Abstract
Reproducing kernel Hilbert space (RKHS) methods provide a unified context for solving a wide variety of statistical modelling and function estimation problems. We consider two such problems: We are given a training set {yi, ti, i = 1, … , n}, where yi is the response for the ith subject, and ti is a vector of attributes for this subject. The value of yi is a label that indicates which category it came from. For the first problem, we wish to build a model from the training set that assigns to each t in an attribute domain of interest an estimate of the probability pj(t) that a (future) subject with attribute vector t is in category j. The second problem is in some sense less ambitious; it is to build a model that assigns to each t a label, which classifies a future subject with that t into one of the categories or possibly “none of the above.” The approach to the first of these two problems discussed here is a special case of what is known as penalized likelihood estimation. The approach to the second problem is known as the support vector machine. We also note some alternate but closely related approaches to the second problem. These approaches are all obtained as solutions to optimization problems in RKHS. Many other problems, in particular the solution of ill-posed inverse problems, can be obtained as solutions to optimization problems in RKHS and are mentioned in passing. We caution the reader that although a large literature exists in all of these topics, in this inaugural article we are selectively highlighting work of the author, former students, and other collaborators.
1. Introduction
For this article we define “soft classification” as the problem of classifying an object, based on a vector t of its attributes, into one of two or more categories, where it is desired to estimate the probability pj(x) that the correct identification is category j. We define “hard classification” as the problem of classification where these probabilities are not of primary interest, for example, in cases where the object is easily classifiable by a human. An example of the first problem might be: Given the medical record of a subject, consisting of age, sex, blood pressure, body mass index, cholesterol level, and other relevant variables, provide the 10-year probability of a heart attack. Here k = 2 (heart attack or not), and one might want a soft classification model for use in “evidence-based” medicine. The physician can refer to this model and tell a patient, for example, that if they change some of these variables by specific amounts they reduce the estimated 10-year risk of a heart attack by so much. Examples of the second problem include character and speech recognition and the recognition of objects in images. In these cases the potential for highly accurate classification is there (because humans can do it), but an estimate of the probability of correct classification is generally of interest primarily as a tool in evaluating or comparing the efficacy of competing classifiers. There are, of course, intermediate situations for which the probability of correct classification is mostly high, but it is desired to identify examples for which the classification may be dubious. Other examples include cases for which the number of potential variables available for classification is much higher than the number of samples available for study, i.e., gene expression data. In this case it might be desired to estimate a probability, but there is not enough information to do it accurately. A modest number of statistical tools are available in the literature for soft classification, whereas hard classification techniques are ubiquitous. Descriptions of many of the latter techniques may be found in the Proceedings of the Neural Information Processing Society available at http://nips.djvuzone.org.
Together with colleagues and students I have spent a number of years developing tools for soft classification based on solving optimization methods in reproducing kernel Hilbert spaces (RKHS, to be described). The hard classification technique known as support vector machines (SVMs; refs. 1–3) has recently become popular in the classification literature and in practice. Although the original arguments deriving the SVM were quite different, it is well known that they may be obtained as solutions to optimization problems in RKHS (see refs. 4–6).
In this inaugural article we compare and contrast hard and soft classification methods that are obtained as solutions to optimization problems in RKHS with reference (essentially only) to published and in-progress work by the present author and collaborators despite the fact that there are many other important papers. The focus here is on classification problems with a small number of well defined categories, as opposed to problems such as speech or handwriting recognition.
We will describe RKHS in terms that do not require understanding of Hilbert spaces. Then we will identify the optimization problems of interest, those for penalized likelihood (soft classification) and SVMs (hard classification), give some examples, and comment on problems of computing and tuning the methods and the comparative advantages and disadvantages of both. Other related (hard) classification methods will be noted also, as will some of the other problems that can be solved via optimization problems in RKHS.
2. RKHS
We first need to define positive definite functions. Let 𝒯 be an index set; for example 𝒯 may be the unit interval or the unit cube, Euclidean d-space, or a finite set, say {1, 2, … , N}. Let s, t ∈ 𝒯. K(s, t) is said to be (strictly) positive definite if, for any ℓ and any distinct t1, … , tℓ ∈ 𝒯, the ℓ × ℓ matrix with j, k entry K(tj, tk) is (strictly) positive definite, that is, ∑j,k ajakK(tj, tk) > 0 for any nonzero a1, … , aℓ. Fix t∗ ∈ 𝒯 and let Kt∗(s) be the function of s defined by Kt∗(s) ≡ K(t∗, s). If 𝒯 = {1, … , N}, then this function is simply an N vector; if 𝒯 is the unit interval, then this function is defined on the unit interval, etc. It is a theorem (see ref. 7) that 〈Kt∗, Kt∗∗〉 = K(t∗, t∗∗) defines an inner product and hence a distance (norm) on the class of all finite linear combinations of functions of this form. (This relationship is also the source of the term “reproducing kernel.”) Thus this class of functions has a geometry, which includes the notion of orthogonal projections. ∥f − g∥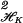 = 〈f, f〉 −2 〈f, g〉 + 〈g, g〉 and the reader may think of 〈f, g〉/∥f∥ℋK∥g∥ℋK as the cosine of the angle between f and g. The (uniquely determined) collection of all finite linear combinations of these functions and their limits in this norm constitute an RKHS, which is uniquely determined by K. Hereafter this space will be called ℋK (see refs. 7 and 8). The geometry of projecting one element of this space onto a subspace of ℋK spanned by a finite number n of elements of ℋK proceeds just as it would in projecting an element in Euclidean space onto an n dimensional subspace using all the known inner products.
= 〈f, f〉 −2 〈f, g〉 + 〈g, g〉 and the reader may think of 〈f, g〉/∥f∥ℋK∥g∥ℋK as the cosine of the angle between f and g. The (uniquely determined) collection of all finite linear combinations of these functions and their limits in this norm constitute an RKHS, which is uniquely determined by K. Hereafter this space will be called ℋK (see refs. 7 and 8). The geometry of projecting one element of this space onto a subspace of ℋK spanned by a finite number n of elements of ℋK proceeds just as it would in projecting an element in Euclidean space onto an n dimensional subspace using all the known inner products.
