Abstract
A high-throughput assay for enzyme activity has been developed that is reaction independent. In this assay, a small-molecule yeast three-hybrid system is used to link enzyme catalysis to transcription of a reporter gene in vivo. Here we demonstrate the feasibility of this approach by using a well-studied enzyme-catalyzed reaction, cephalosporin hydrolysis by the Enterobacter cloacae P99 cephalosporinase (β-lactam hydrolase, EC 3.5.2.6). We show that the three-hybrid system can be used to read out cephalosporinase activity in vivo as a change in the level of transcription of a lacZ reporter gene and that the wild-type cephalosporinase can be isolated from a pool of inactive mutants by using a lacZ screen. The assay has been designed so that it can be applied to different chemical reactions without changing the components of the three-hybrid system. A reaction-independent high-throughput assay for protein function should be a powerful tool for protein engineering and enzymology, drug discovery, and proteomics.
Enzymes are able to catalyze a broad range of chemical transformations not only with impressive rate enhancements but also with both regio- and stereoselectivity and so are attractive candidates as practical alternatives to traditional small-molecule catalysts. With applications as diverse as chemical synthesis, reagents for commercial products and biomedical research, and even therapeutics, there is a great demand for enzymes with both improved activity and novel catalytic function (1, 2). In theory, the properties of an enzyme can be altered by rational design; however, rational design is greatly hindered in practice by the complexity of protein function. With advances in molecular biology the possibility has arisen that an enzyme with the desired catalytic property can instead be isolated from a large pool of protein variants. Recently directed evolution has been used successfully to modify the substrate (3) or cofactor specificity (4) of an existing enzyme. These experiments, however, are limited to reactions that are inherently screenable or selectable, reactions where the substrate is a peptide (5, 6) or the product is fluorescent (4) or an essential metabolite (3, 7, 8). What are needed now to realize the power of directed evolution experiments are screening and selection strategies that are general, strategies that do not limit the chemistry and that can readily be adapted to a new target reaction.
Early success with assays based on binding to transition-state analogs and suicide substrates convinced researchers that it should be possible to engineer proteins to catalyze a broad range of reactions (9), but it was difficult to translate binding events into read-outs for enhanced catalytic activity. Recently, attention has turned to direct selections for catalytic activity. While strategies ranging from in vitro fluorescence assays to physically linking the enzyme to its substrate have all recently been reported (10–15), a general solution to this problem is yet to emerge.
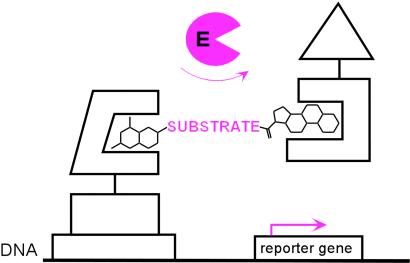
In vivo complementation, in which an enzyme is selected on the basis of its ability to complement an essential activity that has been deleted from a wild-type cell, has proven to be one of the most powerful approaches to enzyme evolution (3, 7). However, complementation is limited to natural reactions that are selectable. The challenge then was to devise a complementation strategy that would be reaction independent. Our approach is to use the yeast three-hybrid assay to link enzyme catalysis to transcription of a reporter gene in vivo (Fig. 1). As a first step, we had previously designed a yeast three-hybrid system in which a dimeric small molecule bridges a DNA-binding domain–receptor fusion protein and an activation domain–receptor fusion protein (16). By dimerizing the two fusion proteins by means of the receptors, the small molecule effectively reconstitutes the transcriptional activator, turning on transcription of a downstream reporter gene. We envisioned that this system could be used as a read-out for enzyme-catalyzed bond cleavage reactions simply by replacing the chemical linker between the two small molecules with the bond to be cleaved and adding an enzyme as a fourth component to the system. Cleavage of the bond between the two small molecules by an active enzyme would disrupt reconstitution of the transcriptional activator and, hence, transcription of the reporter gene. We chose cephalosporin hydrolysis by a cephalosporinase (β-lactam hydrolase, EC 3.5.2.6) as a model reaction around which to develop the system (17). Here we present results showing that our yeast three-hybrid system can be used to link cephalosporinase activity to transcription of a lacZ reporter gene in vivo. In addition, we show that a lacZ screen can be used in a high-throughput format to isolate the wild-type cephalosporinase enzyme from a pool of inactive mutants.
Fig 1.
