiation region with the mutant BglII-AccI fragments. pQMAD contains an ∼6-kb XbaI-HindIII chloroplast DNA fragment with the aadA gene (Goldschmidt-Clermont, 1991) inserted in the EcoRV site between the petA and petD genes to serve as a selectable marker for chloroplast transformation. As controls, plasmids containing the mutations A-AUU/AUG and A-AUU/AUU were constructed in the same way using the appropriate pHcRV1.6 site-directed mutants as described previously (Chen et al., 1995).
Construction of atpB Transforming Plasmids
The plasmid pAtpB-EP1.8 contains the 5′ end of the atpB gene on a 1.8-kb EcoRI-PstI subfragment of BamHI fragment 10 of the Chlamydomonas chloroplast genome (Harris, 1989) inserted into pBluescript SK−. Site-directed mutagenesis was performed on pAtpB-EP1.8 to produce mutations at the initiation region of atpB. In each case, an SspI site was introduced upstream of the initiation codon by mutating the nucleotides at −12 and −15. After mutagenesis, larger plasmids carrying these mutations were obtained by replacing the EcoRI-ClaI fragment of the plasmid pDAAD with a mutant EcoRI-ClaI fragment. pDAAD contains the aadA gene (Goldschmidt-Clermont, 1991) inserted in the KpnI site of the inverted repeat downstream of the atpB gene (Rott et al., 1996).
Chlamydomonas Strains, Growth Conditions, and Chloroplast Transformation
P17 (Stern et al., 1991) is a wild-type Chlamydomonas strain derived from CC373 (mt+, ac-u-c-2-21; Shepherd et al., 1979) by bombardment with the atpB gene. P17 was used as the recipient strain to create all of the petA mutants and the atpB mutants with an AUG initiation codon. CC373 was used to create all of the atpB mutants with an AUU initiation codon. The ΔpetA strain was provided by Francis-André Wollman (Institute de Biologie Physico-Chimique, Paris, France) (Kuras and Wollman, 1994). A-UAA/AUG* and A-UAA/AUU* (Chen et al., 1995) and B-ACU/AUG and B-ACU/AUU were described previously (Rimbault et al., 2000). Cells were grown in Tris-acetate-phosphate (TAP) medium or minimal medium at 25°C (Harris, 1989) under constant light (70 μE·m−2·sec−1). To test temperature sensitivity, petA and atpB AUG strains were grown at 35°C in TAP medium.
petA and atpB mutations were introduced into the chloroplast genome by tungsten particle bombardment (Kindle et al., 1991). Transformants expressing the aadA cassette were selected on TAP medium containing spectinomycin (100 μg/mL).
Polymerase Chain Reaction and DNA Sequencing
Polymerase chain reaction (PCR) was used to screen spectinomycin-resistant chloroplast transformants for the presence of the restriction site indicative of the mutation of interest, as described previously (Higgs et al., 1998). To screen putative petA transformants, the initiation region was amplified with primers petA2 and petA5 (Chen et al., 1995). The resulting 680-bp petA PCR product was digested with SspI and analyzed by gel electrophoresis. Putative atpB transformants were screened in a similar manner using primers DBS2 and NS1B (Levy et al., 1997) to amplify a 1017-bp fragment containing the initiation region.
After site-directed mutagenesis, the desired mutants were identified by manual sequencing of plasmid DNA using the dideoxy chain terminator method (Sambrook et al., 1989). Chloroplast transformants that had gained the SspI site were verified by direct sequencing of the PCR products used for restriction analysis. PCR products were sequenced either manually by the dideoxy chain terminator method from low-melting-point agarose gels (Chang et al., 1993) or by cycle sequencing with the ABI Prism 310 Big Dye terminator kit (PE-Applied Biosystems, Foster City, CA).
