Abstract
The construction of artificial networks of transcriptional control elements in living cells represents a new frontier for biological engineering. However, biological circuit engineers will have to confront their inability to predict the precise behavior of even the most simple synthetic networks, a serious shortcoming and challenge for the design and construction of more sophisticated genetic circuitry in the future. We propose a combined rational and evolutionary design strategy for constructing genetic regulatory circuits, an approach that allows the engineer to fine-tune the biochemical parameters of the networks experimentally in vivo. By applying directed evolution to genes comprising a simple genetic circuit, we demonstrate that a nonfunctional circuit containing improperly matched components can evolve rapidly into a functional one. In the process, we generated a library of genetic devices with a range of behaviors that can be used to construct more complex circuits.
Keywords: biocomputation, molecular evolution, repressor, gene regulation, random mutagenesis
Theoretical and experimental studies of simple synthetic genetic circuits (1–5) are elucidating the operating principles of more elaborate genetic regulatory networks in living cells. Artificial genetic circuits may also have applications in controlling cellular behavior for biotechnology or medicine. However, the few experimental demonstrations published to date underscore the difficulty of designing and implementing genetic circuits in living cells. Building a genetic circuit in vivo requires tedious optimization of many often poorly understood parameters of protein–DNA interactions and mRNA and protein stabilities, among others. Although presenting daunting challenges for engineering design, biological systems also offer a uniquely powerful design feature: the ability to evolve and be optimized under the pressure of natural or artificial selection. We are developing evolutionary strategies for constructing synthetic genetic circuits, an approach we believe to be generally applicable for optimizing individual “devices” as well as the circuits that are assembled from those devices. Thus circuit engineering in cells can tolerate designs that are bad in some features (such as matching gate characteristics), because we can force the system to evolve to produce the desired outcome. This important feature can help compensate for the designer's profound ignorance of sequence–function relationships in programming cellular behavior.
Different evolutionary strategies can be applied to genetic circuits. Recent work by Guet et al. (4) described an innovative application of biological circuit design in which the connectivity of a simple network was shuffled to observe emergence of various logic functions. Although screening random networks may yield unexpected network architectures for a given target function and even unexpected functions for given components, this approach will likely be less useful for forward engineering of genetic circuits that exhibit specific desired behaviors, primarily due to the combinatorial explosion of possibilities that accompanies increased network size and complexity. The design strategy we envision as being most effective for engineering functional circuits combines rational and evolutionary strategies, an example of which we describe in this report. First, using well-understood engineering principles, the designer assembles a circuit to fulfill the design criteria from a set of devices with well-characterized device physics (6). However, because the behavior of biological components inside living cells is highly context-dependent, the actual circuit performance will likely differ from the design predictions, often resulting in a poorly performing or nonfunctional circuit. The second phase of biological circuit engineering is tuning in vivo, using laboratory evolution methods that have proven extremely effective for molecular optimizations (7).
The objective of the present work is 2-fold. First, we demonstrate how directed evolution can rapidly tune unoptimized circuits by generating a functional circuit from two improperly matched genetic logic gates. Second, we show that the circuit outlined in Fig. 1 can serve as a platform for evaluating the characteristics of a genetic device, in this case an inverter, and that directed evolution creates a set of devices with varying characteristics. This pool of devices will be useful for the forward engineering of other biological circuits (6, 8).
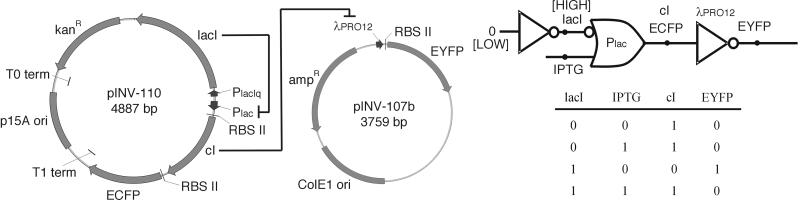
Fig 1.