Chemical complementation. A reaction-independent complementation assay for enzyme catalysis based on the yeast three-hybrid assay. A heterodimeric small molecule bridges a DNA-binding domain–receptor fusion protein and an activation domain–receptor fusion protein, activating transcription of a downstream reporter gene in vivo. Enzyme catalysis of either cleavage or formation of the bond between the two small molecules can be detected as a change in transcription of the reporter gene. The assay can be applied to new chemical reactions simply by synthesizing small molecules with different substrates as linkers and adding an enzyme as a fourth component to the system.
Materials and Methods
Chemical Synthesis.
A complete description of the synthesis of the methotrexate-cephem-dexamethasone (Mtx-Cephem-Dex) substrate, together with full experimental details and compound characterization, is given in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Biological Methods.
Standard methods for molecular biology in Saccharomyces cerevisiae and Escherichia coli were used (18, 19). A more complete description of the general methods for molecular biology, plasmid construction, and enzyme purification is given in Supporting Text in the supporting information.
Yeast Strain Construction.
Strains and plasmids used in this study are listed in Table 1 in the supporting information on the PNAS web site. The genes encoding the LexA-DHFR and B42-GR fusion proteins (DHFR, dihydrofolate reductase; GR, glucocorticoid receptor) under control of the GAL1 promoter were integrated into the yeast strain FY251 at the chromosomal loci ade4 and ade2, respectively, to give yeast strain V947Y. These improvements to the Mtx-Dex yeast three-hybrid system will be described more fully in a separate publication (K.B., G.S.-J., and V.W.C., unpublished work). For the 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) plate assays, yeast strain V947Y was transformed with the lacZ reporter plasmid pMW112 (2μ, URA3, kanR) and vector pVC167 containing no enzyme (empty vector), pVC172 encoding P99 cephalosporinase, or pCB827 encoding P99 Ser-64 → Ala cephalosporinase. The negative control strain (see Fig. 3B, column 1) was made by transforming yeast strain FY251 with the plasmid pMW112, the P99 cephalosporinase expression vector pVC172, and the LexA-DHFR and B42 expression plasmids pKB521 and pMW102, respectively. For the liquid o-nitrophenyl β-d-galactopyranoside (ONPG) assays, yeast strains were made by mating yeast strain V947Y to yeast strain FY250 transformed with the plasmid pMW112 and the pVC167 vector containing no enzyme, pVC172 encoding P99 cephalosporinase, or pCB827 encoding P99 Ser-64 → Ala cephalosporinase. Strains were mated on YPD (yeast extract/peptone/dextrose) plates and then streaked onto synthetic complete medium plates lacking histidine, uracil, tryptophan, and leucine to select for mated diploid strains.
Fig 3.
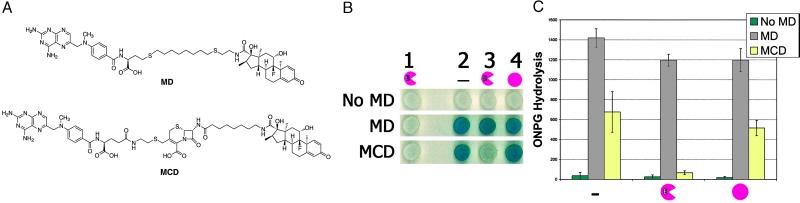
Chemical complementation links enzyme catalysis to reporter gene transcription. (A) Structures of the Mtx-Dex (MD) and Mtx-Cephem-Dex (MCD) heterodimers. (B) X-Gal plate assays of cephalosporinase-dependent Mtx-Cephem-Dex-induced lacZ transcription. Yeast strains containing a lacZ reporter gene were grown on X-Gal indicator plates with or without Mtx-linker-Dex molecules as indicated. Columns 1–4 correspond to yeast strains containing a LexA DNA-binding domain fusion protein, a B42 activation domain fusion protein, and enzyme, as follows: Column 1, LexA-DHFR, B42, P99 cephalosporinase. Plates in column 1 lack GR and are used as negative controls. Column 2, LexA-DHFR, B42-GR, no enzyme; column 3, LexA-DHFR, B42-GR, P99 cephalosporinase; and column 4, LexA-DHFR, B42-GR, P99 Ser-64 → Ala cephalosporinase. The rows correspond to individual X-Gal plates, which have different small molecules as indicated: No MD, No Mtx-Dex; MD, 1 μM Mtx-Dex; MCD, 10 μM Mtx-Cephem-Dex. (C) ONPG liquid assays. Yeast strains expressing the LexA-DHFR and B42-GR fusion proteins and containing a lacZ reporter gene and expressing no enzyme (left), P99 cephalosporinase (center), or P99 Ser-64 → Ala cephalosporinase (right) were grown in liquid culture and assayed for β-galactosidase activity with ONPG as a substrate. The liquid culture contained small molecules as indicated. The assays were done in triplicate. ONPG hydrolysis rates are reported as nmol/min per mg of total protein, and the error bars for the specific activity correspond to the standard deviation from the mean. Strains containing the active P99 cephalosporinase showed an 8-fold decrease in the level of lacZ transcription relative to strains containing the inactive Ser-64 → Ala variant.
