Much of modern biological research is concerned with identifying genes and the protein products of genes involved in cellular processes: determining how, when, and where they are involved in specific biochemical processes. The tools by which these aims are achieved can be roughly divided into two types: those that are specific to the study of individual or classes of proteins versus those with more general and broad utility. General tools are methods that allow for rapid inference of function of a gene product, either from the mRNA or the proteins it codes for, for any particular molecule or class of molecules. These are increasingly in demand, particularly those that can be applied to entire genomes or large subsets of the genes contained therein. At the same time, generality comes at a cost. By their nature, general tools do not usually provide high-quality information about the function of a gene and may even mislead, particularly when applied across large numbers of genes. Examples include DNA microarrays and multidimensional separation-MS and yeast two-hybrid strategies that detect protein–protein interactions or complexes (1–9). Although these approaches can, respectively, provide information on whether and to what extent a given gene is being transcribed in a defined condition and with which proteins the protein gene product is interacting with, they cannot provide any insight into other crucial questions. For instance, many proteins are enzymes that catalyze biochemical reactions. In the case of novel genes of unknown function it is not easy to definitively determine whether they are enzymes nor what reactions they may catalyze. In this issue of PNAS Baker et al. (10) present a proof-of-principle study on another approach that might fulfill this need, but unlike the approaches described previously, the strategy is applied in intact, living cells. Here we discuss the significance of this distinction, the historical context of the development of the method, and potential applications to the problems of studying gene function and to another class of problems: designing proteins that embody desired characteristics or function of potential industrial, therapeutic, or fundamental interest.
An assay for detection of an active form of a cephalosporinase of Enterobacter cloacae called P99 is presented.
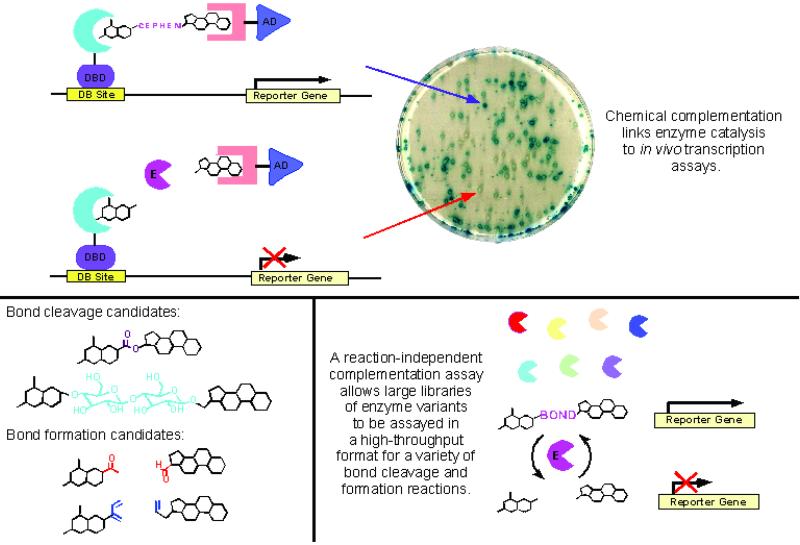
The essential elements of the strategy Baker et al. (10) describe are variations on two successful and popular technologies that have emerged in the last 10 years, namely the yeast three-hybrid assay and protein “dimerizer” technology (11–20). The basis of their approach is described in Fig. 1. In the general concept, a chimeric molecule (the dimerizer) that contains three components is synthesized: one moiety that is a high-affinity ligand for one protein, one that is a ligand for another protein, and a linker that contains a substrate for some specific or general class of enzymes. The dimer is introduced into yeast containing a two-hybrid transcriptional reporter assay system consisting of the two proteins to which the dimerizer binds, fused to complementary DNA binding and RNA polymerase activating domains, respectively. The dimerizer binds to the two proteins simultaneously, allowing for transcription of a reporter gene whose presence can be detected by enzymatic assays. The two dimerizing proteins are dihydrofolate reductase and glucocortocoid receptor ligand-binding domains. These are fused to LexA DNA binding and B42 RNA polymerase activation domains, respectively. In the specific case presented by Baker et al. (10), an assay for detection of an active form of a cephalosporinase of Enterobacter cloacae called P99 is presented. Thus, the dimerizer consists of methotrexate linked via a thioether to the β-lactam cephalosporin and in turn, to dexamethasone by a peptide bond (Mtx-cephem-Dex). When these proteins are expressed in the budding yeast Saccharomyces cerevisiae grown on medium containing an appropriate concentration of the dimerizer, simultaneous binding of dihydrofolate reductase-LexA to the Mtx moiety and of glucocortocoid receptor-B42 to the Dex moiety of Mtx-cephem-Dex, results in reconstitution of an active LexA promoter and transcription of the β-galactosidase reporter gene. The activity of the β-galactosidase gene product is detected by using substrates (5-bromo-4-chloro-3-indolyl β-d-galactoside and o-nitrophenyl-d-β-galactoside) that are converted to colored or fluorescent products by the β-galactosidase. Detection of β-lactamase activity is based on a loss of expression of β-galactosidase if the β-lactone ring is cleaved by a lactamase, resulting in expulsion of the leaving group at the C3′ position of the cephalosporin and therefore disintegration of the dimerizer induced dihydrofolate reductase-LexA-glucocortocoid receptor-B42 complex. Thus, a loss of activity indicates that a lactamase activity is present in the cell. Baker et al. tested the system by screening an artificial library of P99 the cephalosporinase containing either active WT or inactive mutant forms and demonstrate that selection for active enzyme can be reliably made.