The plasmid diagram shows the implementation of the present circuit. Plasmid pINV-110 constitutively expresses the LacI repressor, which inhibits transcription from the Plac promoter in the absence of IPTG. The expression of the CI repressor and ECFP fluorescent marker is controlled by Plac, which is inducible by externally added IPTG. Repressor CI acts on λPRO12 on pINV-107b to repress the transcription of the EYFP gene, the output fluorescence indicator. The two plasmids contain different origins of replication as well as different antibiotic resistance genes, which allow them to be maintained stably within a single cell. The logic diagram (Upper Right) represents the logical representation of the same biochemical circuit. The Plac promoter comprises an IMPLIES logic gate with respect to the two inputs LacI and IPTG and the output CI, whose truth table is shown below the diagram. The output of the IMPLIES gate, CI, is the input to the inverter based on the λPRO12 promoter, ultimately controlling expression of the fluorescent output, EYFP. Note that the levels of EYFP output are the inverse of the input CI in the truth table. In this study, we targeted mutations to the CI protein.
Materials and Methods
Plasmids.
Plasmids pINV-110 and pINV-112-R3 encode λ repressor CI and enhanced cyan fluorescent protein (ECFP) under the control of lac promoter Plac (Fig. 1) (8, 9). Plasmid pINV-112-R3 differs from pINV-110 in the ribosome-binding site (RBS) sequence located upstream of cI coding region, resulting in weaker translation efficiency. The ECFP, which is not used in this study, serves to report the expression level of the coexpressed cI gene. These plasmids contain a kanamycin resistance marker and the p15A replication origin. Plasmid pINV-107 (ampicillin-resistant, ColE1 replication origin) encodes enhanced yellow fluorescent protein (EYFP) under the control of the synthetic λ right promoter (λPRO12). Plasmid pINV-107b is essentially identical to pINV-107 described previously (8, 9), except for a single point mutation that removes a BsaI restriction site for cloning purposes. Plasmid pINV-107b-gfp was constructed from pINV-107b by exchanging the EYFP gene with a variant of green fluorescent protein (GFPuv from pGFPuv, CLONTECH).
Enzymes and Reagents.
All restriction enzymes, Vent DNA polymerase, T4 DNA ligase, and T4 polynucleotide kinase were purchased from New England Biolabs. TaqDNA polymerase was obtained from Promega. Competent cells were prepared by using the Z-Competent Escherichia coli Transformation Kit (Zymo Research, Orange, CA) with DH5α strain. Isopropyl β-d-thiogalactoside (IPTG) was from ICN. LB (Difco) was used for bacterial cultures. All other reagents were of the highest quality available.
Library Construction and Screening.
High-fidelity PCR was performed by using Vent DNA polymerase to amplify the region of plasmid pINV-110 outside the cI coding region with primers 5′-AGGTGGGGTCTCTTCTGGTCGGCGCATAGCTG-3′ and 5′-AGGTGGGGTCTCACAATTCCACACAACATACGA-3′. The resulting fragment containing two BsaI sites introduced in the primers was phosphorylated by T4 polynucleotide kinase followed by circularization using T4 DNA ligase to form pINV-110-lib. Two primers, 5′-AGGTGGGGTCTCAATTGTGAGCGGATAACAATGGATAAC-3′ and 5′-AGGTGGGGTCTCCCAGAACACCTTGCCGATCAGC-3′, were used to amplify the cI gene by using pINV-110 as the template. Error-prone PCR reactions were performed as described (10, 11), with varying MnCl2 concentrations to adjust the mutation rate. The library was constructed by ligating the BsaI digests of pINV-110-lib and error-prone PCR products. The ligation mixture was transformed into competent DH5α cells harboring pINV-107b-gfp and plated on LB plates containing appropriate antibiotics. Second-generation libraries were constructed similarly, using the mutants obtained from the first round as the template in the error-prone PCR reaction.
Screening was performed by visual observation of GFPuv fluorescence of the colonies on media plates placed over a UV-transilluminator (VWR Scientific, West Chester, PA) at 365 nm. Depending on the growth conditions (with or without IPTG), fluorescent or nonfluorescent colonies were picked and transferred to another plate or grown in liquid culture for further analysis.
