Abstract
The TruGene human immunodeficiency virus type 1 (HIV-1) genotyping kit/OpenGene DNA sequencing system (Bayer HealthCare, Tarrytown, NY) reliably produced clinically acceptable resistance profiles for reverse transcriptase and protease inhibitors on patient samples diluted to ∼100 copies/ml following extraction with the QIAamp viral RNA minikit (QIAGEN Inc., Valencia, CA). One modification of the standard protocol was made to guarantee PCR amplification: a centrifugation step to concentrate virus was added before RNA extraction. For genotypic antiretroviral resistance testing, no significant differences in the identification and sensitivity of detection for codon mutations, base mutations, and multibase sites were found between the original and diluted samples.
Genotypic antiretroviral resistance testing (GART) is a routine tool in the management of human immunodeficiency virus (HIV) patients on highly active antiretroviral therapy (2). Commercial assays require ≥1,000 copies/ml, but noncommercial, modified commercial, and proprietary GART assays have successfully detected resistance at much lower levels (1, 3, 4, 6-8). Since 2001, the Infectious Diseases Laboratory at the Veterans Affairs Medical Center in Washington, D.C., has performed genotypic testing using the TruGene HIV type 1 (HIV-1) genotyping kit/OpenGene DNA sequencing system (Bayer HealthCare LLC, Tarrytown, NY). We observed that this commercial assay was consistently able to genotype clinical samples well below 1,000 copies/ml. We designed this study to validate genotypic testing at very low viral loads because an earlier recognition of resistance to antiretroviral agents may well enhance the clinician's opportunity to alter therapy, prevent the emergence of added resistance, and better preserve future treatment options.
(Results of this study were presented in part at the 45th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, D.C., 16 to 19 December 2005.)
MATERIALS AND METHODS
Peripheral blood was drawn by venipuncture into EDTA tubes, kept at room temperature, and centrifuged within 4 h of drawing at 1,000 × g for 15 min. Plasma was stored at −80°C and thawed in a 10 to 15°C water bath for 15 min before use. Twenty consecutive clinical GART specimens were chosen for the presence of HIV RNA with multiple mutations leading to complex resistance profiles and with >1,000 copies/ml (median, 13,933 copies/ml; range, 1,032 to 345,560 copies/ml; Versant HIV-1 RNA 3.0 assay [bDNA]; Bayer HealthCare). The samples were diluted in human serum-based dilution matrix (AcroMetrix Corporation, Benecia, CA) or HIV-negative plasma to ∼100 copies/ml (median, 136 copies/ml; range, <75 to 254 copies/ml) (Table 1). The diluted samples were stored and thawed in the same manner as the original samples before processing.
TABLE 1.
Versant HIV-1 RNA 3.0 Assay (bDNA) results and GART specimen handling for 20 original versus diluted clinical samplesa
| Sample | Originalb copies/ml | Dilutedc,d copies/ml |
|---|---|---|
| 1 | 14,656 | 163 |
| 2 | 9,350 | 183 |
| 3 | 10,874 | 139 |
| 4 | 4,278 | 254 |
| 5 | 28,603 | 244 |
| 6 | 8,322 | 136 |
| 7 | 4,709 | 126 |
| 8 | 64,558 | 136 |
| 9 | 345,560 | 124 |
| 10 | 1,032 | 177 |
| 11 | 118,970 | 209 |
| 12 | 13,210 | 152 |
| 13 | 33,931 | 116 |
| 14 | 19,354 | 127 |
| 15 | 6,819 | 112 |
| 16 | 95,475 | 143 |
| 17 | 8,361 | 90 |
| 18 | 58,402 | 101 |
| 19 | 1,127 | <75 |
| 20 | 44,555 | 136 |
GART was performed by using QiaAmp viral RNA minikit extraction followed by TruGene HIV-1 genotyping kit PCR amplification. Three TruGene-certified technologists processed and analyzed specimens.
Viral-load determinations were performed with two kit lots in 10 runs; GART was performed with two TruGene kit lots in 6 runs.
