Abstract
The ability to photoregulate enzyme activities could provide important new opportunities for development of diagnostic assays, sequential bioprocessing, and lab assays in both traditional and microfluidic formats. We show here that the photoinduced changes in the size and hydration of a “smart” polymer chain coil can be used to regulate substrate access and enzyme activity when conjugated to the enzyme at a specific point just outside the active site. The photoresponsive polymers thus serve jointly as antennae and actuators that reversibly respond to distinct optical signals to switch the polymer–enzyme conjugates on and off, and work when the conjugate is free in solution or when immobilized on magnetic beads.
Keywords: smart polymer, photoswitch, biotechnology, bioMEMs , bioconjugates
Nature has evolved many sophisticated photoresponsive systems, such as vision, photosynthesis, and photomorphogenesis (1–4). These native photoresponsive systems are generally composed of a photosensitive element, which captures optical signals and converts them to physicochemical signals, and a second functional element (e.g., a protein domain), which senses the physicochemical signals and exhibits new output functions. A common feature is a photochromic molecule (chromophore) embedded in a biomolecular matrix. The absorbed light activates a chemical transformation in the chromophore (e.g., photoisomerization), which subsequently controls the conformation and/or assembly of the surrounding biomolecule or biomembrane. These biological systems provide models by which photoswitches for reversibly controlling biomolecular recognition events can be developed to use device friendly photoirradiation signals.
Photoswitches could be used in a number of biotechnology applications, such as photocontrolled enzymatic bioprocessing, phototriggered targeted drug delivery systems, and photocontrolled separation/recovery systems in bioMEMs formats. In general, there are two fundamental classes of photoswitchable biomolecules that have been developed (5): single-cycle and multicycle photoswitches. Single-cycle photoswitches are activated by the removal of protective groups on photoirradiation (6–9). Multicycle photoswitches have also been reported for controlling enzyme activities and ligand binding activities through direct photoisomerization of chromophores, indirect control of chromophore binding and unbinding, and by control of substrate access to immobilized enzymes in responsive hydrogels (10–16). Site-specific incorporation of photochromes into an enzyme represent a related approach (17).
We have been developing an approach to controlling protein activity that utilizes “smart” polymers as molecular switches. These stimuli-responsive polymers undergo a reversible change in size and hydrophobicity in response to external stimuli such as temperature and/or pH. We have previously demonstrated that the change in polymer physical properties through small external changes in temperature or pH can be used to reversibly control biotin and biotinylated-macromolecule access to, and/or release from, smart polymer-streptavidin bioconjugates (18–21). In this report, we have developed a photoresponsive polymer switch that serves as a molecular antennae and actuator to reversibly turn the enzyme endoglucanase 12A (EG 12A) activity on and off in response to distinct wavelengths of light (Fig. 1).
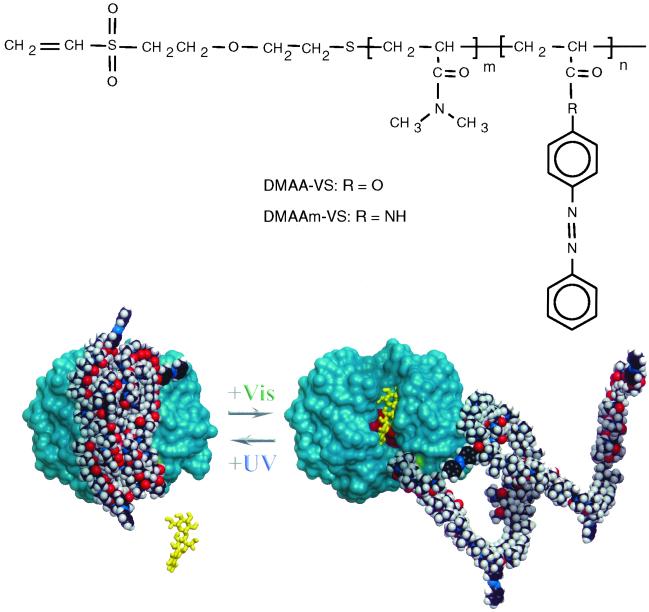
Fig 1.