Circuit Performance Measurements.
Detailed circuit measurements were performed in liquid cultures. Mutants that were identified by screening were isolated as plasmids (pINV-110 mutants) and cotransformed into DH5α cells with pINV-107b. The EYFP reporter was chosen for its strong fluorescence and spectral separation from ECFP, whereas GFPuv was conveniently visualized on plates for high-throughput screening purposes. The cells were initially grown to stationary phase and diluted 250-fold into 1 ml of fresh LB medium containing varying amounts of IPTG and appropriate antibiotics. The cells were grown for 5.5 h at 37°C to log phase and diluted 250-fold into 1 ml of fresh medium. The cell cultures were harvested at log phase (OD600 ≈ 0.25) after 6 h of growth, centrifuged, and suspended in 150 μl of PBS, pH 7.5. The cells were transferred to a 96-well microplate in which EYFP fluorescence (500 nm excitation, 530 nm emission, 530 nm cutoff) was measured by using a fluorescence microplate reader (Molecular Devices, SpectraMAX Gemini XS). The fluorescence data were normalized against cell densities, which were measured by using a microplate reader (Molecular Devices, SpectraMAX 250) at 600 nm. Measurements of the behavior of the circuits in response to varying IPTG levels are summarized in the transfer curve, where each point on the curve represents fluorescence data from three independent cultures of the same circuit under the same induction conditions.
Results and Discussion
Genetic Circuit.
Fig. 1 shows a circuit whose ultimate output, the concentration of EYFP, is inversely related to the level of an externally added inducer, IPTG. The figure illustrates the genetic circuit, its corresponding logic circuit, and a logic function truth table for different input combinations. In the logic circuit diagram, concentrations of DNA-binding proteins and effector molecules represent signal values that serve as inputs and outputs to logic gates. Logic gate computation is performed via transcriptional regulation where the level of an input DNA-binding regulatory protein controls the level of an output protein expressed from a corresponding promoter.
In this circuit, the LacI protein is constitutively expressed from PlacIQ and represses the Plac promoter. Plac transcriptional activity is controlled by modulating the concentration of an externally added inducer, IPTG, that binds to LacI and interferes with LacI-Plac binding. The LacI/IPTG/Plac interactions constitute a logic gate that performs the IMPLIES logic function, with LacI (which in this case is always HIGH) and IPTG as the two inputs and the coexpressed CI and ECFP as the outputs. The IMPLIES function is equivalent to the logic statement “¬LacI ∨IPTG,” where ¬ denotes the NOT function and ∨ denotes the OR function. The first two columns in Fig. 1's truth table show the different input cases for the IMPLIES function, whereas the third column (CI) illustrates the output of this operation. As shown, the CI output is normally HIGH except for the case where LacI is HIGH and IPTG is LOW. In this particular circuit, only the last two cases are encountered, because LacI is always HIGH.
The CI output of the IMPLIES gate controls expression of the λPRO12 promoter. The promoter is derived from bacteriophage λ right promoter (λPR) where one of the three operator sequences (OR3) is deleted. The λPRO12 promoter is repressible by λ repressor CI as is λPR. The CI/λPRO12 repressor/promoter system is the logical analog of an inverter (NOT operation) in the sense that the concentration of EYFP (output) is inversely related to the concentration of CI (input) due to CI transcriptional repression of the λPRO12 promoter. Overall, without IPTG the CI level should be LOW and EYFP level should be HIGH; adding IPTG should increase CI levels and in turn decrease EYFP levels. Due to the cooperative binding of CI to λPRO12, one would expect an inverse sigmoidal relationship between CI and λPRO12 expression.
For this circuit to function correctly, the logic gate that performs the IMPLIES logic operation must be matched properly to the downstream logic gate that performs the NOT operation. Biochemically, this means that the protein output of the first gate (HIGH or LOW) must fall within the protein concentration range that the second gate interprets as the same logic level. Tuning device interfaces to achieve this matching is not trivial given the complex biological phenomena that govern this behavior. This problem becomes more acute as the number of different devices required to construct more complex circuits increases. In fact, when even this relatively simple circuit was first built, it did not function as intended: no EYFP was expressed at any IPTG input concentration (Fig. 2, pINV-110) due to a mismatch between the two gates. In the absence of external inducer, expression of CI from the leaky Plac was sufficient to repress λPRO12. Thus the LOW output of the IMPLIES gate was not properly mapped to the LOW input of the inverter and was interpreted as HIGH instead (9).