Viral-load determinations were performed with one kit lot in three runs; GART was performed with one TruGene kit lot in seven runs.
Dilution Matrix (DM) was used as diluent for samples 1 to 14, but amplification failed in 2, 5, 12, 13, and 14. The diluent was switched to HIV-negative plasma, and samples 5, 15, 16, and 18 were amplified but not 17, 19, and 20. Viral dilutions of sample 2 in DM and samples 12, 13, 14, 17, 19, and 20 in HIV-negative plasma were concentrated by centrifugation before extraction. Amplification was then successful.
Viruses in the diluted sample were only when PCR amplification failed concentrated: one milliliter was centrifuged at 23,500 × g and 4°C for 75 min in a 2-ml microtube (PN 72.693.005; Sarstedt Inc., Newton, NC), and 800 μl of supernatant was removed. The manual extraction of 140 μl of original, diluted, or diluted-centrifuged samples was performed using the materials and Mini Spin protocol provided by the QIAamp viral RNA minikit (QIAGEN Inc., Valencia, CA) and absolute ethanol (catalog no. E7023; Sigma-Aldrich, St. Louis, MO).
For GART, the manufacturer's directions for the TruGene HIV-1 genotyping kit and OpenGene DNA sequencing system were also followed. The kit required diethyl pyrocarbonate-treated water (catalog no. 9916; Ambion Inc., Austin, TX). Codons 4 to 99 of the protease (PR) gene and codons 38 to 247 of the reverse transcriptase (RT) gene of the pol region of the HIV-1 genome were amplified by PCR and sequenced. After base call editing by two technologists (all Jump to Position options were checked), a TruGene HIV-1 resistance report was produced using the Bayer GuideLines 9.0 rules. The OpenGene genetic fingerprint was used to confirm specimen identity and the absence of cross-contamination as an adjunct to the negative control. Resistance interpretations for seven protease inhibitors (PI) were determined by the presence or absence of 20 specific PR resistance codons. Resistance interpretations for seven nucleoside and nucleotide reverse transcriptase inhibitors (NRTI/NtRTI) and three nonnucleoside reverse transcriptase inhibitors (NNRTI) were based on 22 and 14 RT resistance codons, respectively. The report's four interpretation choices were (i) no evidence of resistance, (ii) possible resistance, (iii) resistance, and (iv) insufficient evidence.
Statistical analysis.
Continuous data from 20 original and diluted samples were analyzed by two-tailed, paired sample t test with significance taken at P values <0.05. Correlations were performed for the same 20 paired samples.
RESULTS
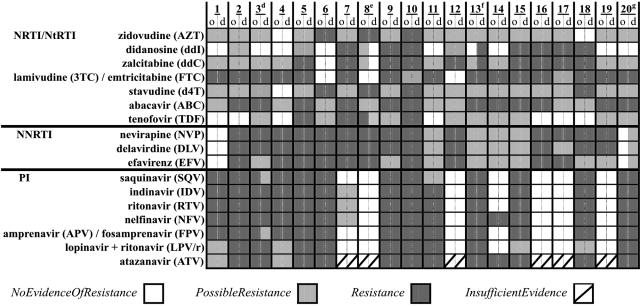
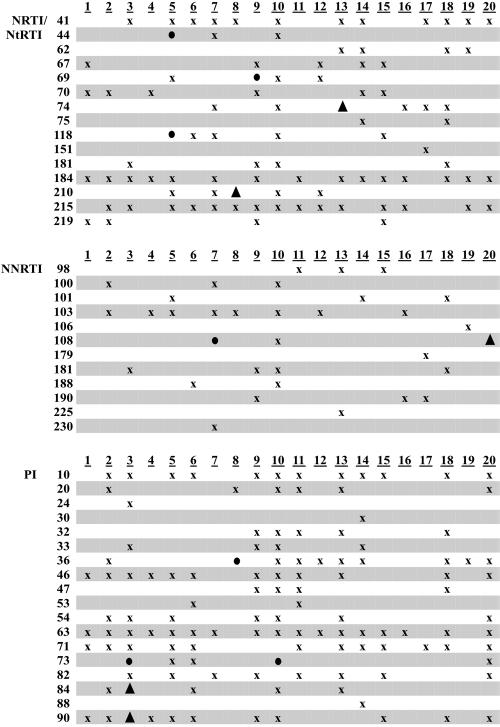
All 20 dilutions were successfully amplified and sequenced. The complete resistance profiles matched for 16 of the paired samples (Fig. 1). The remaining four pairs collectively had two PI, five NRTI/NtRTI, and three NNRTI discordances based on mutations at PR gene codons 84 and 90 and RT gene codons 74, 108, and 210 (Fig. 1 and 2). These crucial mutations were identified in both original and diluted specimens (Fig. 1).