Schematic model for the photoresponsive enzyme switch. The photoresponsive copolymer compositions are shown with the end-modified vinyl sulfone terminus for thiol-specific conjugations. The 3D model of EG 12A displays the relative locations of position 55 (colored green with schematic polymer coil attached) and the catalytic active site residues D99, E116, and E200 shown in red. The ONPC substrate is shown schematically to show the orientation of the active site groove. The polymer coil is shown as a 10-kDa chain with a distribution of nine DMA monomers to one azobenzene monomer.
Materials and Methods
Materials.
4-hydroxyazobenzene and 4-aminoazobenzene (Aldrich) were recrystallized from ethanol and water, and dried in vacuo. N,N-dimethyl acrylamide (DMA) (Fluka), diethyl ether, acryloylchloride, triethylamine, 2-mercaptoethanol (Aldrich) were purified by distillation under reduced pressure. 2,2′-azobisbutyronitrile (J. T. Baker, Phillipsburg, NJ) was recrystallized from methanol. Ethanol, dimethylformamide, tetrahydrofuran, methylene chloride, potassium ter-butoxide, 4-phenylazomaleinanil, divinylsulfone, 4-phenylazomaleinanil and EDTA (Aldrich) were used as received. Restriction enzymes XbaI and BglII were purchased from New England Biolabs. QIA Prep Spin Plasmid kit (Qiagen, Valencia, CA) and QuikChange Site-Directed Mutagenesis kit (Stratagene) were used as received. Butyl Sepharose (Amersham Pharmacia), bis-Tris propane, ammonium sulfate, o-nitrophenyl-β-d-cellobioside (ONPC) (Sigma), hydroxyethyl cellulose medium Viscosity (Fluka), Tris(2-carboxyethyl)-phosphine hydrochloride (TCEP), bicinchoninic acid protein assay kit, Magna Bind Streptavidin, EZ-Link sulfo-NHS-LC-LC-biotin, 4′-hydroxyazobenzene-2-carboxylic acid (Pierce), Ultrafree Biomax (Millipore), Precast gel (NOVEX, San Diego) and Kaleidoscope MW marker (Bio-Rad) were used as received. All other reagents were of analytical grade.
Synthesis of Azobenzene-Containing Monomer.
Two photoresponsive monomers, N-4-phenylazophenyl acrylamide (AZAAm) and 4-phenylazophenyl acrylate (AZAA), were synthesized that have different linkages between the azobenzenes and polymerizable groups. 4-Aminoazobenzene or 4-hydroxyazobenzene (0.2 mol) and triethylamine (0.26 mol) were dissolved in diethyl ether (200 ml). Acryloylchloride (0.24 mol) dissolved in 80 ml of diethyl ether was added dropwise at 0°C under nitrogen atmosphere with stirring. The reaction mixture was allowed to come to room temperature and stirred for 4 h. The reaction solution was filtered to remove triethylammonium chloride, washed with water and evaporated. The product was recrystallized from ethanol–water mixture and dried in vacuo. The structures were confirmed by 1H-NMR (Spectrospin and Bruker, dpx200) in perdeuterated DMSO. AZAAm: δ = 5.7–6.7 (m, 3H), 7.4–7.8 (m, 5H), 7.8–8.2 (m, 4H); AZAA: δ = 6.1–6.7 (m, 3H), 7.3–7.7 (m, 5H), 7.8–8.2 (m, 4H).
Synthesis of Photoresponsive Polymer.