Fig 1.
A reaction-independent complementation assay for detecting enzyme activities of genes or artificial libraries of genes. (Upper) A specific “three-hybrid” assay for detecting lactamase activity. The two dimerized proteins consist of dihydrofolate reductase (rose) and glucocortocoid receptor ligand-binding domains (blue). These are fused to LexA DNA binding and B42 RNA polymerase activation domains and will reconstitute active RNA polymerase complex to transcribe the reporter gene β-galactosidase when the two proteins are noncovalently ligated via the dimerizer dexamethasone-cephem-methotrexate molecule. (Lower Left) Examples of ligation or cleavage reactions that could be used in the assay. (Lower Right) Application of the assay to protein engineering. Figure was provided by Debleena Sengupta and Virginia Cornish (Columbia University, New York).
The Baker technology represents a first step in creating generalized detectors of enzyme activities in cells. It is a potentially valuable tool in proteomic and protein engineering for the discovery of novel enzymatic activities. There are, as the authors point out, many variations on this theme that could be devised. The dimerizer could contain general or specific substrates for many cleavage or ligation reactions, aimed at different classes of enzymes. Previously, a system in bacteria had been described based on AraC chimera dimerizer-regulated transcription to screen for dehydratase activity (21). The different basis of the dimerizer chemistry make these approaches complementary and might allow the screening of a larger set of enzymatic reactions. Second, reporter assays that are not limited to a cellular compartment or specific cell type could be used. For instance, a recent example of a fluorescence resonance energy transfer (FRET) assay for detecting specific protein kinase activities has been described, and such a FRET-based assay could also be used with a dimerizer system (22, 23). Equally, several protein fragment complementation assays could be used. These are designed to detect protein–protein interactions but small molecule-induced dimerization of proteins have been demonstrated with these techniques, and in addition to detecting enzyme activities, some of these assays could be used to sublocalize these precisely inside the cells (24–28). However, one important limitation of enzyme activity assays for proteomic or drug discovery is that the enzymatic activity has to be studied in a cell model devoid or weakly displaying similar enzymatic activity for the screen to be easily tractable. The combination of these technologies in different cell types could expand the nature of enzymatic reactions assayable.
Elegant strategies to allow for in vitro covalent tagging of specific enzymes with, for examples, biotin and fluorescent dyes have been developed. Ben Cravatt and coworkers at Scripps (29–34) have developed a number of experimental strategies that allow for identification of protease and other enzyme activities in different cell types under different circumstances such as in different types of cancer cells. As demonstrated, it allowed for phenotyping of these cells for invasiveness. With increased knowledge of the chemistry of diverse enzymatic activities, one could conceivably develop compounds for every biologically catalyzed reaction. Thus, the strategy described by Baker et al. (10) would be complementary to these other approaches in a proteomic perspective.