We describe RKHS at the above level of abstraction to emphasize how general the concept is, but in the sequel we will only be concerned with a couple of examples. A simple example is a kernel K associated with the space of periodic functions on [0, 1], which integrate to 0 and have square integrable second derivative. It is K(s, t) = B2(s)B2(t)/(2!)2 − B4(|s − t|)/4!, where s, t ∈ [0, 1], and Bm is the mth Bernoulli polynomial (see ref. 8) where other spline kernels may be found. The square norm is known to be ∫ (f"(s))2ds, and the Kt∗ are splines. Another popular kernel is the Gaussian kernel, K(s, t) =exp(−(1/σ2)∥s − t∥2) defined for s, t in Euclidean d space, Ed, where the norm in the exponent is the Euclidean norm. Elements of this space are generated from functions of the form ft∗(s) =exp(−(1/σ2)∥s − t∗∥2), t∗ ∈ Ed. The square norm of a function in the RKHS associated with this space involves division of the function's squared Fourier transform by the Fourier transform of the Gaussian function, but we do not need to know that here. However, it is useful to know that tensor sums and products of positive definite functions are also positive definite, which allows building positive definite functions on tensor products of all kinds of domains. The trivial case in 𝒯 = Ed is K(s, t) = s′t, and then ℋK is the d-dimensional space of homogeneous linear functions, and ∥Kt∗∥
(f"(s))2ds, and the Kt∗ are splines. Another popular kernel is the Gaussian kernel, K(s, t) =exp(−(1/σ2)∥s − t∥2) defined for s, t in Euclidean d space, Ed, where the norm in the exponent is the Euclidean norm. Elements of this space are generated from functions of the form ft∗(s) =exp(−(1/σ2)∥s − t∗∥2), t∗ ∈ Ed. The square norm of a function in the RKHS associated with this space involves division of the function's squared Fourier transform by the Fourier transform of the Gaussian function, but we do not need to know that here. However, it is useful to know that tensor sums and products of positive definite functions are also positive definite, which allows building positive definite functions on tensor products of all kinds of domains. The trivial case in 𝒯 = Ed is K(s, t) = s′t, and then ℋK is the d-dimensional space of homogeneous linear functions, and ∥Kt∗∥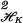 = ∥t∗∥
= ∥t∗∥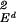 . The original SVMs were linear and corresponded to this case.
. The original SVMs were linear and corresponded to this case.
We are now ready to write a (very special case of) a general lemma about optimization problems in RKHS (6).
Lemma.
Given observations {yi, ti, i = 1, 2, … , n}, where yiis a real number and ti ∈ 𝒯, and given K and (possibly) given some particular functions{φ1, … , φM} on 𝒯, find f of the form f(s) = ∑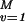 dvφv(s) + h(s) where h ∈ ℋK to minimize
dvφv(s) + h(s) where h ∈ ℋK to minimize
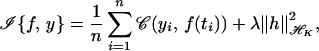 |
where 𝒞 is a convex function off(ti). It is assumed that the minimizer of𝒞(yi, f(ti)) in the span of the {φv} is unique. Then the minimizer ofℐ{f, y} has a representation of the form
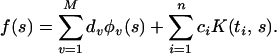 |
The coefficient vectors d = (d1, … , dM)′ and c = (c1, … , cn)′ are found by substituting Eq. 2 into the first term in Eq. 1 and using the fact that ∥∑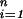 ciK(ti,⋅)∥
ciK(ti,⋅)∥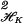 = c′Knc, where Kn is the n × n matrix with i, jth entry K(ti, tj). The minimization generally has to be done numerically by an iterative descent method except in the case where 𝒞 is quadratic in f, in which case a linear system has to be solved.
= c′Knc, where Kn is the n × n matrix with i, jth entry K(ti, tj). The minimization generally has to be done numerically by an iterative descent method except in the case where 𝒞 is quadratic in f, in which case a linear system has to be solved.
When K(⋅, ⋅) is a smooth function of its arguments and n is large, it has been found that excellent approximations to the minimizer of Eq. 1 for various 𝒞 can be found with functions of the form
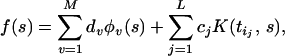 |
where the ti1, … , tiL are a relatively small subset of t1, …, tn, thus reducing the computational load. The ti1, … , tiL may be chosen in various ways: as a random subset, by clustering the ti and selecting from each cluster (9), or by a greedy algorithm (as for example in ref. 10), depending on the problem.
3. Penalized Likelihood Estimation: Two Categories
We first consider only two categories, or outcomes, labeled 1 and 0; for example, 1 is the outcome that a subject gets a disease and 0 is the outcome that they do not. Let p(t) be the probability that a subject with attribute vector t ∈ 𝒯 gets the disease and define the log odds ratio f(t) = log[p(t)/(1 − p(t)]. The likelihood function for data {yi, ti} is p(ti) if yi = 1 and (1 − p(ti)) if yi = 0, which may be written p(ti)yi(1 − p(ti))1−yi. Substituting f(ti) into the likelihood function, we obtain the negative log likelihood
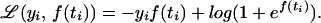 |
Given a training set {yi, ti, i = 1, … n}, an estimate for f and hence p may be obtained by setting 𝒞(yi, f(ti)) in Eq. 1 to be ℒ(yi, f(ti)) of Eq. 4. This kind of penalized likelihood estimate goes back at least to ref. 11.
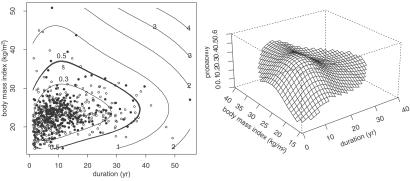
Let t ∈ [0, 1]d, the unit cube, and let t = (x1, …, xd) and ti = (xi1, … , xid). Under general conditions we can expand any integrable f(t) as follows: f(t) = μ + ∑α fα(xα) + ∑α<β fαβ(xα, xβ) + ⋯ + fαβ…(x1, … , xd), where the components satisfy side conditions that make them identifiable and generalize the usual side conditions for parametric ANOVA, that is, all averages with respect to each xα are 0 (see ref. 12). The series is truncated in some manner. By using weighted tensor sums and products of reproducing kernels, the theory above may be generalized to replace λ∥f∥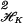 in Eq. 1 by ∑α λα Jα(fα) + ∑α<βλαβJαβ(fαβ) + ⋯, where Jα, Jαβ, … are square norms in the component subspaces. Details may be found in refs. 8, 12, and 13. The λα, λαβ, … are smoothing parameters to be chosen. The right panel in Fig. 1 gives the estimated 4-year risk of progression of diabetic retinopathy based on data from the Wisconsin Epidemiologic Study of Diabetic Retinopathy (WESDR) (14). The attribute vector is t = (x1, x2, x3) = (duration of diabetes, glycosylated hemoglobin, body mass index), and was observed in a group of n = 669 subjects at the start of the study, and the outcome (progression or not) was observed at the 4-year followup and coded as y = 1 or 0. The plot was based on the model f(x1, x2, x3) = μ + f1 (x1) + d2x2 + f3(x3) + f13(x1, x3). The figure is plotted for x2 fixed at its median. The software that produced this analysis can be found in the code grkpack (15). The method for choosing smoothing parameters behind this plot is discussed in ref. 12 (see also ref. 16). A later method, which is used in some subsequent work, is called generalized approximate cross validation (GACV) (17, 18) and will be discussed further in Section 8. An important problem is the selection of terms that are to be retained in the model. This was done in an informal manner in ref. 12. A recent approach to the problem of selecting terms is discussed in refs. 19 and 20.