Two photoresponsive polymers with hydroxyl termini, DMAAm and DMAA, were synthesized by free-radical copolymerization of DMA with AZAAm or AZAA in dimethylformamide at 60°C for 20 h, using 2-mercaptoethanol as a chain transfer agent and 2,2′-azobisbutyronitrile as an initiator (monomer concentration = 2 mol/liter). The ratios of DMA/AZAAm or AZAA/2-mercaptoethanol/2,2′-azobisbutyronitrile were varied to change the molecular weights and the photoresponsive phase transition temperatures. The products were purified by precipitation in diethyl ether three times and dried in vacuo. The contents of AZAAm or AZAA incorporated in the copolymers were determined by 1H-NMR (Spectrospin and Bruker, dpx200), comparing the ratio of aromatic and aliphatic hydrogens. The number average molecular weights (Mn) of the copolymers were determined by gel permeation chromatography, (Waters, Styragel HR3 and HR4) in tetrahydrofuran, using polystyrene standards. The hydroxyl terminus of DMAA or DMAAm was converted to a vinylsulfone (VS) group for conjugation to a sulfhydryl group in proteins. DMAA or DMAAm was dissolved in 20 ml of methylenechloride with 0.03 g of potassium tert-butoxide and 100 μl of divinylsulfone (DVS/OH = 10:1 molar ratio). The solution was stirred for 12 h at room temperature under nitrogen atmosphere.
Lower Critical Solution Temperature (LCST) Measurement.
The LCST of the polymer was determined as the temperature at 10% of the maximum absorbance at 600 nm. The polymer concentration was 2 mg/ml in 50 mM sodium acetate buffer (PB), pH 5.5. Two light sources were used to compare the polymer LCST under UV vs. visible (VIS) illumination. A UV Cure Lamp (Thorlabs, Newton, NJ) and Fiber-Lite Illuminator (Edmund Scientific, Barrington, NJ) were used with two bandpass filters (UG-1 for UV, VG-1 for VIS, Edmund Scientific). Thus, the UV light had a peak at 350 nm with a spectral band ranging from 300 to 400 nm, and the VIS light had a peak at 520 nm with a cutoff of 420 nm.
Site-Directed Mutagenesis of EG 12A.
The EG 12A mutant N55C was constructed by the QuikChange site-directed mutagenesis method (Stratagene). The QuikChange product was digested with DpnI and transformed into Top10F′ cells (Stratagene). The transformed cells were selected on agar plates with carbenicillin, and grown in LB medium. The mutated gene was isolated in a miniprep protocol (QIAprep, Qiagen). The following primers (Integrated DNA Technologies Inc., Coralville, IA) were used for the site-directed mutagenesis with the plasmid pGPTpyrG1 (22) containing the EG 12A gene inserted between unique cloning sites, XbaI and BglII (the mutated codons are underlined): 5′-p-GTGGTCCGGCGGCCAGTGCAACGTCAAGTCGTACC-3′ and 5′-p-GGTACGACTTGACGTTGCACTGGCCGCCGGACCAC-3′. The site-directed mutation was confirmed by the ABI sequencing method.
Expression of the EG 12A Mutant.
Plasmid pGPTpyrG1 with the mutated EG 12A gene was transformed into Aspergillus niger to express the site-directed mutant. The expression plasmid utilizes the strong glucoamylase promoter and A. niger strain dgr246p2 (22). Transformants were selected on media lacking uridine and then screened for high remazol brilliant blue carboxymethylcellulose activity in a shake flask culture. The best producers in shake flasks were grown in 15-liter fermentors.
Purification and Characterization of the N55C EG 12A Mutant.
The fermentation broth was treated with endoglycosidase H (Endo H) to prevent the heterogeneous glycosylation of the expressed EG 12A. Endo H (9 mg/ml) was added to the fermentation broth to a final dilution of 50 to 1 mix and incubated at 37°C overnight. Ammonium sulfate (AS) was added to 0.5 M with stirring, and subsequently centrifuged for 10 min. The pellet was discarded. Butyl Sepharose (20 ml) beads in a drip column were equilibrated with 0.5 M AS in 50 mM bis-Tris propane, pH 5.5. A 15-ml aliquot of the enzyme-treated supernatant was loaded onto the column and washed with 3 volumes of the equilibration solution. The EG 12A mutant was eluted from the beads by 50 mM bis-Tris propane, pH 5.5, without AS. The fractions that had absorbance at 280 nm were collected. The EG 12A was exchanged into 50 mM sodium acetate buffer (AB) pH 5.5 and concentrated to a 1 mg/ml concentration (Millipore Ultrafree membrane; molecular mass cutoff = 5 kDa). The site-directed mutation of N55C was confirmed by using a peptide mapping method, in which the proteins were hydrolyzed by specific proteases and the fragments were analyzed by LC-MS for the cysteine alteration.