How can these new methods aid in large-scale gene discovery? We think that the process of ontologically defining each gene of an organism could be envisioned in a three-step strategy going from general to more specific considerations. The first step consists of linking the gene of interest to all of the cellular processes it might be involved in, as inferred, for example, from the complete set of physical interactions map obtained from a large-scale genomic screen with methods such as yeast two-hybrid assays and MS or combinations of DNA and protein microarray data (2, 4–6, 35–38). The next step consists of the validation of the functional inference made with the results of the large-scale experiments. Generally speaking these can be attained by comparing the phenotype observed, whether through classical genetic approaches, RNAi, or newly emerging technologies, between the gene products thought to be involved in interactions network with the gene under study and this gene. This kind of comparison could then be expanded to the pattern observed in subcellular localizations and expression profiles and to genes involved in the inferred functional class, but that have not been shown to interact with the gene of interest. However, and, this is the last step, the addition of more specific assays for assigning function to genes would be a huge step toward completion of this goal. The new enzyme discovery tool described by Baker et al. would add an extra and more specific functional inference in an obvious way to this scheme.
We think the potential use of the method described by Baker et al. (10) to protein engineering is very promising. Effectively, in the search for novel enzymatic activity, the general strategy includes two distinct methods: one that can generate a diverse and large set of sequences and the other that allows for isolation of the sequences producing polypeptides displaying the sought-after characteristics. Obviously, an ideal way to screen for an enzyme activity is to express the library in a cell in which expression of library members with the desired characteristics confers growth capabilities in a given condition, in a specific medium or at nonpermissive growth temperature, for example. In the case where there is no such assay, the methods described by Baker et al. would be perfectly suited for screening novel enzymatic activities, but of course, with the same inherent limitations that we discussed for functional inference of gene products.
One way or another and for whatever purpose, we can hope to see the development of biological research tools that at once can be applied generally while at the same time provide specific information. But all of these technologies need to be challenged to check whether they can be useful for addressing real scientific questions. We are seeing that the development of novel strategies or combinations of smart technologies that already exist are creating new opportunities to explore the details of gene function in finer detail.
See companion article on page 16537.
References
- 1.Fields S. & Song, O. (1989) Nature 340, 245-246. [DOI] [PubMed] [Google Scholar]
- 2.Ito T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., Sakaki, Y., Zhang, J., Ma, Y., Taylor, S. S. & Tsien, R. Y. (2001) Proc. Natl. Acad. Sci. USA 98, 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lashkari D. A., DeRisi, J. L., McCusker, J. H., Namath, A. F., Gentile, C., Hwang, S. Y., Brown, P. O. & Davis, R. W. (1997) Proc. Natl. Acad. Sci. USA 94, 13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uetz P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. (2000) Nature 403, 623-627. [DOI] [PubMed] [Google Scholar]
- 5.Ho Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180-183. [DOI] [PubMed] [Google Scholar]
- 6.Gavin A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141-147. [DOI] [PubMed] [Google Scholar]
- 7.Boulton S., Gartner, A., Reboul, J., Vaglio, P., Dyson, N., Hill, D. E. & Vidal, M. (2002) Science 295, 127-131. [DOI] [PubMed] [Google Scholar]
- 8.Roberts C. J., Nelson, B., Marton, M. J., Stoughton, R., Meyer, M. R., Bennett, H. A., He, Y. D., Dai, H., Walker, W. L., Hughes, T. R., et al. (2000) Science 287, 873-880. [DOI] [PubMed] [Google Scholar]
- 9.Chu S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P. O. & Herskowitz, I. (1998) Science 282, 699-705. [DOI] [PubMed] [Google Scholar]