Isolation of Nucleic Acids and Filter Hybridization
Plasmid DNA was isolated from Escherichia coli by alkaline lysis miniprep (Sambrook et al., 1989; Zhou et al., 1990) or using a Qiagen (Valencia, CA) plasmid miniprep kit. Total Chlamydomonas DNA and RNA isolation, electrophoresis, blotting, and hybridization were performed as described previously (Drager et al., 1998). α-32P-dCTP–labeled linear, double-stranded DNA probes were made as described previously (Drager et al., 1998). The 680-bp petA2/petA5 PCR product was used to detect the petA transcript, and the 1-kb DBS2/NS1B PCR product was used to detect atpB mRNA. The petD transcript was detected with an ∼900-bp PCR product using the primers WS4 and WS6 (Chen et al., 1993), and atpH was detected using the 291-bp AflIII-ClaI fragment from plasmid pR8 (Drapier et al., 1998). Detection and quantification of 32P labeling were performed using a PhosphorImager or Storm Scanner and ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Total Protein Preparation and Immunoblotting
Total proteins were prepared as described previously (Drager et al., 1998). SDS-PAGE, electroblotting, and reaction with antibodies were performed as described previously (Higgs et al., 1998). Antibodies raised against the Chlamydomonas chloroplast ATP synthase β-subunit (1:2000 dilution) were provided by F.-A. Wollman, and the anti-cytochrome f antibody (1:4000 dilution) was described previously (Chen et al., 1995). After incubation with the primary antibodies, blots were either reacted with 125I-protein A or reacted with an anti-rabbit IgG horseradish peroxidase conjugate antibody (Promega; 1:2500 dilution) and visualized using a PhosphorImager (Molecular Dynamics) or by enhanced chemiluminescence (Durant, 1990), respectively. Quantification was performed with the ImageQuant software and/or by comparison with a dilution series of wild-type protein. Each immunoblot procedure was performed multiple times with multiple protein preparations and several independent transformants for each mutation.
GenBank Accession Number
The GenBank accession number for the Chlamydomonas fMet-tRNA is X13870. The mature tRNA is from positions 3757 to 3829.
Acknowledgments
We thank Xuemei Chen, Karen Kindle, Olivier Vallon, Rob Drager, David Higgs, and other members of the Stern laboratory for useful discussions. This work was supported by National Science Foundation Grants MCB-9723274 and MCB-9896397 to D.B.S. D.E. was supported by a Howard Hughes Medical Institute Predoctoral Fellowship.
References
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Blowers, A.D., Ellmore, G.S., Klein, U., and Bogorad, L. (1990). Transcriptional analysis of endogenous and foreign genes in chloroplast transformants of Chlamydomonas. Plant Cell 2, 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonham-Smith, P.C., and Bourque, D.P. (1989). Translation of chloroplast-encoded messenger RNA: Potential initiation and termination signals. Nucleic Acids Res. 17, 2057–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C., Kwok, S.F., Bleecker, A.B., and Meyerowitz, E.M. (1993). Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 262, 539–544. [DOI] [PubMed] [Google Scholar]
- Chen, X., Kindle, K., and Stern, D. (1993). Initiation codon mutations in the Chlamydomonas chloroplast petD gene result in temperature-sensitive photosynthetic growth. EMBO J. 12, 3627–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Kindle, K.L., and Stern, D.B. (1995). The initiation codon determines the efficiency but not the site of translation initiation in Chlamydomonas chloroplasts. Plant Cell 7, 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drager, R.G., Girard-Bascou, J., Choquet, Y., Kindle, K.L., and Stern, D.B. (1998). In vivo evidence for 5′→3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J. 13, 85–96. [DOI] [PubMed] [Google Scholar]
- Drapier, D., Suzuki, H., Levy, H., Rimbault, B., Kindle, K.L., Stern, D.B., and Wollman, F.-A. (1998). The chloroplast atpA gene cluster in Chlamydomonas reinhardtii: Functional analysis of a polycistronic transcription unit. Plant Physiol. 117, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron, M., Rahire, M., and Rochaix, J.-D. (1982). Sequence of the chloroplast 16S rRNA gene and its surrounding regions of Chlamydomonas reinhardtii. Nucleic Acids Res. 10, 7609–7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube, S.K., Marcker, K.A., Clark, B.F., and Cory, S. (1968). Nucleotide sequence of N-formyl-methionyl-transfer RNA. Nature 218, 232–233. [DOI] [PubMed] [Google Scholar]