Conjugation of Photoresponsive Polymers to the EG 12A Mutant.
Conjugations of N55C to DMAA-VS or DMAAm-VS were carried out site-specifically by reacting the vinyl sulfone group of the polymer with the sulfhydryl group of N55C in 50 mM PB, pH 8.0. To reduce disulfide bonds to sulfhydryl groups, Tris(2-carboxyethyl)-phosphine hydrochloride at 10-fold molar excess to the N55C mutant was added and incubated for 20 min at room temperature. A 50-fold molar excess of the polymer to the mutant was then added to obtain high conjugation efficiency, with overnight reaction at 4°C. The buffer was exchanged to 50 mM AB (pH 5.5) at 4°C using an ultrafiltration membrane (Ultrafree, Millipore). The conjugate was then separated from the unconjugated protein by thermally induced precipitation at 52°C. The unconjugated mutant was retained in the supernatant. The precipitate was resuspended in 50 mM AB, pH 5.5. The thermally induced precipitation was repeated three times to remove the unconjugated mutants completely. The conversion of the conjugates was quantitated with the bicinchoninic acid protein assay (Pierce).
Conjugation of 4-Phenylazomaleinanil (AZ-MI) to the EG 12A Mutant.
To investigate the polymer effect on the photoswitching activity, a single azobenzene moiety was directly conjugated to the N55C mutant as a control. N55C (200 μg) was exchanged into 100 mM sodium phosphate buffer (pH 7.0) using the Ultrafree membrane (Millipore). A 10-fold molar excess of Tris(2-carboxyethyl)-phosphine hydrochloride (TCEP) was added to the EG 12A solution to reduce the disulfide bonds to sulfhydryl groups. A 50-fold excess of AZ-MI was dissolved in dimethylformamide (10% volume of the total conjugation solution), and mixed with the EG 12A solution (EG 12A/TCEP/AZ-MI = 1:10:50). The conjugation was carried out at 4°C overnight with gentle rotation. After the reaction, the buffer was exchanged to 50 mM AB, pH 5.5. The conjugate was separated from the unconjugated AZ-MI with a polyacrylamide desalting column (Econopac 10PG, Bio-Rad).
Immobilization of the Conjugates on Magnetic Beads.
The conjugates were biotinylated by reacting with sulfo-NHS-LC-LC-biotin. The conjugate was dissolved in 100 mM sodium phosphate buffer, pH 7.4 at 4°C, at a concentration of 200 μg/ml. A 10-fold molar excess of sulfo-NHS-LC-LC-biotin, dissolved in deionized water immediately before use, was added to the solution at room temperature. The solution was gently rotated at 4°C for 5 h. The biotinylated conjugate was purified from unconjugated biotin by ultrafiltration using Biomax-5 (molecular mass cutoff = 5 kDa), followed by thermal precipitation at 52°C. The degree of biotinylation was determined by using the 4′-hydroxyazobenzene-2-carboxylic acid (HABA) assay. Briefly, 0.5 ml of mixture of avidin (20 μM) and HABA (600 μM) was added to a quartz cuvette. Absorbance at 500 nm was measured on a UV-VIS spectrophotometer, and the concentration of the biotinylated protein solution was determined from the calibration curve of a known concentration of biotin solutions. The biotinylated conjugate was immobilized on streptavidin magnetic beads. A 1-mg aliquot of magnetic beads was mixed with 20 μg of conjugate in 1 ml of 50 mM AB (pH 5.5) containing 50 mM NaCl, 5 mM EDTA, and 0.2 wt % of BSA. The quantity of the immobilized conjugate was determined by depletion of the conjugate from the supernatant. The immobilized conjugates were separated from the free polymer by washing the beads 10 times by 1 ml of 50 mM AB, pH 5.5.