β-Galactosidase Assays.
Yeast strains expressing the appropriate fusion proteins and reporter genes were assayed for β-galactosidase activity by using standard X-Gal plate or liquid ONPG assays. X-Gal plate assays were done as previously reported (16), except that the plates lacked histidine, uracil, tryptophan, and leucine, and contained 0.5% galactose, 1.5% glucose, and 2% raffinose and no small molecule, 1 μM Mtx-Dex, or 10 μM Mtx-Cephem-Dex. For the liquid ONPG assays, each yeast strain was inoculated into 3 ml of synthetic complete medium lacking histidine, uracil, tryptophan, and leucine and containing 0.5% galactose, 1.5% glucose, and 2% raffinose and no small molecule, 1 μM Mtx-Dex, or 10 μM Mtx-Cephem-Dex and grown for 2 days at 30°C. Each culture was divided into three 1-ml aliquots for the ONPG assay. Cells were washed with ice-cold PBS, pH 7, and then lysed by using acid-washed glass beads (vortex 10 min, 4°C, glass bead disruption buffer). A 20-μl sample of the soluble portion of the cell lysate was assayed at 37°C for β-galactosidase activity in 990 μl of Z-buffer with 200 μl of 4 mg/ml ONPG as substrate. Total protein concentration of the soluble portion of the lysate was determined by using a Bradford assay (Bio-Rad) according to the manufacturer's suggestions.
Other Enzyme Activity Assays.
P99 cephalosporinase activity with nitrocefin was detected by an increase in A486 (ɛ = 20,300 cm−1⋅M−1). The activity was assayed in a solution (500 μl) of 10 mM sodium phosphate buffer, pH 7.0, containing nitrocefin (50 μM final assay concentration from a 100 mM stock solution in dimethyl formamide). Reactions were performed at 25°C and started with the addition of 10 nM enzyme (250 μl of a 20 nM solution in assay buffer).
P99 cephalosporinase activity with the Mtx-Cephem-Dex substrate was detected by using a continuous assay for reduction of Ellman's reagent. In this assay, the cleavage of the Mtx-Cephem-Dex by the P99 cephalosporinase is detected as an increase in A412 caused by the reduction of 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB, Ellman's Reagent) (ɛ412 = 13,600 cm−1⋅M−1) by the Mtx-thiol leaving group. P99 cephalosporinase activity was assayed in a solution (500 μl) of 10 mM sodium phosphate buffer (pH 7.0) containing Mtx-Cephem-Dex (50 μM final concentration from a 10 mM stock solution in dimethyl formamide) and DTNB (50 μM final concentration from a 10 mM stock solution in dimethyl formamide). Reactions were performed at 25°C and started with the addition of 100 nM enzyme (250 μl of a 200 nM solution in assay buffer). The Guggenheim method was used to determine that reaction of the Mtx-thiol with DTNB was not the rate-determining step (20).
Results and Discussion
Selection Scheme and Model Reaction.
We set out to devise a reaction-independent complementation assay using a small-molecule yeast three-hybrid system to link enzyme catalysis to transcription of a reporter gene in vivo (Fig. 1). We based this assay on a Mtx-Dex yeast three-hybrid system recently reported by our laboratory (16). In this system, a heterodimeric Mtx-Dex small molecule dimerizes DHFR, which binds Mtx with high affinity, and the hormone-binding domain of the GR, which binds Dex. DHFR is fused to the DNA-binding protein LexA and GR to the B42 activation domain, such that Mtx-Dex effectively reconstitutes the artificial transcriptional activator LexA-B42 and increases transcription of a downstream reporter gene (Fig. 2B) (16). To adapt this system to read out enzyme catalysis of bond cleavage reactions, we planned to replace the linkage between Mtx and Dex with the substrate for the reaction and add an enzyme as a fourth component to the system. Enzyme catalysis would then be detected as cleavage of the Mtx-substrate-Dex molecule and a decrease in transcription of the reporter gene. This approach is general because it can be applied to new reactions simply by synthesizing Mtx-Dex molecules with different substrates as chemical linkers. Genetic assays are high-throughput because large pools of proteins can be sorted on the basis of clear changes in cellular phenotype.