- Durant, I. (1990). Light-based detection on biomolecules. Nature 346, 297–298. [DOI] [PubMed] [Google Scholar]
- Eckhardt, H., and Luhrmann, R. (1981). Recognition by initiator transfer ribonucleic acid of a uridine 5′ adjacent to the AUG codon: Different conformational states of formylatable methionine-accepting transfer ribonucleic acid at the ribosomal peptidyl site. Biochemistry 20, 2075–2080. [DOI] [PubMed] [Google Scholar]
- Fargo, D.C., Zhang, M., Gillham, N.W., and Boynton, J.E. (1998). Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol. Gen. Genet. 257, 271–282. [DOI] [PubMed] [Google Scholar]
- Ganoza, M.C., Fraser, A.R., and Neilson, T. (1978). Nucleotides contiguous to AUG affect translational initiation. Biochemistry 17, 2769–2775. [DOI] [PubMed] [Google Scholar]
- Ganoza, M.C., Sullivan, P., Cunningham, C., Hader, P., Kofoid, E.C., and Neilson, T. (1982). Effect of bases contiguous to AUG on translation initiation. J. Biol. Chem. 257, 8228–8232. [PubMed] [Google Scholar]
- Ganoza, M.C., Marliere, P., Kofoid, E.C., and Louis, B.G. (1985). Initiator tRNA may recognize more than the initiation codon in mRNA: A model for translational initiation. Proc. Natl. Acad. Sci. USA 82, 4587–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1991). Transgenic expression of aminoglycoside adenine transferase in the chloroplast: A selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res. 19, 4083–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, G., Mielke, C., Hollatz, I., Blocker, H., and Frank, R. (1990). RNA primary sequence or secondary structure in the translational initiation region controls expression of two variant interferon-beta genes in Escherichia coli. J. Biol. Chem. 265, 17627–17636. [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego: Academic Press). [DOI] [PubMed]
- Harris, E.H., Burkhart, B.D., Gillham, N.W., and Boynton, J.D. (1989). Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: Correlation of genetic and physical maps of the chloroplast genome. Genetics 123, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.H., Boynton, J.E., and Gillham, N.W. (1994). Chloroplast ribosomes and protein synthesis. Microbiol. Rev. 58, 700–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, D.C., Kuras, R., Kindle, K.L., Wollman, F.A., and Stern, D.B. (1998). Inversions in the Chlamydomonas chloroplast genome suppress a petD 5′ untranslated region deletion by creating functional chimeric mRNAs. Plant J. 14, 663–671. [DOI] [PubMed] [Google Scholar]
- Higgs, D.C., Shapiro, R.S., Kindle, K.L., and Stern, D.B. (1999). Small cis-acting sequences that specify secondary structures in a chloroplast mRNA are essential for RNA stability and translation. Mol. Cell. Biol. 19, 8479–8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, T., and Sugiura, M. (1996). Cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: Development of an in vitro translation system from tobacco chloroplasts. EMBO J. 15, 1687–1695. [PMC free article] [PubMed] [Google Scholar]
- Hirose, T., Kusumegi, T., and Sugiura, M. (1998). Translation of tobacco chloroplast rps14 mRNA depends on a Shine-Dalgarno-like sequence in the 5′-untranslated region but not on internal RNA editing in the coding region. FEBS Lett. 430, 257–260. [DOI] [PubMed] [Google Scholar]
- Hui, A., and de Boer, H.A. (1987). Specialized ribosome system: Preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. USA 84, 4762–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, A., Hayflick, J., Dinkelspiel, K., and de Boer, H.A. (1984). Mutagenesis of the three bases preceding the start codon of the beta-galactosidase mRNA and its effect on translation in Escherichia coli. EMBO J. 3, 623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle, K.L., Richards, K.L., and Stern, D.B. (1991). Engineering the chloroplast genome: Techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 88, 1721–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, T.A. (1985). Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, R., and Wollman, F.-A. (1994). The assembly of cytochrome b6/f complexes: An approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 13, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, H., and Maliga, P. (2001). Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol. 125, 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunez-Otero, J. (1993). rRNA-mRNA complementarity: Implications for translation initiation. Trends Biochem. Sci. 18, 406–408. [DOI] [PubMed] [Google Scholar]