Catalytic Activity Assay.
The catalytic activites of the EG 12A conjugates were measured using the chromogenic ONPC as a model substrate at 45°C under UV or VIS irradiation. ONPC is hydrolyzed by EG 12A to generate o-nitrophenol, which is detected by absorption at 405 nm. To elucidate the mechanism of the stimuli-response switches, the kinetic parameters Km and kcat were determined by Lineweaver–Burk analysis assuming Michaelis–Menten conditions. The activity of EG 12A-polymer conjugate was measured in 50 mM sodium acetate buffer, pH 5.5. ONPC (2–10 mM) and the conjugate (100 nM) solutions were preincubated at each temperature. The reaction was initiated by addition of ONPC to the EG 12 samples, followed by incubation at the same temperature. A 100-μl aliquot was taken from the reaction solution, and the reaction was terminated with the addition of 50 μl of 200 mM glycine buffer, pH 10. The absorbance at 405 nm was measured in a microtiter plate reader (Benchmark, Bio-Rad). The concentration of o-nitrophenol was calculated by using the extinction coefficient at 405 nm.
Results and Discussion
The model enzyme chosen for this work was the commercially important endoglucanase 12A (abbreviated here as EG 12A, although formally classified as Cel12A) from Trichoderma reesei that catalyzes the hydrolysis of internal β (1–4) linkages of cellulose (23). The structure of EG 12A has been recently reported at 1.9 Å resolution (22). EG 12A displays the conserved glycoside hydrolase fold of the clan-C family 12, with a carboxylic acid trio at the active site. The Asn-55 position was chosen as appropriately solvent exposed and on the outer edge of the concave cellulose binding cleft (Fig. 1) for covalent coupling to the photoresponsive polymers. The N55C site-directed mutant was subsequently constructed and produced in A. niger by using an established expression system.
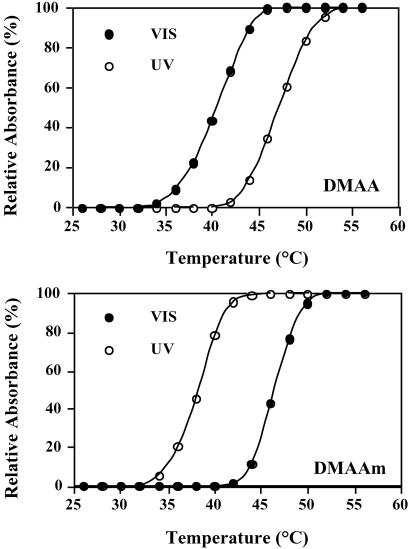
We designed two different but related photoresponsive polymers based on copolymers of DMA with azophenyl-containing monomers (24). The DMA-co-4-phenylazophenyl acrylate (DMAA) and DMA-co-N-4-phenylazophenyl acrylamide (DMAAm) copolymers display inverse phase transitions in response to UV and VIS light irradiation. They were synthesized by free-radical copolymerization of DMA with AZAAm and AZAA (Fig. 1). The polymers are photoresponsive and also display temperature-responsive LCST behavior. Their LCSTs are shifted in opposite directions under UV vs. VIS illumination (Fig. 2). The Mn of DMAA used in this study was 10.5 kDa with 6.0 mol % of AZAA and the Mn of DMAAm was 9.6 kDa with 5.5 mol % of AZAAm. These copolymer compositions were chosen because their UV/VIS phase transitions occurred within the desired temperature range of ≈40–45°C, where EG 12A has high activity and thermal stability. At these temperatures, the DMAA polymer exists as a soluble, extended coil under far UV illumination (≈350 nm), and collapses into a compact, hydrophobic conformation under VIS illumination (≈420 nm), whereas the DMAAm polymer switches between these two states in the opposite way.
Fig 2.