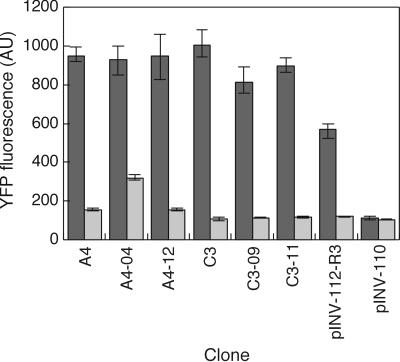
Fig 2.
Switching characteristics of selected mutants. The output YFP levels were measured at 0.1 μM (LOW input, dark bars) and 1,000 μM IPTG (HIGH input, light bars). pINV-112-R3 is the rationally engineered mutant with an altered RBS. pINV-110 is the nonfunctional circuit used as the starting circuit for directed evolution.
Weiss et al. described the rational “debugging” of this circuit (8, 9). This involved multiple site-directed mutagenesis experiments to modify the RBS and the operator sequences, guided by quantitative simulations of the circuit using published kinetic rates for λ repressor and λPR interactions. The simulations computed a variety of steady-state input/output relations for the CI/λPRO12 inverter, mapping CI input levels to corresponding λPRO12 output expression levels. Given the flat and always LOW response of the original circuit, Weiss examined mechanisms to modify the circuit to provide the desired inverse sigmoidal curve. Solutions included engineering a weaker RBS upstream of the cI gene as well as mutations in the OR1 operator site of the λPRO12 promoter to weaken repressor binding. For instance, with a weaker RBS, fewer CI translation events take place when the leaky Plac is fully repressed by a HIGH LacI, resulting in a sufficiently low level of the strong CI repressor and thereby allowing EYFP expression. This labor-intensive process rectified the mismatched gate connection and resulted in functional in vivo circuits (8, 9). We decided to explore a directed evolution approach to debugging genetic circuits, starting from Weiss' nonfunctional construct.
The circuit of Fig. 1 serves as a platform for evaluating the behavior of the second gate, in this case an inverter. On substituting the CI repressor and λPRO12 with other repressor and repressible promoter pairs, for example, the system can be used to characterize the transfer curve of an arbitrary digital logic inverter. By accumulating these data for various inverters and their mutants, we aim to facilitate the future design of more complex genetic circuits: we can choose a set of matching genetic devices that constitute a circuit from a genetic tool box. Thus, obtaining mutant devices with a wide range of device characteristics is a further goal of our directed evolution experiments.
Circuit Engineering by Directed Evolution.
Directed evolution by sequential rounds of random mutagenesis or recombination and high-throughput screening for desired characteristics has proven effective for solving difficult biological design problems, including modifying individual proteins (12) as well as whole metabolic pathways (13, 14). By limiting mutations to a specific region, we can rapidly test how that part of the circuit contributes to overall circuit function. The RBS of the circuit in Fig. 1 was modified by Weiss, but the CI protein was not engineered because the functional consequences of amino acid substitutions of proteins are very difficult to predict, even with structural information. Here, we targeted random mutations to the cI gene and its RBS to determine whether functional circuits could result from mutations in this single gene and its RBS.
Approximately 50% of the colonies in the library of circuits mutated in this fashion were fluorescent in the absence of IPTG, where the parental construct (pINV-110) was clearly nonfluorescent. This change in phenotype was expected with mutations that inactivate the CI protein and make it unable to repress transcription of the reporter gene from λPRO12. To identify the subset of functional mutants that still retain the ability to turn off the output signal on IPTG induction, fluorescent colonies from the first screen were transferred to the second plate containing 800 μM IPTG. Between 5% and 10% of the colonies were nonfluorescent in this second screen.