FIG. 1.
TruGene HIV-1 resistance report interpretations for 20 original (o; HIV-1 viral load: median, 13,933 copies/ml; range, 1,032 to 345,560 copies/ml; bDNA) versus diluted (d; HIV-1 viral load: median, 136 copies/ml; range, <75 to 254 copies/ml; bDNA) clinical samples. GuideLines 9.0 rules for interpretations were released by Bayer in December 2004. Superscript d, discordant SQV and APV/FPV interpretations due to PR I84V and L90M mutations detected in the original sample; superscript e, discordant ddI, ddC, and TDF interpretations due to RT L210W mutation detected in original sample; superscript f, discordant ddI and ddC interpretations due to RT L74V mutation detected in diluted sample; superscript g, discordant NVP, DLV, and EFV interpretations due to RT V108I mutation detected in the diluted sample.
FIG. 2.
Distribution of TruGene HIV-1 resistance report codon mutations. Twenty clinical samples were analyzed by GuideLines 9.0 rules released by Bayer December 2004. x, results of original and diluted samples matched; ▴, results did not match, resistance interpretation affected; •, results did not match, resistance interpretation not affected. The following resistance codon mutations were not found: NRTI/NtRTI, RT gene codons 65, 77, 78, 88, 115, 116, and 161; NNRTI, RT gene codons 227 and 236; PI, PR gene codons 48 and 50.
The total numbers of RT and PR codon mutations (580) and base mutations (1,066) were determined from the results of both original and diluted GART assays and were designated “all” for correlation purposes. In both the original and diluted samples, 559 codon mutations were detected. The difference between the means (0.00 with a 95% confidence interval [CI] of −0.77 to 0.77 and P = 1.000) was not significant. The correlations (r2) of codon mutations between “all” versus original and “all” versus diluted samples were 0.984 and 0.968, respectively. The numbers of base mutations detected in the original and diluted samples were 1,009 and 1,021, respectively. The difference between the means (−0.60 with a 95% CI of −2.25 to 1.05 and P = 0.455) was not significant. The r2 values of base mutations between “all” versus original and “all” versus diluted samples were 0.939 and 0.925, respectively. Measures of concordance for GART parameters are presented in Table 2 and were similar to values we obtained in replicate testing of undiluted samples with >1,000 copies/ml (unpublished data).
TABLE 2.
Summary of GART resistance interpretation and codon and base percent concordances for 20 originala versus dilutedb samples using the TruGene HIV-1 genotyping kit and OpenGene DNA sequencing system
| Drug class or protein | Concordance (%) (n) of:
|
||||
|---|---|---|---|---|---|
| Resistance interpretationsc | Codon mutationsd | All codonsd | Base mutationse | All basese | |
| NRTI/NtRTI | 96.4 (140) | 94.5 (91)f | 98.9 (440) | 95.5 (112)f | 99.6 (1,320) |
| NNRTI | 95.0 (60) | 93.9 (33)f | 99.3 (280) | 91.7 (36)f | 99.6 (840) |
| PI | 98.6 (140) | 95.9 (123) | 98.8 (400) | 94.7 (133) | 99.4 (1,200) |
| RT | 93.4 (303) | 99.5 (4,200) | 91.1 (652) | 99.5 (12,600) | |
| PR | 92.1 (277) | 99.0 (1,920) | 89.4 (414) | 99.2 (5,760) | |
HIV-1 viral load: median, 13,933 copies/ml; range, 1,032 to 345,560 copies/ml (bDNA).