Fig 2.
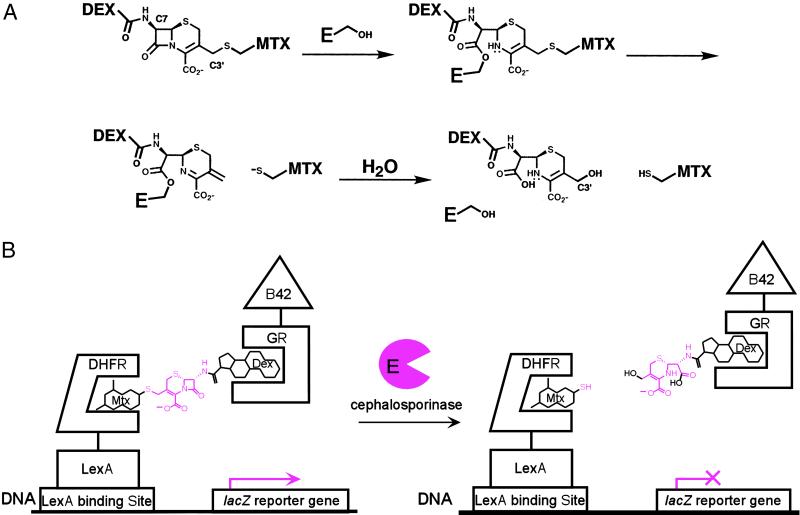
Cephalosporinase model reaction. Cephalosporin hydrolysis provides a simple cleavage reaction to demonstrate the complementation strategy. (A) Cephalosporin hydrolysis by a cephalosporinase enzyme. Cephalosporinases are serine-protease enzymes and catalyze the hydrolysis of cephalosporin antibiotics by means of an acyl-enzyme intermediate. Hydrolysis of the β-lactam bond in Mtx-Cephem-Dex results in expulsion of the leaving group at the C3′ position of the cephem core, effectively breaking the bond between Mtx and Dex. (B) Cephalosporin hydrolysis by the cephalosporinase enzyme disrupts transcription of a lacZ reporter gene. The Mtx-Cephem-Dex substrate dimerizes a LexA DNA-binding domain-dihydrofolate reductase (LexA-DHFR) and a B42 activation domain-glucocorticoid receptor (B42-GR) fusion protein, activating transcription of a lacZ reporter gene. Addition of active cephalosporinase enzyme results in cleavage of the Mtx-Cephem-Dex substrate and disruption of lacZ transcription.
We chose cephalosporin hydrolysis by the Enterobacter cloacae P99 cephalosporinase as a simple cleavage reaction (17) to demonstrate the selection strategy (Fig. 2). Cephalosporins are β-lactam antibiotics, and cephalosporinases are the bacterial resistance enzymes that hydrolyze and, therefore, inactivate these antibiotics. The cephalosporinase enzyme is well characterized biochemically and structurally (21, 22), and the synthesis of cephem compounds is established (23). We chose to incorporate Mtx and Dex at the C3′ and C7 positions, respectively, of the cephem core. Cleavage of the β-lactam bond in cephalosporins results in expulsion of the leaving group at the C3′ position, effectively breaking the bond between Mtx and Dex (Fig. 2A). Thus, the Mtx-Cephem-Dex substrate should dimerize the transcriptional activator, causing transcription of the reporter gene in the yeast three-hybrid assay. When the cephalosporinase enzyme is expressed, however, the cephem linkage should be cleaved, and protein dimerization and transcription of the reporter gene should be disrupted.
Mtx-Cephem-Dex Substrate.