- Levy, H., Kindle, K.L., and Stern, D.B. (1997). A nuclear mutation that affects the 3′ processing of several mRNAs in Chlamydomonas chloroplasts. Plant Cell 9, 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran, W., Wollman, F.A., and Vallon, O. (2000). Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b(6)f complex. Plant Cell 12, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, T., Matsuo, M., and Matsuda, Y. (1991). Structural analysis and expression during dark-light transitions of a gene for cytochrome f in Chlamydomonas reinhardtii. Plant Cell Physiol. 32, 863–872. [Google Scholar]
- Mayfield, S.P., Cohen, A., Danon, A., and Yohn, C.B. (1994). Translation of the psbA mRNA of Chlamydomonas reinhardtii requires a structured RNA element contained within the 5′ untranslated region. J. Cell Biol. 127, 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, J.E.G., and Brimacombe, R. (1994). Prokaryotic translation: The interactive pathway leading to initiation. Trends Genet. 10, 402–407. [DOI] [PubMed] [Google Scholar]
- Monde, R.A., Schuster, G., and Stern, D.B. (2000). Processing and degradation of chloroplast mRNA. Biochimie 82, 573–582. [DOI] [PubMed] [Google Scholar]
- Morelle, G., Frank, R., and Meyerhans, A. (1991). Restructuring the translation initiation region of the human parathyroid hormone gene for improved expression in Escherichia coli. Biochim. Biophys. Acta 1089, 320–324. [DOI] [PubMed] [Google Scholar]
- Nickelsen, J., Fleischmann, M., Boudreau, E., Rahire, M., and Rochaix, J.-D. (1999). Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell 11, 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenbuhl, F., and Nickelsen, J. (2000). Cis- and trans-acting determinants for translation of psbD mRNA in Chlamydomonas reinhardtii. Mol. Cell. Biol. 20, 8134–8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimbault, B., Esposito, D., Drapier, D., Choquet, Y., Stern, D., and Wollman, F.A. (2000). Identification of the initiation codon for the atpB gene in Chlamydomonas chloroplasts excludes translation of a precursor form of the beta subunit of the ATP synthase. Mol. Gen. Genet. 264, 486–491. [DOI] [PubMed] [Google Scholar]
- Rott, R., Drager, R.G., Stern, D.B., and Schuster, G. (1996). The 3′ untranslated regions of chloroplast genes in Chlamydomonas reinhardtii do not serve as efficient transcriptional terminators. Mol. Gen. Genet. 252, 676–683. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Chen, X., Kindle, K.L., and Stern, D.B. (1994). Function of the Chlamydomonas reinhardtii petD 5′ untranslated region in regulating the accumulation of subunit IV of the cytochrome b6/f complex. Plant J. 6, 503–512. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed, Vol. 2. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Shepherd, H.S., Boynton, J.E., and Gillham, N.W. (1979). Mutations in nine chloroplast loci of Chlamydomonas affecting photosynthetic functions. Proc. Natl. Acad. Sci. USA 76, 1353–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. (1998). Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, D.B., Radwanski, E.R., and Kindle, K.L. (1991). A 3′ stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell 3, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, D.B., Higgs, D.C., and Yang, J. (1997). Transcriptional and translational activities of chloroplasts. Trends Plant Sci. 2, 308–315. [Google Scholar]
- Sugiura, M., Hirose, T., and Sugita, M. (1998). Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 32, 437–459. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K., and Subramanian, A.R. (2000). The plastid ribosomal proteins: Identification of all the proteins in the 50S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275, 28466–28482. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K., von Knoblauch, K., and Subramanian, A.R. (2000). The plastid ribosomal proteins: Identification of all the proteins in the 30S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275, 28455–28465. [DOI] [PubMed] [Google Scholar]
- Zerges, W. (2000). Translation in chloroplasts. Biochimie 82, 583–601. [DOI] [PubMed] [Google Scholar]
- Zerges, W., Girard-Bascou, J., and Rochaix, J.D. (1997). Translation of the chloroplast psbC mRNA is controlled by interactions between its 5′ leader and the nuclear loci TBC1 and TBC3 in Chlamydomonas reinhardtii. Mol. Cell. Biol. 17, 3440–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C., Yang, Y., and Jong, A.Y. (1990). Mini-prep in ten minutes. Biotechniques 8, 172–173. [PubMed] [Google Scholar]