HIV-1 viral load: median, 136 copies/ml; range, <75 to 254 copies/ml (bDNA).
GuideLines 9.0 rules released by Bayer December 2004 for the analysis of seven NRTI/NtRTI, three NNRTI, and seven PI antiretroviral class drugs. A total of five NRTI/NtRTI, three NNRTI and two PI resistance interpretations were discordant.
210 reverse transcriptase gene codons were analyzed per test including 22 NRTI/NtRTI and 14 NNRTI resistance gene codons, and 96 protease gene codons were analyzed per test including 20 PI resistance gene codons. The median number of all codon mutations per clinical sample was 28 (range, 18 to 49). A total of 5 NRTI/NtRTI, 2 NNRTI, 5 PI, 20 reverse transcriptase, and 22 protease gene codons were discordant.
Base calls of original versus diluted samples were judged concordant if they agreed on the presence or absence of mutations from the wild type. The median number of all base mutations per clinical sample was 51 (range, 40 to 73). A total of 5 NRTI/NtRTI, 3 NNRTI, 7 PI, 58 reverse transcriptase, and 44 protease base calls were discordant.
Four RT Y181C mutations counted in both RT drug classes as per GuideLines 9.0 rule.
Heterozygosity is defined as the presence of multiple bases at one genomic site. For a heterozygote to be recognized or called by the OpenGene program, each base must account for at least 20% of the total signal (Fig. 3). Of the 254 multibase sites identified, 172 and 165 heterozygotes were called in the original and diluted samples, respectively (Fig. 4). The difference between the means (0.35 with a 95% CI of −4.37 to 3.07 and P = 0.808) was not significant. There were also discordant mutant base calls at 102 sites, with more base mutations detected in the diluted samples (57) than in the original samples (45). As a final observation, the five base sites that were the source of the 10 resistance interpretation mismatches were reviewed, and no uncalled signals of mutant bases were visible below the 20% threshold.
FIG. 3.
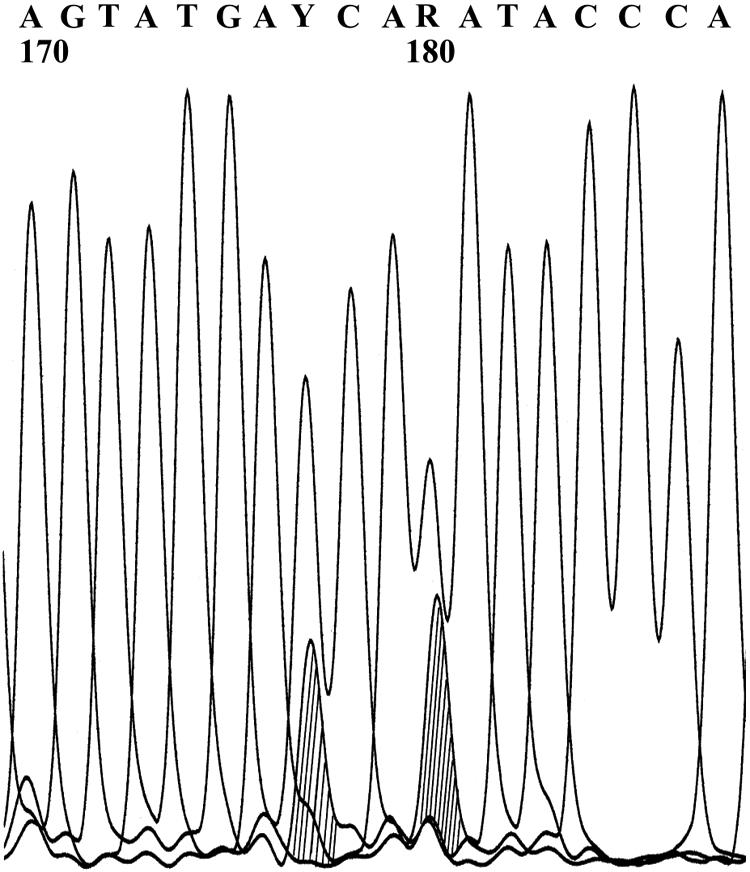
Curve Viewer results for HIV-1 protease gene reverse nucleotide sequence, sites 170 to 187. A, adenosine; G, guanine; T, thymine; C, cytosine. At base 177 is heterozygote Y (C or T), and at base 180 is heterozygote R (A or G). The sequence was obtained with the Bayer TruGene HIV-1 genotyping kit and OpenGene DNA sequencing system. The original four-color picture was converted to black and white in OpenGene with shading added at sites 177 and 180.