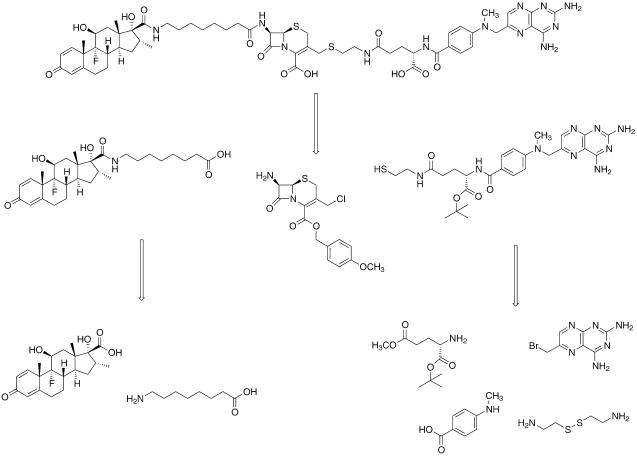
While previous work with the yeast two- and three-hybrid assays suggests that the linker between Mtx and Dex should not affect the transcription read-out, we first wanted to test our assumption that Mtx-Cephem-Dex would retain the ability to activate transcription in our yeast three-hybrid system. In addition, we wanted to carry out in vitro kinetic studies to be sure that Mtx-Cephem-Dex is an efficient substrate for the cephalosporinase enzyme. The first step was to synthesize the Mtx-Cephem-Dex substrate. The original synthesis of the Mtx-Dex heterodimer had been designed to facilitate the inclusion of different chemical linkers between the Mtx and Dex portions of the molecule (16). Thus, the commercial cephem intermediate 7-amino-3-chlormethyl-3-cephem-4-carboxylic acid p-methoxybenzyl ester (ACLE) could be readily incorporated into this synthesis as shown in Scheme . This synthesis relies on first building the Mtx and Dex halves of the molecule and then coupling them to the chemical linker. A thiol analog of Mtx was synthesized so that the final cephem substrate would have a thioether linkage at the C3′ position. The Mtx synthesis began with a differentially protected glutamic acid derivative. The α-carboxylate of γ-methyl glutamate was protected as the t-butyl ester by using t-butyl acetate under acidic conditions. The protected glutamate intermediate was then coupled to N-methyl-p-aminobenzoic acid by using standard peptide coupling conditions. The γ-methyl ester was hydrolyzed and then coupled to cystamine by using benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBOP). 6-Bromomethylpteridine was added to the molecule, and then the disulfide was reduced to the free thiol by using tributylphosphine. A derivative of Dex was prepared with a free terminal carboxylic acid. Dex was cleaved oxidatively by using sodium periodate as reported, and the resulting carboxylic acid was coupled to t-butyl-1-amino-8-hexanoate. The t-butyl protecting group was cleaved with trifluoroacetic acid (TFA) to give the free terminal carboxylic acid. The Dex carboxylate was added to the cephem core first, coupling to the C7 amine of ACLE by using dicyclohexylcarbodiimide. Then the thiol analog of Mtx was reacted at the C3′ position of the cephem by using a simple SN2 displacement reaction. Finally, the t-butyl and p-methoxybenzyl protecting groups were removed with neat TFA, and the product was purified by using C18 reversed-phase high-pressure liquid chromatography.
Scheme 1.
Met-Cephem-Dex retrosynthetic analysis.
Using standard β-galactosidase activity assays both on plates and in liquid culture, we showed that Mtx-Cephem-Dex can activate lacZ transcription in a yeast strain containing the LexA-DHFR and B42-GR fusion proteins and a lacZ reporter gene (Fig. 3 B and C and data not shown). In these assays, the Mtx-Cephem-Dex substrate was simply added to the growth medium, and the extracellular concentration of the Mtx-Dex compounds ranged from 0.010 to 10 μM. As can be seen, lacZ transcription in the strain expressing LexA-DHFR and B42-GR is small-molecule dependent (Fig. 3B, column 2, and Fig. 3C). Only background levels of β-galactosidase activity were detected when Mtx-Cephem-Dex was omitted from the growth medium or either DHFR or GR was deleted from the three-hybrid fusion proteins (Fig. 3B, row 1 and column 1, Fig. 3C, and data not shown).
Having shown that Mtx-Cephem-Dex is an efficient dimerizer in vivo, we wanted to confirm that it is a good substrate for the cephalosporinase enzyme by using purified enzyme in vitro. The P99 cephalosporinase was subcloned into a T7 expression system with a C-terminal His6-tag, overexpressed, and purified by using a nickel-affinity resin. Because Mtx has a strong absorbance at 264 nm, turnover could not be determined on the basis of the change in the absorbance at 264 nm upon cleavage of the lactam bond as is standard (17). Thus, a coupled assay using 5,5′-dithiobis(2-nitrobenzoic acid) (Ellman's reagent) was developed for measuring Mtx-Cephem-Dex hydrolysis. Upon cleavage of the β-lactam bond and expulsion of the Mtx thiol from the C3′ position of the cephem, the Mtx thiol reduces Ellman's reagent, leading to an increase in the absorbance at 412 nm. When this coupled assay was used, the P99 cephalosporinase was shown to turn Mtx-Cephem-Dex over with a specific activity of 0.062 ± 0.003 μmol/min per mg of enzyme. By comparison, one of the best substrates for the E. cloacae P99 cephalosporinase, nitrocefin, is turned over with a specific activity of 167 ± 29 μmol/min per mg of enzyme.
Enzyme Catalysis Linked to Transcription Read-Out.