DNA sequencing of the circuits from these colonies revealed many solutions to the evolutionary challenge. Notable was an abundance of mutants that acquired a stop codon inside the cI gene. The CI repressor consists of an N-terminal DNA-binding domain and a C-terminal dimerization domain. The stop codons all appeared in the C-terminal domain. These mutations are expected to disrupt the cooperativity and reduce the affinity of repressor–operator binding, thereby enabling the circuit output to switch states as desired. As drastic a solution as it may seem, elimination of much of the C-terminal domain still results in a protein that retains its ability to bind DNA as well as a weak tendency to dimerize through the intact N-terminal domain (15, 16). Thus the truncated protein remains biochemically active in the context of our system. Another set of mutations disrupted the start codon or appeared upstream of cI coding region where the RBS resides. These mutations probably reduce the translation efficiency by shifting the translation start position and/or reducing the ribosome binding, a strategy analogous to the one adopted by Weiss for rational engineering (8).
Mutations that delete a significant portion of the CI protein and mutations outside the protein coding region can yield functional and potentially useful genetic devices. We wished, however, to identify which mutations in full-length CI would also yield a functional circuit. These protein mutants may allow finer tuning of protein–DNA interactions compared with engineering the RBS and/or operator sequences. Clones A4 and C3 were chosen from the first set of functional circuits for further optimization in a second generation of evolution. Clones A4 and C3 contain mutations only in the CI coding sequence and no mutations in the regulatory sequences. For the second round, the first screen was performed on 800 μM IPTG plates to isolate nonfluorescent clones, because we expected that many mutations would generate nonfunctional CI protein, which would still appear positive if we screened for HIGH signal on plates not containing IPTG. The nonfluorescent colonies were transferred to fresh plates without IPTG, and 15 fluorescent colonies derived from each parent were isolated for DNA sequencing and circuit performance analysis. Sequence data revealed that all 30 coded for full-length protein. Six were identical to their respective parents and were not analyzed further. All other clones contained one to four amino acid substitutions in the CI protein relative to their parents. Many synonymous mutations, mutations that code for the same amino acids, were also found. It is possible that these mutations affect circuit behavior by changing transcription or translation efficiency.
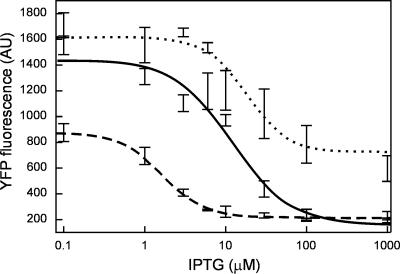
As mentioned above, the circuit of Fig. 1 serves as a platform for measuring the device physics of the cI-λPRO12 inverter. To measure quantitative characteristics of the evolved devices, experiments were conducted in liquid culture to determine the levels of HIGH and LOW output states under 0.1 μM (LOW) and 1,000 μM (HIGH) IPTG input, respectively. Data are shown in Fig. 2 for six selected mutants along with nonfunctional pINV-110 and previously (rationally) engineered functional pINV-112-R3. All of the clones tested exhibit higher EYFP levels under 0.1 μM IPTG compared with pINV-112-R3, which may be advantageous in many applications. Also apparent is the variation in the HIGH and LOW levels with respect to the same input levels of IPTG, indicating the presence of devices with different characteristics. Full transfer curves of the inverters in clones A4–04 and C3, along with pINV-112-R3, were determined across a range of IPTG input concentrations. Shown in Fig. 3, the data reveal various threshold levels at which the inverters switch from HIGH to LOW state, emphasizing that a variety of behaviors can be generated.
Fig 3.
Transfer curves (output vs. input) of selected clones containing the genetic circuit. Dotted line: A4–04, solid line: C3, dashed line: pINV-112-R3. The curves were fitted to a Hill function: y = a/(1 + bxn) + c, where x is IPTG concentration. The Hill coefficients n obtained are 1.7, 1.2, and 1.6 for A4–04, C3, and pINV-112-R3, respectively.