FIG. 4.
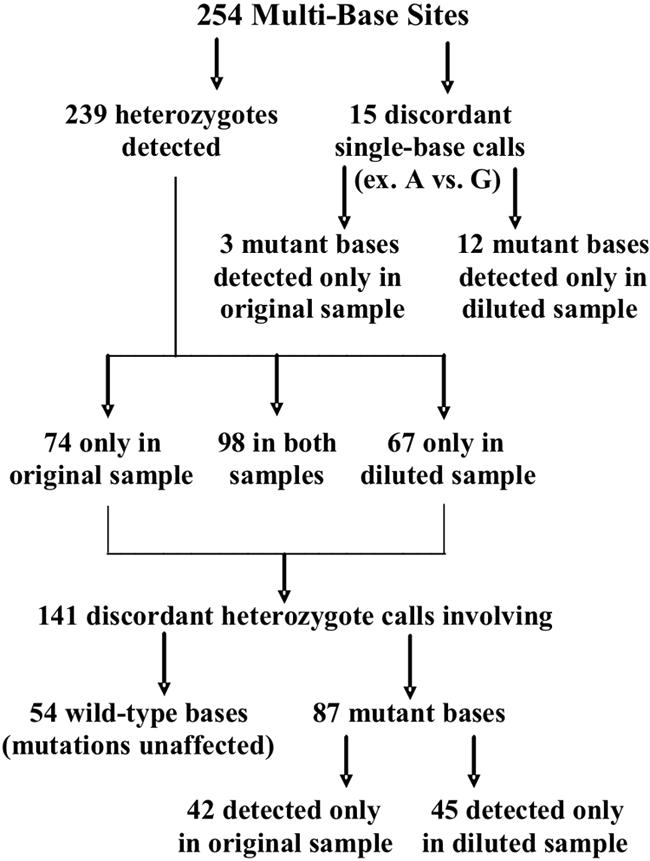
Distribution of heterozygote and mutant base detection for 20 original versus diluted clinical samples. Multibase sites were identified from the paired GART results; median, 11.5 multibase sites per clinical sample; range, 1 to 29 multibase sites per clinical sample. GART was performed by using the Bayer TruGene HIV-1 genotyping kit and OpenGene DNA sequencing system.
DISCUSSION
The materials and methods used in this study were based on QIAGEN RNA extraction followed by TruGene GART with one modification of the standard protocol. An optional centrifugation step was added to concentrate virions prior to extraction. Although developed independently, this procedure is similar to one previously described (R. M. Lloyd, Jr., R. Schuurman, H. L. Stang, T. De Groot, L. M. Hough, D. A. Burns, R. L. Mathis, and P. M. Feorino, Accuracy and reproducibility of ultra low-copy HIV viral genotyping, QIAGEN News customer application article, 2000; J. Lawrence, R. M. Lloyd, Jr., M. Hough, P. M. Feorino, and M. A. Thompson, 7th Conf. Retrovir. Opportun. Infect., abstr. 795, 2000). The latter reported genotyping 46/46 plasma samples containing HIV RNA at 50 to 400 copies/ml.
One unexpected result was observed in this study. Using only the standard protocol, samples diluted in dilution matrix failed amplification 5/14 times and after switching to HIV-negative plasma 3/7 times (Table 1). Adding the centrifugation step corrected this problem. However, in the prior 3 years, no modification was needed to amplify 26/27 clinical samples at <1,000 copies/ml (median, 562 copies/ml; range, <50 to 998 copies/ml). Altering the matrix by diluting the original clinical samples may have decreased viral RNA recovery during extraction, reducing the amount of template available for amplification (5).