Having shown that the Mtx-Cephem-Dex substrate retained the ability to activate transcription in vivo and was cleaved effectively by the cephalosporinase enzyme in vitro, the next step was to add the enzyme to the yeast three-hybrid system and find conditions where we could observe enzyme-dependent transcription. Toward this end, the yeast three-hybrid strain was re-engineered so that the expression of the transcriptional activator fusion proteins and the enzyme could be regulated independently. The LexA-DHFR and B42-GR proteins were placed under the control of the fully regulatable GAL1 promoter; the P99 cephalosporinase, under the repressible MET promoter. In addition, the enzyme was expressed from a plasmid containing a spectinomycin-resistance marker, whereas the transcriptional activator fusion proteins were expressed from vectors containing kanamycin-resistance markers, to facilitate isolation of the plasmid encoding the enzyme at the end of a screen or selection. Finally, no ampicillin markers were used because these markers encode β-lactamase enzymes, which, if expressed, could cleave the Mtx-Cephem-Dex substrate. First, we established independently that the cephalosporinase was being expressed in an active form in the yeast cells by using nitrocefin, a known chromogenic substrate for the cephalosporinase (24) (data not shown). Then, using standard β-galactosidase assays on X-Gal plates, we developed conditions where expression of the P99 cephalosporinase disrupted Mtx-Cephem-Dex-mediated lacZ transcription (Fig. 3B, column 3). These results were confirmed by using quantitative assays in liquid culture with ONPG (Fig. 3C) and established that the three-hybrid assay can be used to detect cephalosporinase activity.
A number of experiments were carried out to confirm that the change in transcription of the reporter gene was in fact caused by enzyme turnover. First, we showed that enzyme-dependent disruption of the transcription read-out was cephem dependent by comparing the levels of lacZ transcription with Mtx-Dex and Mtx-Cephem-Dex. Using standard lacZ transcription assays both on plates and in liquid culture, we determined the levels of lacZ transcription in yeast strains expressing different LexA and B42 fusion proteins, enzymes, and a lacZ reporter gene (Fig. 3 B and C). In these assays, no small molecule, Mtx-Dex with a noncleavable linker at 1 μM in the medium, or Mtx-Cephem-Dex with the cleavable cephem linker at 10 μM was used. As can be seen, lacZ transcription in the strain expressing LexA-DHFR and B42-GR is small-molecule dependent (Fig. 3B, column 2, and Fig. 3C). Expression of the wild-type cephalosporinase enzyme disrupts this small-molecule-induced transcription activation when the cells are grown in the presence of Mtx-Cephem-Dex, but not Mtx-Dex (Fig. 3B, column 3, and Fig. 3C). Importantly, expression of the cephalosporinase enzyme has little effect on the levels of Mtx-Dex-activated lacZ transcription (Fig. 3B, row 2, and Fig. 3C).
Another important control is to establish that disruption of lacZ transcription is due to turnover of the cephem linkage and not simply sequestration of the Mtx-Cephem-Dex substrate by the cephalosporinase enzyme. To address this question, we compared the activity of the wild-type cephalosporinase enzyme in this assay with that of an inactive mutant. For the inactive variant, a mutant enzyme in which alanine replaced the active-site serine nucleophile was prepared. Cephalosporinases are serine-protease enzymes, and Ser-64 is the active-site serine known to be essential for turnover of the cephem substrate (21). In contrast with the wild-type cephalosporinase, there was no detectable change in the levels of lacZ transcription for cells expressing the Ser-64 → Ala mutant enzyme (Fig. 3B, columns 3 and 4, and Fig. 3C). The optimal difference in signal between the active and inactive enzyme was observed when the cells were grown in 0.5% galactose, 1.5% glucose, and 134 μM methionine. Under these conditions, the three-hybrid fusion proteins are expressed at low levels, and the enzyme is maximally expressed. Together, these results establish that the change in transcription of the lacZ reporter gene is due to enzyme-catalyzed turnover of the Mtx-Cephem-Dex substrate.
High-Throughput lacZ Screen.