in Eq. 1 by ∑α λα Jα(fα) + ∑α<βλαβJαβ(fαβ) + ⋯, where Jα, Jαβ, … are square norms in the component subspaces. Details may be found in refs. 8, 12, and 13. The λα, λαβ, … are smoothing parameters to be chosen. The right panel in Fig. 1 gives the estimated 4-year risk of progression of diabetic retinopathy based on data from the Wisconsin Epidemiologic Study of Diabetic Retinopathy (WESDR) (14). The attribute vector is t = (x1, x2, x3) = (duration of diabetes, glycosylated hemoglobin, body mass index), and was observed in a group of n = 669 subjects at the start of the study, and the outcome (progression or not) was observed at the 4-year followup and coded as y = 1 or 0. The plot was based on the model f(x1, x2, x3) = μ + f1 (x1) + d2x2 + f3(x3) + f13(x1, x3). The figure is plotted for x2 fixed at its median. The software that produced this analysis can be found in the code grkpack (15). The method for choosing smoothing parameters behind this plot is discussed in ref. 12 (see also ref. 16). A later method, which is used in some subsequent work, is called generalized approximate cross validation (GACV) (17, 18) and will be discussed further in Section 8. An important problem is the selection of terms that are to be retained in the model. This was done in an informal manner in ref. 12. A recent approach to the problem of selecting terms is discussed in refs. 19 and 20.
Fig 1.
(Left) Data. (Right) Estimate of 4-year risk of progression of diabetic retinopathy as a function of body mass index and duration for glycosylated hemoglobin fixed at its median (from ref. 12).
This penalized likelihood method provides an estimate pλ for p which is known to converge to the true p as the sample size becomes large, under various conditions, including a good choice of the λ (see refs. 13, 21, and 22).
4. SVM Classification: Two Categories
Suppose that we have two categories as before, but we are interested only in making a decision as to which category a future object with attribute vector t∗ is in. It will be convenient for this purpose to change the coding. We let yi = 1 if the ith subject or object from the training set is in the first category, and yi = −1 if it is in the second category. It can be shown that with this coding the negative log likelihood function becomes
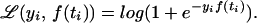 |
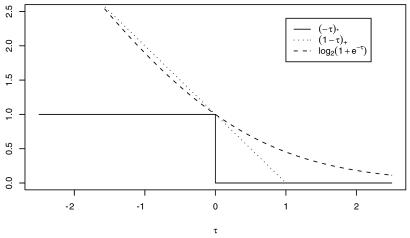
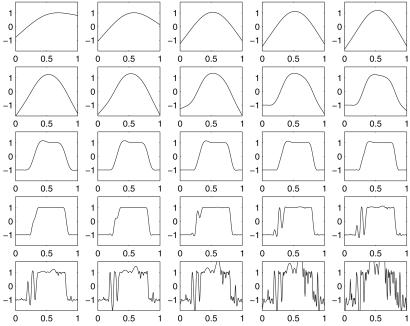
Fig. 2 gives a plot of log(1 + e−τ) as a function of τ. Now we are not interested in the complete function p(t) but only in obtaining enough information to make a classification decision. In the so-called standard case where the costs of both kinds of misclassification are equal and the training set is representative of the population to be classified in the future, the optimal classifier for a subject with attribute vector t∗ is determined by whether p(t∗) is greater or less than 1/2, equivalently, whether f(t∗) is positive or negative. A member of the training set with label yi will be classified correctly by fλ(ti) if τi ≡ yifλ(ti) is positive and incorrectly if τi is negative. Thus, if only classification is of interest, one could consider letting 𝒞(yi, f(ti)) of Eq. 1 be the ∗ function [−yif(ti)]∗, where [τ]∗ = 1 if τ > 0 and 0 otherwise. Then the first term on the right-hand side of Eq. 1 would simply count the number of misclassified subjects in the training set. [−τ]∗ is plotted in Fig. 2. However, ℐ of Eq. 1 with 𝒞 the ∗ function can have multiple minima and is difficult to compute, because it is not convex. Define the “plus” function (τ)+ = τ if τ > 0 and 0 otherwise. The so-called hinge function (1 − τ)+ is also plotted in Fig. 2 and is seen to be the closest convex upper bound to the ∗ function that has a slope of −1 at 0. Setting 𝒞(yi, f(ti)) in Eq. 1 to be (1 − yif(ti))+ and setting M = 1 and φ1 (s) = 1 in Eq. 2 results in the optimization problem, the minimizer of which is known as an SVM. In the last few years the SVM has become very popular in the machine learning community for classification problems, because in practice it has been empirically observed to provide excellent classification results in a wide variety of applications. It is to be noted that once the reproducing kernel K(s, t) is defined, the nature of the domain of s and t is not relevant to the calculations, because only values of K(s, t) are needed to define the SVM. Thus problems where s and t are in a high dimensional space while only a small number of samples are available may be treated, at least mathematically, in a unified manner. Up until recently a theoretical understanding of what the SVM was actually estimating was not available. Recently Lin (23) has shown that if the RKHS is sufficiently rich, then under certain circumstances the SVM is estimating the sign of (p − 1/2), equivalently, the sign of f. This is exactly what you need to classify according to Bayes rule. This is illustrated in Fig. 3. To obtain Fig. 3, Lin let p(t) = Pr(Y = 1|t) = 2t, for t ∈ [0, 0.5] and 1 − 2t for t ∈ [0.5, 1]. Thus sign[p(t) − 1/2] = 1, t ∈ (0.25, 0.75), −1, otherwise, which is also the desired sign of f = log[p/(1 − p)]. n = 257 equally spaced values of ti on the unit interval were selected, and yi was generated randomly to be 1 with probability p(ti) and −1 with probability 1 − p(ti). A sufficiently rich spline kernel was chosen to compute the 25 SVMs in Fig. 3 for 25 values of log2λ from −1 to −25, left to right, top to bottom. It can be seen that for log2λ in the neighborhood of −13 to −18, the estimate is very close to signf. λ may be chosen by the GACV method for SVMs (see ref. 24 and Section 8).