The LCST behavior under UV and VIS photoirradiation of the DMAA and DMAAm polymers used to construct EG 12A conjugates. The polymer concentration was 2 mg/ml in 100 mM phosphate buffer, pH 7.2.
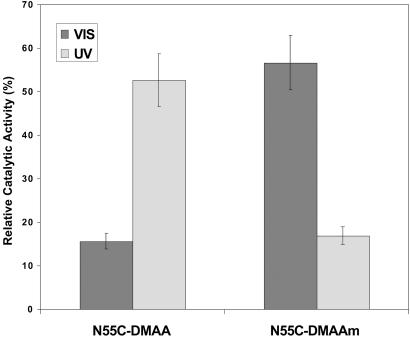
These opposite photoinduced phase transitions of DMAA and DMAAm were used to engineer molecular switches that turn “on” EG 12A as DMAA becomes hydrated and expands with VIS light, and “off” with UV light as the DMAA coil becomes hydrophobic and collapses, with the opposite responses for DMAAm. The hydroxyl termini of DMAA and DMAAm were converted to (VS) groups for conjugation to the engineered sulfhydryl group of the N55C EG 12A mutant. The photoinduced changes in enzyme activity of the conjugates were evaluated under isothermal conditions at 45°C (Fig. 3), where the free polymers (DMAA and DMAAm) exhibited high photosensitivity. With a model substrate, ONPC, the N55C EG 12A mutant displayed a small change in Km (33.9 mM vs. 19.5 mM for wild-type) and no significant change in kcat (13.6 sec−1 vs. 12.8 sec−1). As hypothesized, the N55C-DMAA conjugate was active under UV light and turned off under VIS light, whereas the N55C-DMAAm conjugate was active under VIS light and turned off under UV light (Fig. 3). These enzyme activity responses correlate with the different photoinduced phase transitions of the conjugated polymers.
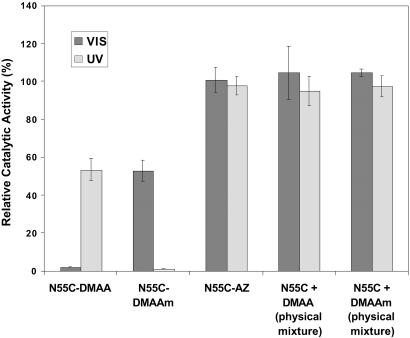
Fig 3.
Photoinduced activity changes of N55C-DMAA, N55C-DMAAm, and N55C-AZ conjugates, along with control physical mixtures of the polymers and N55C EG 12A. The activity was measured for 100 nM conjugates using 8 mM ONPC as a substrate in 50 mM sodium acetate buffer, pH 5.5, at 45°C, and normalized to the activity of the unconjugated N55C EG 12A.
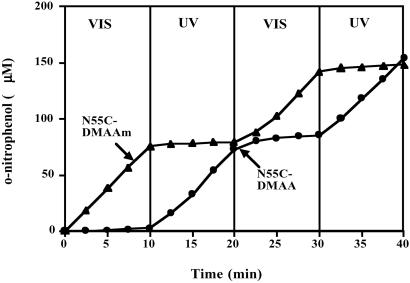
The conjugates both displayed almost a complete lack of activity when the polymers were in their collapsed state, and when the UV or VIS light stimulated the rehydration and expansion of the polymer coil, the conjugates displayed ≈60% of the activity of the unconjugated enzyme. These results reflect the steric blocking of substrate access when the polymer is collapsed, and 60% recovery of substrate access when the polymer is in the rehydrated, expanded state. This 40% loss of activity when the polymer is in the expanded state is not caused by protein denaturation as cycling does not result in any further loss in activity (see below) and the Tm of the conjugate is ≈55–60°C, well above the assay temperature. As a control, the AZ monomer equivalent was conjugated directly to the free thiol of N55C. This EG 12A-AZ conjugate did not exhibit photoresponsive enzyme activity, further demonstrating that the steric change in the size of the polymer chain coil is responsible for the photoinduced changes in the activity of the polymer-EG 12A conjugates (Fig. 3). The photoswitching was reversible and could be cycled between the active state and the off state (Fig. 4). The response time of the enzyme conjugates was less than the time needed to switch the lights (which was ≈ 1 min). In addition, the on and off states were stable for a few hours in the absence of direct light irradiation (80% retention at 1 h at 40°C), demonstrating that the thermal reconversion rates of the polymer are relatively slow.