- 10.Baker K., Bleczinski, C., Lin, H., Salazar-Jimenez, G., Sengupta, D., Krane, S. & Cornish, V. W. (2002) Proc. Natl. Acad. Sci. USA 99, 16537-16542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belshaw P. J., Spencer, D. M., Crabtree, G. R. & Schreiber, S. L. (1996) Chem. Biol. 3, 731-738. [DOI] [PubMed] [Google Scholar]
- 12.Briesewitz R., Ray, G. T., Wandless, T. J. & Crabtree, G. R. (1999) Proc. Natl. Acad. Sci. USA 96, 1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clackson T., Yang, W., Rozamus, L. W., Hatada, M., Amara, J. F., Rollins, C. T., Stevenson, L. F., Magari, S. R., Wood, S. A., Courage, N. L., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 10437-10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Licitra E. J. & Liu, J. O. (1996) Proc. Natl. Acad. Sci. USA 93, 12817-12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivera V. M., Wang, X., Wardwell, S., Courage, N. L., Volchuk, A., Keenan, T., Holt, D. A., Gilman, M., Orci, L., Cerasoli, F., Jr., et al. (2000) Science 287, 826-830. [DOI] [PubMed] [Google Scholar]
- 16.Rollins C. T., Rivera, V. M., Woolfson, D. N., Keenan, T., Hatada, M., Adams, S. E., Andrade, L. J., Yaeger, D., van Schravendijk, M. R., Holt, D. A., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 7096-7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosen M. K., Amos, C. D. & Wandless, T. J. (2000) J. Am. Chem. Soc. 122, 11979-11982. [Google Scholar]
- 18.Sengupta D. J., Zhang, B. L., Kraemer, B., Pochart, P., Fields, S. & Wickens, M. (1996) Proc. Natl. Acad. Sci. USA 93, 8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spencer D. M., Wandless, T. J., Schreiber, S. L. & Crabtree, G. R. (1993) Science 262, 1019-1024. [DOI] [PubMed] [Google Scholar]
- 20.Vogel K. W., Briesewitz, R., Wandless, T. J. & Crabtree, G. R. (2001) Adv. Protein Chem. 56, 253-291. [DOI] [PubMed] [Google Scholar]
- 21.Firestine S. M., Salinas, F., Nixon, A. E., Baker, S. J. & Benkovic, S. J. (2000) Nat. Biotechnol. 18, 544-547. [DOI] [PubMed] [Google Scholar]
- 22.Ting A. Y., Kain, K. H., Klemke, R. L. & Tsien, R. Y. (2001) Proc. Natl. Acad. Sci. USA 98, 15003-15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J., Ma, Y., Taylor, S. S. & Tsien, R. Y. (2001) Proc. Natl. Acad. Sci. USA 98, 14997-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galarneau A., Primeau, M., Trudeau, L. E. & Michnick, S. W. (2002) Nat. Biotechnol. 20, 619-622. [DOI] [PubMed] [Google Scholar]
- 25.Michnick S. W., Remy, I., Campbell Valois, F.-X., Valée-Belisle, A. & Pelletier, J. N. (2000) Methods Enzymol. 328, 208-230. [DOI] [PubMed] [Google Scholar]
- 26.Pelletier J. N., Campbell Valois, F. & Michnick, S. W. (1998) Proc. Natl. Acad. Sci. USA 95, 12141-12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remy I., Wilson, I. A. & Michnick, S. W. (1999) Science 283, 990-993. [DOI] [PubMed] [Google Scholar]
- 28.Remy I., Pelletier, J. N., Galarneau, A. & Michnick, S. W. (2001) in Protein–Protein Interactions: A Molecular Cloning Manual, ed. Golemis, E. A. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 449–475.
- 29.Adam G. C., Cravatt, B. F. & Sorensen, E. J. (2001) Chem. Biol. 8, 81-95. [DOI] [PubMed] [Google Scholar]
- 30.Adam G. C., Sorensen, E. J. & Cravatt, B. F. (2002) Nat. Biotechnol. 20, 805-809. [DOI] [PubMed] [Google Scholar]
- 31.Cravatt B. F. & Sorensen, E. J. (2000) Curr. Opin. Chem. Biol. 4, 663-668. [DOI] [PubMed] [Google Scholar]
- 32.Jessani N., Liu, Y., Humphrey, M. & Cravatt, B. F. (2002) Proc. Natl. Acad. Sci. USA 99, 10335-10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kidd D., Liu, Y. & Cravatt, B. F. (2001) Biochemistry 40, 4005-4015. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y., Patricelli, M. P., Cravatt, B. F., West, M. W. & Hecht, M. H. (1999) Proc. Natl. Acad. Sci. USA 96, 14694-14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu H., Bilgin, M., Bangham, R., Hall, D., Casamayor, A., Bertone, P., Lan, N., Jansen, R., Bidlingmaier, S., Houfek, T., et al. (2001) Science 293, 2101-2105. [DOI] [PubMed] [Google Scholar]
- 36.MacBeath G. & Schreiber, S. L. (2000) Science 289, 1760-1763. [DOI] [PubMed] [Google Scholar]
- 37.Ideker T., Thorsson, V., Ranish, J. A., Christmas, R., Buhler, J., Eng, J. K., Bumgarner, R., Goodlett, D. R., Aebersold, R. & Hood, L. (2001) Science 292, 929-934. [DOI] [PubMed] [Google Scholar]
- 38.Lee T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799-804. [DOI] [PubMed] [Google Scholar]