Fig 2.
SVM, likelihood, and misclassification functions compared.
Fig 3.
fλ for 25 values of log2λ from −1 to −25, left to right, top to bottom. The SVM tends toward the sign of log[p/(1 − p)] when tuned well, here for log2λ ≈ −18. [Reproduced with permission from ref. 23 (Copyright 2002, Kluwer).]
5. Penalized Likelihood Estimation: Multiple Categories
Suppose now that there are k + 1 possible outcomes, with k > 1. Let pj(t), j = 0, 1, … , k be the probability that a subject with attribute vector t is in category k, ∑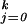 pj(t) = 1. The following approach was proposed in ref. 25: Let fj(t) =log[pj(t)/p0(t)], j = 1, … , k. Then
pj(t) = 1. The following approach was proposed in ref. 25: Let fj(t) =log[pj(t)/p0(t)], j = 1, … , k. Then
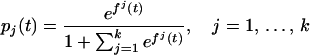 |
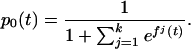 |
The class label for the ith subject is coded as yi = (yi1, … , yik), where yij = 1 if the ith subject is in class j and 0 otherwise. Letting f = (f1, … , fk) the negative log likelihood can be written as
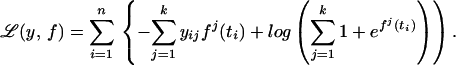 |
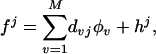 |
where the hj can have an ANOVA decomposition as in Section 3. Generally the fj will have the same terms, with their own d, c. Then λ∥h∥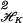 in Eq. 1 is replaced by
in Eq. 1 is replaced by
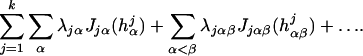 |
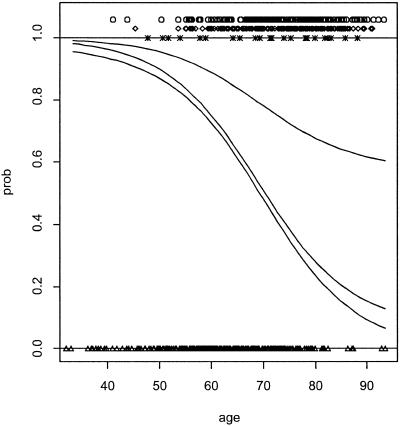
Fig. 4 is based on 10-year mortality data of a group of n = 646 subjects from the WESDR study. Their age (x1), glycosylated hemoglobin (x2), and systolic blood pressure (x3) were (among other things) recorded at baseline, and they were divided into four categories with respect to their status after 10 years, as 0 = alive, 1 = died of diabetes, 2 = died of heart disease, and 3 = died of other causes. Each of the fj, j = 1, 2, 3 was modeled as fj(x1, x2, x3) = μj + f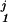 (x1) + f
(x1) + f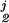 (x2) + f
(x2) + f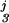 (x3) + f
(x3) + f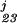 (x2, x3). The pj, j = 0, … , 3 were estimated by minimizing ℐ(y, f) = Eq. 8 + Eq. 9, and the multiple smoothing parameters were estimated by the GACV for polychotomous data (25). For the figure, x2 and x3 were set at their medians, and the differences between adjacent curves, from bottom to top, are probabilities for categories 0, 1, 2, and 3, respectively.
(x2, x3). The pj, j = 0, … , 3 were estimated by minimizing ℐ(y, f) = Eq. 8 + Eq. 9, and the multiple smoothing parameters were estimated by the GACV for polychotomous data (25). For the figure, x2 and x3 were set at their medians, and the differences between adjacent curves, from bottom to top, are probabilities for categories 0, 1, 2, and 3, respectively.
Fig 4.
Ten-year risk of mortality by cause as a function of age and two other risk factors, glycosylated hemoglobin and systolic blood pressure at baseline. The other risk factors have been set at their medians for the plot. The differences between adjacent curves (from bottom to top) are probabilities for alive, diabetes, heart attack, and other causes, respectively. The data are plotted as triangles (alive, on the bottom), crosses (diabetes), diamonds (heart attack), and circles (other). (Reproduced from ref. 25 with permission.)
6. SVMs: Multiple Categories
Thus far, we have been assuming that the observational or training data {yi, ti} is a representative sample from the population of interest. We return to the classification-only problem, now with several categories. If the cost of misclassification is the same for each of the categories of interest, then the optimum classifier would choose category k if pk(t) is larger than pℓ(t) for each ℓ not equal to k. We first consider the so-called standard case, where the cost of misclassification is the same for each category, and as before the training sample is representative. In the next section we will consider the nonstandard case, for which these conditions are relaxed. Many classification problems involve more than two categories, and most authors use some version of one-versus-many or multiple pairwise comparisons (see refs. 1 and 26 for example). Recently a generalization of the SVM that treats all categories simultaneously and symmetrically has been obtained in ref. 27, called the multicategory SVM (MSVM). In the MSVM we have k categories labeled as j = 1, … , k. The class label yi will be coded as a k dimensional vector with 1 in the jth position if example i is in category j and −(1/k−1) otherwise. For example yi = (1, −(1/k−1),…, −(1/k−1)) indicates that the ith example is in category 1. We define a k-tuple of separating functions f(t) = (f1(t), … , fk(t)), with each fj = dj + hj with hj ∈ ℋK, and which will be required to satisfy a sum-to-zero constraint, ∑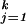 fj(t) = 0, for all t in 𝒯. Note that unlike the estimate of Section 5, all categories are treated symmetrically.
fj(t) = 0, for all t in 𝒯. Note that unlike the estimate of Section 5, all categories are treated symmetrically.