Fig 4.
Sequential photoswitching of the activity of the N55C-DMAA and N55C-DMAAm conjugates. The product (o-nitrophenol) concentration was measured for 100 nM conjugates with free polymer present, using ONPC (8 mM) as a substrate 45°C in 50 mM sodium acetate buffer, pH 5.5.
Immobilized enzymes are also used for many bioprocessing applications. To test the activity of the EG 12A conjugates when immobilized, they were biotinylated and bound onto streptavidin magnetic beads, followed by intensive washing to separate the free polymer. The DMAA- and DMAAm-EG 12A conjugates displayed similar photoresponsive properties when immobilized on the magnetic beads (Fig. 5). Kinetic analysis of the catalytic activity data provide evidence that the underlying mechanism responsible for the photosensitivity is again steric blocking of substrate access. Although Km values depend strongly on the photostate of the DMAA and DMAAm polymers, the kcat values are largely unaltered. This finding is consistent with a steric blocking effect that results from the large change in polymer size between the extended and collapsed states that are photoinduced in the designed temperature range. However, in this immobilized enzyme application, there was some retention of activity when the polymer was switched to the collapsed state, in contrast to the results seen above with free, nonimmobilized enzyme. The catalytic site of EG 12A is a very “open” structure because of the need to accommodate the cellulose substrate, and the isolated polymer coils were apparently not large enough to achieve complete blocking. Future developments might thus use larger polymer chains and/or slightly closer attachment sites to the catalytic core residues.
Fig 5.
Photoinduced activity changes of N55C-DMAA and N55C-DMAAm conjugates when immobilized on streptavidin-coated magnetic beads. Conditions are described in Materials and Methods.
The sharp control and reversibility of the smart polymer–enzyme switches demonstrate that this approach is a useful way to control enzyme activity in a wide variety of applications. The enzyme switches work efficiently when the enzymes are free in solution and show excellent reversibility and time responses. Thus, these conjugates might be useful in microfluidic and lab-on-a-chip formats, as well as in the more conventional solution-based enzyme assays and bioprocessing applications. Because they also function in the immobilized state, these conjugates may be used to control the “on” and “off” states of enzyme activity when they are immobilized on beads and in cell sorting applications. Finally, this approach might also provide a new method for keeping enzymes turned off in pro-drug therapeutic applications until they reach their target, where the target could be activated using optical fiber technology.
Acknowledgments
We thank Dr. J. Milton Harris of Shearwater Polymers (Huntsville, AL) for helpful advice, and Dr. Richard To for helping in the construction of the EG 12A mutants. We also thank Dr. David Hyre for the enzyme–polymer model. This work was supported by National Institutes of Health Grant 53771, the University of Washington Office of Technology Transfer, the Washington Research Foundation, and the Washington Technology Center.
Abbreviations
DMA, N,N-dimethyl acrylamide
ONPC, o-nitrophenyl-β-d-cellobioside
AZAA, 4-phenylazophenyl acrylate
AZAAm, N-4-phenylazophenyl acrylamide
VS, vinylsulfone
LCST, lower critical solution temperature
AB, sodium acetate buffer
AZ, 4-phenylazomaleinanil
VIS, visible
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Haupt W. (1983) Philos. Trans. R. Soc. London B 303, 467-478. [Google Scholar]
- 2.Smith H., (1975) Photochrome and Photomorphogenesis (McGraw-Hill, London), pp. 22.