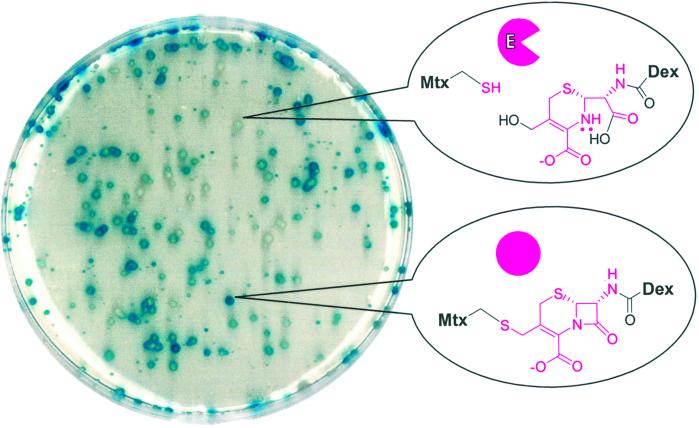
Finally, a lacZ screen was used to isolate the wild-type cephalosporinase from a pool of inactive variants (Fig. 4). A mixture of plasmids encoding the wild-type enzyme (5%) or the Ser-64 → Ala variant (95%) was transformed en masse into a yeast strain expressing the LexA-DHFR and B42-GR fusion proteins and bearing a lacZ reporter plasmid. The resulting transformants were plated on X-Gal plates and screened on the basis of their levels of β-galactosidase expression. Initially, the screen suffered from a high percentage of false positives and false negatives. Integration of the genes encoding the LexA-DHFR and B42-GR fusion proteins, however, stabilized the transcription read-out without significantly affecting the sensitivity of the assay (K.B., G.S.-J., and V.W.C., unpublished work). When the integrated strains were used, ≈5% of the cells showed reduced levels of β-galactosidase expression, as would be expected on the basis of the plasmid ratio. The plasmids encoding the enzyme were extracted from five blue and five white colonies and sequenced. Sequencing confirmed that all five of the blue colonies contained the inactive Ser-64 → Ala mutant enzyme and four of the five white colonies contained the wild-type cephalosporinase enzyme. A secondary lacZ screen with and without Mtx-Dex could rule out all false positives. Further characterization of the integrated strains has confirmed a low false positive rate (K.B., G.S.-J., and V.W.C., unpublished work). The lacZ screen demonstrates that the yeast three-hybrid system can be used reliably to screen libraries of proteins on the basis of catalytic activity.
Fig 4.
High-throughput chemical complementation screen. Active enzyme can be isolated from a pool of inactive mutants. The yeast selection strain was transformed with a 5:95 mixture of plasmids encoding the wild-type active cephalosporinase enzyme and the inactive Ser-64 → Ala cephalosporinase variant, respectively, and then plated onto an X-Gal indicator plate containing 10 μM Mtx-Cephem-Dex. Cells containing the active enzyme could be distinguished on the basis of the levels of X-Gal hydrolysis and hence lacZ transcription.
Conclusion
While this selection strategy has been demonstrated with the cephalosporinase enzyme, its advantage should be its generality. The yeast three-hybrid system should be able to link both bond cleavage and bond formation reactions to transcription of a reporter gene. In the case of bond cleavage reactions, the enzyme should ensure cell survival by cleaving a Mtx-Dex substrate and disrupting transcription of a toxic reporter gene. For bond formation, the enzyme should form a bond between Mtx and Dex, activating transcription of an essential reporter gene. The read-out system, Mtx-Dex, LexA-DHFR, B42-GR, and the reporter gene, can all remain constant while the chemistry changes. Thus, all that needs to be changed for each new reaction is the Mtx-substrate-Dex or Dex-substrate and Mtx-substrate molecules synthesized in the lab and the enzyme library. This assay could be used to engineer glycosyltransferases, aldolases, esterases, amidases, and “diels–alderases,” all with a variety of substrate specificities and regio- and stereoselectivities. By converting the assay to a coupled enzyme assay, it may even be possible to detect oxidases and reductases. In addition to providing a powerful selection for the evolution of enzymes with new activities, there should be many uses for a reaction-independent high-throughput assay for enzyme catalysis. The assay can be used to study enzyme function, either to test hundreds of mutants to identify amino acids important for the catalytic activity of an enzyme or hundreds of different molecules to determine the substrate specificity of an enzyme. Likewise, the assay could be applied to drug discovery by screening libraries of small molecules on the basis of inhibition of enzyme activity and a change in transcription of the reporter gene. A distinct advantage is that a new assay would not have to be developed for each new enzyme target. Finally, this assay should be particularly well suited to proteomics. A battery of Mtx-Dex substrates with different substrates as linkers could be prepared and then used to screen cDNA libraries to identify enzymes that fall into common families, such as glycosidases or aldolases. Because mammalian, as well as yeast, three-hybrid assays are standard, the assays could be carried out in the endogenous cell line, ensuring correct posttranslational modification of the proteins. The key to all of these applications is a robust assay for enzymatic activity.