Let Ljr = 1, r ≠ j, Ljj = 0, j, r = 1, … , k. Let cat(yi) = j if yi is from category j. Then, if yi is from category j, Lcat(yi)r = 0 if r = j and 1 otherwise. Then the MSVM is defined as the vector of functions fλ = (f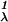 , … , f
, … , f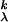 ), with each hk in ℋK satisfying the sum-to-zero constraint, which minimizes
), with each hk in ℋK satisfying the sum-to-zero constraint, which minimizes
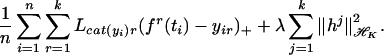 |
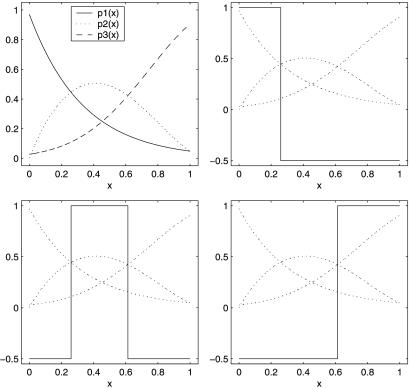
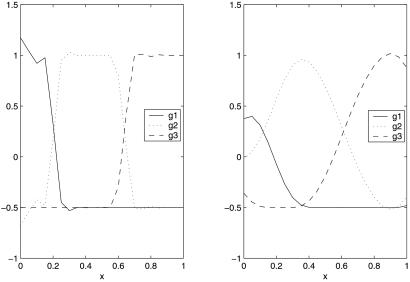
Generalizations of the penalty term are possible if necessary. It can be shown that the k = 2 case reduces to the usual two-category SVM just discussed, and it is shown in ref. 27 that the target for the MSVM is f(t) = (f1(t), … , fk(t)) with fj(t) = 1 if pj(t) is bigger than the other pl(t) and fj(t) = −(1/k−1) otherwise. Fig. 5 describes a simulated example to suggest this result. In Upper Left are given pj(t), j = 1, 2, 3, and in Upper Right, Lower Right, and Lower Left the three optimum fj are superimposed on the pj. The fj take on only the values 1 and −(1/2) ≡ −(1/k−1). For the experiment n = 200, values of ti were chosen according to a uniform distribution on the unit interval, and the class labels were generated according to the pj. Fig. 6 gives the estimated f1, f2, and f3. The Gaussian kernel was used. In Fig. 6 Left λ and σ were chosen with the knowledge of the “right” answer. It is strongly suggestive that the target functions are as claimed. In Fig. 6 Right, both λ and σ2 were chosen by the GACV for the MSVM, given in refs. 28 and 29, which is a generalization of the GACV for the two-category SVM previously given in refs. 24 and 30. For comparison purposes, λ and σ2 were chosen by fivefold cross validation on the MSVM functional (first sum in Eq. 10), with very similar results (not shown). In this example, λ was chosen somewhat larger than its optimum value both by the GACV and the fivefold cross validation, but it can be seen that the implied classifier is quite accurate nevertheless. This is the kind of example where the MSVM will beat a one-vs.-many two-category SVM: category 2 would be missed, because the probability of category 2 is less than the probability of not 2 over a region, although it is the most likely category there.
Fig 5.
Probabilities and optimum fjs for three-category SVM demonstration (from refs. 27 and 29).
Fig 6.
MSVM example. (Left) λ chosen knowing the right answer (from refs. 27 and 29). (Right) Using GACV for the MSVM (courtesy of Yoonkyung Lee).
Two interesting applications of the MSVM appear in refs. 29 and 31 and Y. Lee, G.W., and S. Ackerman, in preparation. The first, also discussed in ref. 28, concerns the application of the MSVM to the classification of a set of cDNA profiles. There were four categories of profiles from small round blue cell tumors (SRBCTs) of childhood. The training set consisted of 63 samples falling into the four categories. Initially, each sample consisted of a vector of expression values for 2,308 genes but was first reduced to the 100 most informative expression values by simple gene-by-gene tests, and then three principal components were extracted from the 100 such that the t vectors are of dimension 3. Scatter plots of the three components suggested that this is a relatively easy classification problem, using only three dimensional vectors extracted from the initial 2,308 dimensional vectors. To study the efficacy of the MSVM, a test set of 20 SRBCT samples and 5 non-SRBCT samples were used. All of the 20 test-sample SRBCTs were classified correctly, and the classification of the 5 non-SRBCTs was ambiguous in the sense that no one of the four components of the MSVM was very large. A measure of the strength of the classification based on how close the MSVM is to a target vector [that is, one with entries 1 and −1/(k − 1)] was obtained in ref. 29, and it indicated that all the classifications of the 5 non-SRBCT samples were weak, along with 3 of the 20 SRBCT classifications. The second example from ref. 29 and Y. Lee, G.W., and S. Ackerman (in preparation) concerns the classification of satellite-observed radiance profiles that contained one of two types of clouds or no clouds. The data came from a simulation system that generates model atmospheric profiles and the response of the observing instrument to them and consisted of 744 profiles, each containing a 12-vector of the responses from 12 different instrument channels. The data set was divided randomly in half for a training set and a test set, and the three-category MSVM was applied to the test set. It was clear there was some modest overlap in data from the three classes. Two cases were tried where the 12-vector was reduced to 2 and then to 5 components nonlinearly, using domain knowledge; then in a third analysis the original 12-vector was used, resulting in a slightly worse test-error rate than the previous two cases. But transforming the 12-vectors into their logs turned out to be the best of the four. Both of these examples illustrate an open question discussed by various authors: How best to choose dimension reducing, possibly nonlinear transformations on the observations to improve classification rates.
7. The Nonstandard MSVM
We now consider the case where the proportion π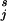 , j = 1, … , k of examples in the training set in each category is not representative of the proportion πj in the population as a whole, and the costs of misclassification are different for different mistakes. Let Cjr be the cost of classifying a j subject as an r, with Cjj = 0. Then the Bayes rule (which minimizes expected cost) is to choose the j for which ∑
, j = 1, … , k of examples in the training set in each category is not representative of the proportion πj in the population as a whole, and the costs of misclassification are different for different mistakes. Let Cjr be the cost of classifying a j subject as an r, with Cjj = 0. Then the Bayes rule (which minimizes expected cost) is to choose the j for which ∑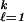 Cℓjpℓ(t) is minimized, where pℓ(t) is the probability that a subject (in the population as a whole) with attribute vector t is in category ℓ. Now let p
Cℓjpℓ(t) is minimized, where pℓ(t) is the probability that a subject (in the population as a whole) with attribute vector t is in category ℓ. Now let p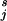 (t) be the probability that a subject in the training set (with proportions π
(t) be the probability that a subject in the training set (with proportions π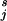 of the different categories) with attribute vector t is in category j. Let now
of the different categories) with attribute vector t is in category j. Let now
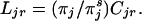 |
One then can show that the optimum classifier chooses the j for which ∑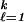 Lℓjp
Lℓjp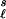 (t) is minimized. The nonstandard MSVM is defined as the vector of functions fλ satisfying the sum-to-zero constraint, which minimizes Eq. 10, with the Ljr there given by Eq. 11. It is shown in ref. 27 that the target of this SVM is: fj(t) = 1 for the j which minimizes ∑
(t) is minimized. The nonstandard MSVM is defined as the vector of functions fλ satisfying the sum-to-zero constraint, which minimizes Eq. 10, with the Ljr there given by Eq. 11. It is shown in ref. 27 that the target of this SVM is: fj(t) = 1 for the j which minimizes ∑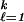 Cℓjpℓ(t) and −(1/k−1) otherwise. Further results and some “toy” examples in the two-category nonstandard case are in ref. 32. Applications of the nonstandard SVM include, for example, evidence-based medical decision making, where different patients might have different personal “costs” with respect to the risks of erroneous treatment decisions. Similarly, both the costs and relative frequencies of the classes in the satellite profile problem discussed previously may be nonstandard, because if the system were to be automated in a numerical weather prediction model, the different kinds of misclassifications may affect the system differently, with more or less costly consequences. When the classes are well separated, taking costs into account will not make much difference, but in modest overlap cases, they can. The GACV for the nonstandard two-category SVM is given in ref. 32 and for the MSVM in ref. 29.