There is a wealth of biochemical and structural data regarding the CI protein (17, 18). The cI gene codes for a 236-residue protein consisting of two domains joined by a 40-residue linker. The N-terminal domain binds to the operator DNA sequences and interacts with RNA polymerase when bound to one of the native bacteriophage promoters, PRM. The N-terminal domain has weak tendency to dimerize: the first crystal structure was solved with the N-terminal domain of CI (residues 1–92) bound to its operator DNA as a dimer (15, 19). The C-terminal domain (residues 132–236) is believed to mediate dimerization as well as cooperative dimer-dimer interaction of CI pairs bound to neighboring operators. The structure of the C-terminal domain has recently been elucidated by x-ray crystallography (20).
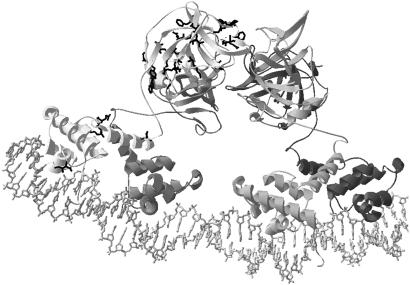
A4 and C3 from the first generation both contain two mutations each in the N-terminal domain (V73I, I54F in A4 and V73I, K67N in C3; mutant sequences are published as supporting information on the PNAS web site, www.pnas.org), which were carried over to the second generation. Of these, only I54F seems to be directly involved in protein–DNA interaction (Fig. 4). Other mutations in the N-terminal domain most likely affect the weak dimerization interaction between the N-terminal domains of the dimer. Most new mutations appeared throughout the C-terminal domain (Fig. 4). Some of the mutations in the C-terminal domain (A152S in C3–05, P158A in A4–04, E233G in C3 and its mutants) coincide with residues previously known to be involved in dimerization (20). Mutations were also observed at positions where dimer–dimer cooperative interaction would be affected (20–22) (e.g., F202V in A4 and its mutants, Q204P in A4–03 and A4–15, Q204R in C3–03, Y210F in C3–14). No mutations were found in the linker.
Fig 4.
The side chains of the mutated amino acid residues found in the evolved circuits are shown in the λ repressor CI tetramer-DNA model (Protein Data Bank ID code ). Mutated residues are shown in only one of the four repressor molecules. Summary of amino acid substitutions found in screened mutants (frequency in parentheses): I54F (11), K67N (7), V73I (18), E74D (1), E89D (1), Y103H (1), V105G (7), Q110L (1), R128G (1), W129R (1), T132A (2), E144G (7), V145A (1), A152S (1), P158A (1), L165F (1), D169A (1), E171G (1), F189V (1), V201A (1), F202V (11), Q204P (2), Q204R (1), Y210F (1), P214A (1), C215G (1), C215R (1), S220T (7), V221G (1), V221A (1), K224E (1), and E233G (7). Note that there are 10 mutants derived from A4 and six mutants from C3 that share common amino acid mutations with their respective parents.
The results indicate that the evolved mutants adjust both protein–DNA and protein–protein interactions to achieve biochemical matching of the two genetic components. However, the specific mutations that lead to a functional circuit would have been difficult to predict, even with the rich structural data available for this system. Although the “rational” debugging approach identified effective solutions that changed repressor–operator affinity and RBS strength (8, 9), the evolutionary experiment rectified the nonfunctional circuit by means of amino acid substitutions that influence CI oligomerization. Using a semiquantitative model of the circuit (9), we simulated the effect of altering the CI dimerization constant and found that adjusting the cooperativity of CI influences circuit performance in a fashion similar to changing the repressor–operator affinity or RBS strength (see supporting information on the PNAS web site). In fact, under appropriate conditions, decreasing CI dimerization can convert a nonfunctional circuit into a functional one, as was observed experimentally.
We have demonstrated that directed evolution can solve a complex in vivo optimization problem involving multiple poorly understood biochemical interactions and parameters. Laboratory evolution is an algorithmic design process, ultimately amenable to automation if the screening criteria and individual operations of mutation and screening can be standardized and described. Genetic circuits, which can often be connected to an easily observable output (e.g., a fluorescent protein) or even a selectable marker, are particularly well-suited to evolutionary optimization. The approach we have outlined should serve as a robust and widely applicable route to obtaining circuits, as well as new genetic devices, that function inside living cells.