- 3.Feher G., Allen, J. P., Okamura, M. Y. & Rees, D. C. (1989) Nature 339, 111-116. [Google Scholar]
- 4.Stryer L. (1986) Am. Rev. Neurosci. 9, 87-119. [DOI] [PubMed] [Google Scholar]
- 5.Willner I. & Rubin, S. (1996) Angew. Chem. Int. Ed. Engl. 35, 367-385. [Google Scholar]
- 6.Pieroni O., Houben, J. L., Fossi, A., Costantino, P. & Ciardelli, F. (1980) J. Am. Chem. Soc. 102, 5913-5915. [Google Scholar]
- 7.Adams S. R., Kao, J. P. Y. & Tsien, Y. (1989) J. Am. Chem. Soc. 111, 7957-7968. [Google Scholar]
- 8.Turner A. D., Pizzo, S. V., Rozakis, G. & Porter, N. A. (1988) J. Am. Chem. Soc. 110, 244-250. [Google Scholar]
- 9.Nargeot J., Nerbonne, J. M., Engels, J. & Lestner, H. A. (1983) Proc. Natl. Acad. Sci. USA 80, 2395-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willner I. & Rubin, S. (1992) J. Am. Chem. Soc. 114, 3150-3151. [Google Scholar]
- 11.Montagnoli G., Monti, S., Nannicini, L. & Felicioli, R. (1976) Photochem. Photobiol. 23, 29-32. [DOI] [PubMed] [Google Scholar]
- 12.Montagnoli G., Monti, S., Nannicini, L., Giovannitti, M. P. & Ristori, M. G. (1978) Photochem. Photobiol. 27, 43-49. [DOI] [PubMed] [Google Scholar]
- 13.Willner I., Rubin, S. & Riklin, A. (1991) J. Am. Chem. Soc. 113, 3321-3325. [Google Scholar]
- 14.Karube I., Nakamoto, Y., Namba, K. & Suzuki, S. (1976) Biochim. Biophys. Acta 429, 975-981. [DOI] [PubMed] [Google Scholar]
- 15.Willner I., Rubin, S., Shatzmiller, R. & Zor, T. (1993) J. Am. Chem. Soc. 115, 8690-8694. [Google Scholar]
- 16.Hohsaka T., Kawashima, K. & Sisido, M. (1994) J. Am. Chem. Soc. 116, 413-414. [Google Scholar]
- 17.Ueda T., Murayama, K., Yamamoto, T., Kimura, S. & Imanishi, Y. (1994) J. Chem. Soc. Perkin. Trans. 1, 225-230. [Google Scholar]
- 18.Bulmus E. V., Ding, Z., Long, C. J., Stayton, P. S. & Hoffman, A. S. (2000) Bioconjugate Chem. 11, 78-83. [DOI] [PubMed] [Google Scholar]
- 19.Stayton P. S., Shimoboji, T., Long, C., Chilkoti, A., Chen, G., Harris, J. M. & Hoffman, A. S. (1995) Nature 378, 472-474. [DOI] [PubMed] [Google Scholar]
- 20.Ding Z., Fong, R. B., Long, C. J., Hoffman, A. S. & Stayton, P. S. (2001) Nature 411, 59-62. [DOI] [PubMed] [Google Scholar]
- 21.Shimoboji T., Ding, Z., Stayton, P. S. & Hoffman, A. S. (2001) Bioconjugate Chem. 12, 314-319. [DOI] [PubMed] [Google Scholar]
- 22.Sandgren M., Shaw, A., Ropp, T. H., Wu, S., Bott, R., Cameron, A. D., Stahlberg, J., Mitchinson, C. & Jones, T. A. (2001) J. Mol. Biol. 308, 295-310. [DOI] [PubMed] [Google Scholar]
- 23.Hakansson U., Fagerstam, L., Pettersson, G. & Andersson, L. (1978) Biochim. Biophys. Acta 524, 385-392. [DOI] [PubMed] [Google Scholar]
- 24.Kröger R., Menzel, H. & Hallensleben, M. L. (1994) Macromol. Chem. Phys. 195, 2291-2298. [Google Scholar]