Cℓjpℓ(t) and −(1/k−1) otherwise. Further results and some “toy” examples in the two-category nonstandard case are in ref. 32. Applications of the nonstandard SVM include, for example, evidence-based medical decision making, where different patients might have different personal “costs” with respect to the risks of erroneous treatment decisions. Similarly, both the costs and relative frequencies of the classes in the satellite profile problem discussed previously may be nonstandard, because if the system were to be automated in a numerical weather prediction model, the different kinds of misclassifications may affect the system differently, with more or less costly consequences. When the classes are well separated, taking costs into account will not make much difference, but in modest overlap cases, they can. The GACV for the nonstandard two-category SVM is given in ref. 32 and for the MSVM in ref. 29.
8. Choosing the Smoothing and Tuning Parameters
Methods for choosing the smoothing/tuning parameters in optimization problems such as Eq. 1 have generated lively interest in statistical and machine learning circles for some time. The λs control the tradeoff between the first term on the right of Eq. 1, the fit to the observations, and the second, the complexity of the solution. For the penalized likelihood situation this is known as the bias-variance tradeoff, and this terminology has migrated to the hard classification case. One of the oldest methods, based on 𝒞(yi, f(ti)) = (yi − f(ti))2 in Eq. 1 is the generalized cross validation (GCV), obtained in refs. 33 and 34. It is targeted at minimizing the predictive mean square error, the mean square difference between the estimate fλ and the (unknown) truth, ftrue, when yi = ftrue(ti) + ɛi, where ɛi is white Gaussian noise with common unknown variance. The GCV may be derived beginning with a leaving-out-one argument and an invariance argument and coincides with leaving-out-one in special circumstances. Theoretical properties have been obtained in various places including refs. 8, 35, and 36. The GACV for Bernoulli (0, 1) data is targeted at the Kullback–Leibler distance from the estimated pλ to the true ptrue, a commonly used measure (but not actually a distance) between two distributions (see ref. 17). Both the GCV and the GACV require the computation of the trace of a difficult to compute matrix when n is large. Randomization techniques for doing this efficiently for the GCV were proposed in refs. 37 and 38 and for the GACV in ref. 18. The GACV for SVMs was obtained by starting with a similar leaving-out-one argument followed by a series of approximations and is targeted at minimizing the expected value of (1 − yfuturefλ)+ in the two-category standard SVM case, where yfuture is a new observation from the population under consideration. The ξα method of ref. 39 is similar to and behaves very much like the GACV (32), although the ξα method is targeted directly at the misclassification rate [−yfuturefλ]∗, whereas the GACV is targeted at (1 − yfuturef)+, an upper bound on the misclassification rate. There are several other methods in the SVM literature that begin with the same leaving-out-one argument or an upper bound for it (see ref. 40) with similar results. The ingredients for these are available when the SVM is computed, so no special calculations are required. Leaving out 1/2, 1/5, 1/10, and other cross-validation procedures for estimating the tuning parameters in SVMs are popular, especially when there is a copious training set available.
9. Which Cost Function?
Returning to the two-category case for ease of exposition, it can be asked when is it better to use the SVM or the penalized likelihood estimate. The penalized likelihood estimate can be used for classification in the standard and nonstandard cases (in the multicategory as well as the two-category case) via the Neyman–Pearson Lemma. If probabilities are desired and several conditions are met, then the penalized likelihood estimate is the more appropriate. These conditions generally include a large data set relative to the number of dimensions and probabilities that are expected to be bounded away from 0 or 1 in regions of interest (because the true f will tend to infinity as p tends to 0 or 1). For classification, the SVM does not suffer from either of these problems and furthermore tends to give a sparse solution, that is, many ci are 0, at the cost of a lack of direct interpretability of classification results that are “weak.” Alternatively, the penalized likelihood estimate can somewhat mitigate these problems for classification by an appropriate thinning of the basis functions. Letting the “cost function” c(τ) be 𝒞(yi, f(ti)) with τ = yif(ti), a hybrid 𝒞 may be defined, which combines features of the SVM and the penalized likelihood estimate by letting c(τ) = ln(1 + e−τ) for −∞ ≤ τ ≤ θ and c(τ) extrapolated linearly for τ > θ by matching c(τ) and c′(τ) at θ and linearly continuing until c(τ) becomes 0, after which it remains 0. With a judicious choice of θ > 0, this c might combine the best properties of the SVM and the penalized likelihood, having the sparsity and ease with p near 0 or 1 of the SVM while estimating the log odds ratio and hence the probability near the intermediate case. Other cs for the classification problem have been proposed by various authors including (1 − τ)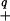 , where q is some power greater than 1. An argument was provided recently for c(τ) linear with a negative slope for τ less than θ, joined smoothly on the right to c(τ) = 1/τ for τ > θ (M. Todd and S. Marron, personal communication). 𝒞(yi, f(ti)) = (yi − f(ti))2, a.k.a. penalized least squares, a.k.a. regularized least squares, a.k.a. ridge regression, long known in the statistics literature for regression problems (see e.g. the references in ref. 34), corresponds to c(τ) = (1 − τ)2 in the case where yi = ±1. Several authors have proposed it in the classification context (T. Poggio, personal communication, and O. Mangasarian, personal communication),† although different names have been attached to it in some cases. Poggio's group found in the cases tried that it compared in classification accuracy with the SVM. It is the easiest to compute, requiring only the solution of a linear system. The extension to the multicategory case is straightforward, replacing (fr(ti) − yir)+ in Eq. 10 by (fr(ti) − yir)2, and imposing the sum-to-zero constraint by requiring the coefficients in the estimates to satisfy a linear system subject to linear equality constraints. A number of other choices of c are discussed in ref. 41, where it is shown for the two-category standard case that under very weak assumptions on c the resulting solution will tend to have the same sign as (p − 1/2).