Supplementary Material
Acknowledgments
This research was supported by National Science Foundation, Biological Information Technology and Storage (BITS) Grant EIA-0130613 and Maxygen Corporation. Y.Y. is supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad.
Abbreviations
IPTG, isopropyl β-d-thiogalactoside
RBS, ribosome-binding site
ECFP, enhanced cyan fluorescent protein
EYFP, enhanced yellow fluorescent protein
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 16516.
References
- 1.Gardner T. S., Cantor, C. R. & Collins, J. J. (2000) Nature 403, 339-342. [DOI] [PubMed] [Google Scholar]
- 2.Elowitz M. B. & Leibler, S. (2000) Nature 403, 335-338. [DOI] [PubMed] [Google Scholar]
- 3.Becskei A. & Serrano, L. (2000) Nature 405, 590-593. [DOI] [PubMed] [Google Scholar]
- 4.Guet C. C., Elowitz, M. B., Hsing, W. & Leibler, S. (2002) Science 296, 1466-1470. [DOI] [PubMed] [Google Scholar]
- 5.Hasty J., McMillen, D., Isaacs, F. & Collins, J. (2001) Nat. Rev. Genet. 2, 268-279. [DOI] [PubMed] [Google Scholar]
- 6.Weiss R., Homsy, G. E. & Knight, T. F., Jr., (1999) Dimacs Workshop on Evolution as Computation (Springer, Princeton), pp. 275–295.
- 7.Arnold F. H. (1998) Acc. Chem. Res. 31, 125-131. [Google Scholar]
- 8.Weiss R. & Basu, S., (2002) First Workshop on Non-Silicon Computing (www.hpcaconf.org/hpca8)..
- 9.Weiss R., (2001) Ph.D. thesis (Massachusetts Institute of Technology)Cambridge, MA.
- 10.Cadwell R. C. & Joyce, G. F. (1992) PCR Methods Appl. 2, 28-33. [DOI] [PubMed] [Google Scholar]
- 11.Cadwell R. C. & Joyce, G. F. (1994) PCR Methods Appl. 3, S136-S140. [DOI] [PubMed] [Google Scholar]
- 12.Farinas E. T., Bulter, T. & Arnold, F. H. (2001) Curr. Opin. Biotechnol. 12, 545-551. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt-Dannert C., Umeno, D. & Arnold, F. H. (2000) Nat. Biotechnol. 18, 750-753. [DOI] [PubMed] [Google Scholar]
- 14.Crameri A., Dawes, G., Rodriguez, E., Jr., Silver, S. & Stemmer, W. P. (1997) Nat. Biotechnol. 15, 436-438. [DOI] [PubMed] [Google Scholar]
- 15.Jordan S. R. & Pabo, C. O. (1988) Science 242, 893-899. [DOI] [PubMed] [Google Scholar]
- 16.Beamer L. J. & Pabo, C. O. (1992) J. Mol. Biol. 227, 177-196. [DOI] [PubMed] [Google Scholar]
- 17.Ptashne M., (1992) A Genetic Switch: Phage Lambda and Higher Organisms (Blackwell, Cambridge, MA).
- 18.Ackers G. K., Johnson, A. D. & Shea, M. A. (1982) Proc. Natl. Acad. Sci. USA 79, 1129-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss M. A., Pabo, C. O., Karplus, M. & Sauer, R. T. (1987) Biochemistry 26, 897-904. [DOI] [PubMed] [Google Scholar]
- 20.Bell C. E., Frescura, P., Hochschild, A. & Lewis, M. (2000) Cell 101, 801-811. [DOI] [PubMed] [Google Scholar]
- 21.Burz D. S. & Ackers, G. K. (1994) Biochemistry 33, 8406-8416. [DOI] [PubMed] [Google Scholar]
- 22.Whipple F. W., Hou, E. F. & Hochschild, A. (1998) Genes Dev. 12, 2791-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.