In conclusion, we confirmed the findings of Lawrence et al. (7th Conf. Retrovir. Opportun. Infect.) by demonstrating that QIAGEN RNA minikit extraction and GART by the TruGene HIV-1 genotyping kit/OpenGene DNA sequencing system could reliably genotype HIV RNA samples of ∼100 copies/ml by following the standard protocol with the addition of a pre-RNA extraction centrifugation step to concentrate virions. In addition, we showed that, from highly drug-experienced patients, the resistance profiles of samples of ∼100 copies/ml matched well the profiles of original samples of >1,000 copies/ml with no significant difference in test performance. Our laboratory's ability to obtain early genotypic results for the HIV patient with persistent, low-level viremia may allow a timelier change in therapy and potentially increase the durability of viral suppression.
Acknowledgments
We thank K. C. Rexroth for technical support.
The Institutional Review Board and the Research & Development Committee at this VA Medical Center reviewed and approved this study. The views expressed in this article are those of the authors and do not reflect the policies of the Department of Veterans Affairs. The authors report no conflicts of interest.
REFERENCES
- 1.Aleman, S., K. Soderbarg, U. Visco-Comandini, G. Sitbon, and A. Sonnerborg. Drug resistance at low viraemia in HIV-1-infected patients with antiretroviral combination therapy. AIDS 16:1039-1044. [DOI] [PubMed]
- 2.Department of Health and Human Services Panel on Clinical Practices. 6. October 2005, posting date. Treatment of HIV infection: guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents. [Online.] http://www.aidsinfo.nih.gov/guidelines/.
- 3.Elbeik T., B. S. Hoo, M. E. Campodonico, J. Dileanis, F. F. Fay, R. L. Bortolozzi, M. S. Benetti, O. H. Fay, N. Marlowe, O. Petrauskene, D. Chernoff, L. Smith, and V. L. Ng. In vivo emergence of drug-resistant mutations at less than 50 HIV-1 RNA copies/ml that are maintained at viral rebound in longitudinal plasma samples from human immunodeficiency virus type-1-infected patients on highly active antiretroviral therapy. J. Hum. Virol. 4:317-328. [PubMed]
- 4.Hermankova, M., S. C. Ray, C. Ruff, M. Powell-Davis, R. Ingersoll, R. T. D'Aquila, T. C. Quinn, J. D. Siliciano, R. F. Siliciano, and D. Persaud. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA 286:196-207. [DOI] [PubMed]
- 5.Kuritzkes, D. R., R. M. Grant, P. Feorino, M. Griswol, M. Hoover, R. Young, S. Day, R. M. Lloyd, C. Reid, G. F. Morgan, and D. L. Winslow. 2003. Performance characteristics of the TRUGENE HIV-1 genotyping kit and the Opengene DNA sequencing systerm. J. Clin. Microbiol. 41:1594-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackie, N., S. Dustan, M. O. McClure, J. N. Weber, and J. R. Clarke. 2004. Detection of HIV-1 antiretroviral resistance from patients with persistently low but detectable viraemia. J. Virol. Methods 119:73-78. [DOI] [PubMed] [Google Scholar]
- 7.Nettles, R. E., T. L. Kieffer, R. P. Simmons, J. Cofrancesco, Jr., R. D. Moore, J. E. Gallant, D. Persaud, and R. F. Siliciano. Genotypic resistance in HIV-1-infected patients with persistently detectable low-level viremia while receiving highly active antiretroviral therapy. Clin. Infect. Dis. 39:1030-1037. [DOI] [PubMed]
- 8.Parkin, N. T., S. D. Deeks, M. T. Wrin, J. Yap, R. M. Grant, K. H. Lee, D. Heeren, N. S. Hellmann, and C. J. Petropoulos. 2000. Loss of antiretroviral drug susceptibility at low viral load during early virologic failure in treatment-experienced patients. AIDS 14:2877-2887. [DOI] [PubMed] [Google Scholar]