, where q is some power greater than 1. An argument was provided recently for c(τ) linear with a negative slope for τ less than θ, joined smoothly on the right to c(τ) = 1/τ for τ > θ (M. Todd and S. Marron, personal communication). 𝒞(yi, f(ti)) = (yi − f(ti))2, a.k.a. penalized least squares, a.k.a. regularized least squares, a.k.a. ridge regression, long known in the statistics literature for regression problems (see e.g. the references in ref. 34), corresponds to c(τ) = (1 − τ)2 in the case where yi = ±1. Several authors have proposed it in the classification context (T. Poggio, personal communication, and O. Mangasarian, personal communication),† although different names have been attached to it in some cases. Poggio's group found in the cases tried that it compared in classification accuracy with the SVM. It is the easiest to compute, requiring only the solution of a linear system. The extension to the multicategory case is straightforward, replacing (fr(ti) − yir)+ in Eq. 10 by (fr(ti) − yir)2, and imposing the sum-to-zero constraint by requiring the coefficients in the estimates to satisfy a linear system subject to linear equality constraints. A number of other choices of c are discussed in ref. 41, where it is shown for the two-category standard case that under very weak assumptions on c the resulting solution will tend to have the same sign as (p − 1/2).
Classification problems may have few to extremely many variables (our examples here are the few variables case), may have few to extremely many observations available for a training set, and may be very easy to fairly difficult to classify; “fairly difficult” may include data from the different classes overlapping and/or atypical samples. It is safe to claim, bolstered by theory as well recent simulation results of various authors, that no one c or 𝒞 is going to dominate all others over the range of classification problems. The choice of K can be important or unimportant depending on the example. The Gaussian kernel appears to be a good general purpose kernel for classification in many examples. Other examples of radial kernels (that is, depending on ∥s − t∥) may be found in ref. 42, at ftp://ftp.stat.wisc.edu/pub/wahba/talks/nips.96/m-c.talk.ps, and elsewhere. Some information related to the sensitivity/insensitivity of solutions to optimization problems in RKHS to various parameters in K may be found in chapter 3 of ref. 8. Especially if ℋK is a space of flexible functions it is necessary to control the bias-variance tradeoff; choices here may well be the most important. This tradeoff involves the λs, but other choices may also be important. When the Gaussian kernel is used with the SVM, the results are generally sensitive to the choice of σ, which therefore must also be tuned.
10. Concluding Remarks
In the last few years there has been an explosion of classification methods and results, both theoretical and practical, that are related to optimization problems in RKHS. SVMs and related methods have become the method of choice in many classification applications as their properties are becoming known. In each problem a cost function, a kernel K, and a tuning method must be selected, along with the sometimes nontrivial problem of choosing a numerical algorithm. The trick of thinning the representers can sometimes be used to assist in the bias-variance tradeoff while at the same time making the calculations easier. Early stopping of iterative methods, which we haven't discussed, can also help in this tradeoff (43, 44). Relative sensitivity of the results to the various choices is an active area of recent research. Penalized likelihood methods for regression and function estimation, with data involving random variables from various known distributions, are older (11). The numerical solution of ill-posed inverse problems, where the data are modeled as yi = ∫ F(ti, s)f(s)ds + ɛi, where F(t, s) is given, ɛi are noise variables, and it is desired to recover f, may proceed by replacing 𝒞(yi, f(ti)) by 𝒞(yi, ∫ F(ti, s)f(s)ds) in Eq. 1. Then the K(ti, ⋅) in Eq. 3 are replaced by (other) so-called representers, which may be found in refs. 6 and 8 (see also refs. 45–47). Other references can be found via the publication list on my web site (www.stat.wisc.edu/∼wahba).
Optimization methods in RKHS have turned out to be useful in a wide variety of problems in statistical model building, machine learning, curve and surface fitting, ill-posed inverse problems, and elsewhere. Technical reports and Ph.D. theses since mid-1993 are available via the TRLIST link on my web site.
Acknowledgments
I owe a great debt to my colleague Yi Lin in the Statistics Department at the University of Wisconsin (Madison) and to my most recent former students (now assistant professors) Yoonkyung Lee (Ohio State University, Columbus) and Hao Helen Zhang (North Carolina State University, Raleigh). I owe much to Drs. Ron Klein and Barbara E. K. Klein of the University of Wisconsin Ophthalmology Department for many fruitful discussions that suggested some of the problems studied here and for making data from the WESDR study available, and to former student Xiwu Lin (now at GlaxoSmithKline) for Fig. 4 and the work behind it. I am grateful to my many other former students who had a hand in earlier problems involving optimization problems in RKHS and to many mentors, including especially my thesis advisor, Manny Parzen, who introduced me to RKHS. This research was supported by National Institutes of Health Grant RO1 EY09946, National Science Foundation Grant DMS 0072292, and National Aeronautics and Space Administration Grant NAGW 10273.
Abbreviations
RKHS, reproducing kernel Hilbert space
SVM, support vector machine
WESDR, Wisconsin Epidemiologic Study of Diabetic Retinopathy
GACV, generalized approximate cross validation
GCV, generalized cross validation
MSVM, multicategory SVM
SRBCT, small round blue cell tumor
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 2, 2000.
Poggio, T., Mukherjee, S., Rifkin, R. & Suykens, J., Foundation of Computational Mathematics 2002 Meeting, Aug. 5–14, 2002, Minneapolis, MN.
References
- 1.Vapnik V., (1998) Statistical Learning Theory (Wiley, New York).
- 2.Scholkopf B. & Smola, A., (2002) Learning with Kernels Support Vector Machines, Regularization, Optimization, and Beyond (MIT Press, Cambridge, MA).
- 3.Cristianini N. & Shawe-Taylor, J., (2000) An Introduction to Support Vector Machines (Cambridge Univ. Press, Cambridge, U.K.).
- 4.Wahba G. (1999) in Advances in Kernel Methods: Support Vector Learning, eds. Scholkopf, B., Burges, C. & Smola, A. (MIT Press, Cambridge, MA), pp. 69–88.