Supplementary Material
Acknowledgments
V.W.C. thanks R. Sauer for allowing her to initiate this project as a postdoctoral fellow in his laboratory and A. Cochran, D. Landry, S. Licht, H. Madhani, A. Mitchell, and 68-5 for their input. We are grateful for financial support for this work from the Beckman Foundation, National Institutes of Health (RO1-GM62867), and Columbia University. V.W.C. is a recipient of a Beckman Young Investigator Award, a Burroughs Wellcome Fund New Investigator Award in the Toxicological Sciences, a Camille and Henry Dreyfus New Faculty Award, and a National Science Foundation Career Award. We thank the Otsuka Chemical Co. (Osaka) for kindly providing the advanced cephem intermediate ACLE.
Abbreviations
Mtx, methotrexate
Dex, dexamethasone
DHFR, dihydrofolate reductase
GR, glucocorticoid receptor
X-Gal, 5-bromo-4-chloro-3-indolyl β-d-galactoside
ONPG, o-nitrophenyl β-d-galactopyranoside
ACLE, 7-amino-3-chlormethyl-3-cephem-4-carboxylic acid p-methoxybenzyl ester
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 16513.
References
- 1.Arnold F. (2001) Nature 409, 253-257. [DOI] [PubMed] [Google Scholar]
- 2.Lin H. & Cornish, V. W. (2002) Angew. Chem. 114, 4580-4606.; (2002) Angew. Chem. Int. Ed. 41, 4402–4425. [DOI] [PubMed] [Google Scholar]
- 3.Yano T., Oue, S. & Kagamiyama, H. (1998) Proc. Natl. Acad. Sci. USA 95, 5511-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joo H., Lin, Z. & Arnold, F. (1999) Nature 399, 670-673. [DOI] [PubMed] [Google Scholar]
- 5.Baum E., Bebernitz, G. & Gluzman, Y. (1990) Proc. Natl. Acad. Sci. USA 87, 10023-10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith T. & Kohorn, B. (1991) Proc. Natl. Acad. Sci. USA 88, 5159-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermes J., Blacklow, S. & Knowles, J. (1990) Proc. Natl. Acad. Sci. USA 87, 696-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kast P., Asif-Ullah, M., Jiang, N. & Hilvert, D. (1996) Proc. Natl. Acad. Sci. USA 93, 5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz P. & Lerner, R. (1995) Science 269, 1835-1842. [DOI] [PubMed] [Google Scholar]
- 10.Koltermann A., Kettling, U., Bieschke, J., Winkler, T. & Eigen, M. (1998) Proc. Natl. Acad. Sci. USA 95, 1421-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsen M., Stephens, D., Griffiths, D., Daugherty, P., Georgiou, G. & Iverson, B. (2000) Nat. Biotechnol. 18, 1071-1074. [DOI] [PubMed] [Google Scholar]
- 12.Pedersen H., Holder, S., Sutherlin, D., Schwitter, U., King, D. & Schultz, P. (1998) Proc. Natl. Acad. Sci. USA 95, 10523-10528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atwell S. & Wells, J. (1999) Proc. Natl. Acad. Sci. USA 96, 9497-9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firestine S., Salinas, F., Nixon, A., Baker, S. & Benkovic, S. (2000) Nat. Biotechnol. 18, 544-547. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y., Patricelli, M. & Cravatt, B. (1999) Proc. Natl. Acad. Sci. USA 96, 14694-14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin H., Abida, W. M., Sauer, R. T. & Cornish, V. W. (2000) J. Am. Chem. Soc. 122, 4247-4248. [Google Scholar]
- 17.Page M., (1992) The Chemistry of the β-Lactams (Chapman & Hall, Glasgow).
- 18.Ausubel F., Brent, R., Kingston, R., Moore, D., Seidman, J., Smith, J. & Struhl, K., (1995) Current Protocols in Molecular Biology (Wiley, New York).
- 19.Adams A., Gottschling, D., Kaiser, C. & Stearns, T., (1998) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Jencks W. P., (1969) Catalysis in Chemistry and Enzymology (McGraw-Hill, New York).
- 21.Galleni M. & Frere, J. (1988) Biochem. J. 255, 119-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobkovsky E., Moews, P. C., Liu, H., Zhao, H., Frere, J. & Knox, J. R. (1993) Proc. Natl. Acad. Sci. USA 90, 11257-11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durckheimer W., Adam, F., Fischer, G. & Kirrstetter, R. (1988) Adv. Drug Res. 17, 61-234. [Google Scholar]
- 24.Plückthun A. & Knowles, J. (1987) J. Biol. Chem. 262, 3951-3957. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.