- 5.Evgeniou T., Pontil, M. & Poggio, T. (2000) Adv. Comput. Math. 13, 1-50. [Google Scholar]
- 6.Kimeldorf G. & Wahba, G. (1971) J. Math. Anal. Appl. 33, 82-95. [Google Scholar]
- 7.Aronszajn N. (1950) Trans. Am. Math. Soc. 68, 337-404. [Google Scholar]
- 8.Wahba G., (1990) Spline Models for Observational Data (Soc. Indust. Appl. Math., Philadelphia), Vol. 59.
- 9.Xiang D. & Wahba, G., (1998) Proceedings of the 1997 ASA Joint Statistical Meetings, Tech. Rep. 982 (Dept. of Stat., Univ. of Wisconsin, Madison), pp. 94–98.
- 10.Luo Z. & Wahba, G. (1997) J. Am. Stat. Assoc. 92, 107-114. [Google Scholar]
- 11.O'Sullivan F., Yandell, B. & Raynor, W. (1986) J. Am. Stat. Assoc. 81, 96-103. [Google Scholar]
- 12.Wahba G., Wang, Y., Gu, C., Klein, R. & Klein, B. (1995) Ann. Statist. 23, 1865-1895. [Google Scholar]
- 13.Gu C., (2002) Smoothing Spline ANOVA Models (Springer, New York).
- 14.Klein R., Klein, B. E. K., Moss, S. E., Davis, M. D. & DeMets, D. L. (1984) Arch. Ophthalmol. 102, 520-526. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y. (1997) Commun. Stat. Simul. Comput. 26, 765-782. [Google Scholar]
- 16.Gu C. (1992) J. Comput. Graph. Stat. 1, 169-179. [Google Scholar]
- 17.Xiang D. & Wahba, G. (1996) Stat. Sin. 6, 675-692. [Google Scholar]
- 18.Wahba G., Lin, X., Gao, F., Xiang, D., Klein, R. & Klein, B. (1999) in Advances in Information Processing Systems 11, eds. Kearns, M., Solla, S. & Cohn, D. (MIT Press, Cambridge, MA), pp. 620–626.
- 19.Zhang H., Wahba, G., Lin, Y., Voelker, M., Ferris, M., Klein, R. & Klein, B., (2001) Proceedings of the ASA Joint Statistical Meetings, Tech. Rep. 1042 (Dept. of Stat., Univ. of Wisconsin, Madison).
- 20.Zhang, H., Wahba, G., Lin, Y., Voelker, M., Ferris, M., Klein, R. & Klein, B. (2000) Tech. Rep. 1059 (Dept. of Stat., Univ. of Wisconsin, Madison).
- 21.Cox D. & O'Sullivan, F. (1990) Ann. Statist. 18, 1676-1695. [Google Scholar]
- 22.Lin Y. (2000) Ann. Statist. 28, 734-755. [Google Scholar]
- 23.Lin Y. (2002) Data Mining Knowl. Discov. 6, 259-275. [Google Scholar]
- 24.Wahba G., Lin, Y. & Zhang, H. (2000) in Advances in Large Margin Classifiers, eds. Smola, A., Bartlett, P., Scholkopf, B. & Schuurmans, D. (MIT Press, Cambridge, MA), pp. 297–311.
- 25.Lin X., (1998) Ph.D. thesis (Univ. of Wisconsin, Madison); Tech. Rep. 1003 (Dept. of Stat., Univ. of Wisconsin, Madison).
- 26.Ramaswamy S., Tamayo, P., Rifkin, R., Mukherjee, S., Yeang, C., Angelo, M., Ladd, C., Reich, M., Latulippe, E., Mesirov, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 15149-15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, Y., Lin, Y. & Wahba, G. (2002) Comput. Sci. 33, in press; Tech. Rep. 1043 (Dept. of Stat., Univ. of Wisconsin, Madison).
- 28.Lee, Y. & Lee, C.-K. (2002) Tech. Rep. 1051 (Dept. of Stat., Univ. of Wisconsin, Madison).
- 29.Lee Y., (2002) Ph.D. thesis (Univ. of Wisconsin, Madison); Tech. Rep. 1062 (Dept. of Stat., Univ. of Wisconsin, Madison).
- 30.Lin Y., Lee, Y. & Wahba, G. (2002) Mach. Learn. 46, 191-202. [Google Scholar]
- 31.Lin, Y., Lee, Y. & Wahba, G. (2002) Tech. Rep. 1063 (Dept. of Stat., Univ. of Wisconsin, Madison).
- 32.Wahba G., Lin, Y., Lee, Y. & Zhang, H. (2002) in Nonlinear Estimation and Classification, eds. Denison, D., Hansen, M., Holmes, C., Mallick, B. & Yu, B. (Springer, New York), pp. 129–148.
- 33.Craven P. & Wahba, G. (1979) Numer. Math. 31, 377-403. [Google Scholar]
- 34.Golub G. H., Heath, M. & Wahba, G. (1979) Technometrics 21, 215-224. [Google Scholar]
- 35.Li K. C. (1986) Ann. Statist. 14, 1101-1112. [Google Scholar]
- 36.Speckman P. (1985) Ann. Statist. 13, 970-983. [Google Scholar]
- 37.Hutchinson M. (1989) Commun. Stat. Simul. 18, 1059-1076. [Google Scholar]
- 38.Girard D. (1989) Numer. Math. 56, 1-23. [Google Scholar]
- 39.Joachims T., (2000) Proceedings of the International Conference on Machine Learning (Morgan Kaufmann, San Mateo, CA).
- 40.Chapelle O., Vapnik, V., Bousquet, O. & Mukherjee, S. (2002) Mach. Learn. 46, 131-159. [Google Scholar]
- 41.Lin, Y. (2002) Tech. Rep. 1044 (Dept. of Stat., Univ. of Wisconsin, Madison).
- 42.Micchelli C. (1986) Constr. Approximation 2, 11-22. [Google Scholar]
- 43.Wahba G. (1987) in Proceedings of the Alpine–U.S. Seminar on Inverse and Ill Posed Problems, eds. Engl, H. & Groetsch, C. (Academic, New York), pp. 37–51.
- 44.Wahba G., Johnson, D., Gao, F. & Gong, J. (1995) Mon. Weather Rev. 123, 3358-3369. [Google Scholar]
- 45.Nychka D., Wahba, G., Goldfarb, S. & Pugh, T. (1984) J. Am. Stat. Assoc. 79, 832-846. [Google Scholar]
- 46.O'Sullivan F. & Wahba, G. (1985) J. Comput. Phys. 59, 441-455. [Google Scholar]
- 47.Wahba G. (1977) SIAM J. Numer. Anal. 14, 651-667. [Google Scholar]