Abstract
Quercetin 2,3-dioxygenase (2,3QD) is the only firmly established copper dioxygenase known so far. Depending solely on a mononuclear Cu center, it catalyzes the breakage of the O-heterocycle of flavonols, producing more easily degradable phenolic carboxylic acid ester derivatives. In the enzymatic process, two C—C bonds are broken and concomitantly carbon monoxide is released. The x-ray structures of Aspergillus japonicus 2,3QD anaerobically complexed with the substrate kaempferol and the natural substrate quercetin have been determined at 1.90- and 1.75-Å resolution, respectively. Flavonols coordinate to the copper ion as monodentate ligands through their 3OH group. They occupy a shallow and overall hydrophobic cavity proximal to the metal center. As a result of a van der Waals contact between the most outward flavonol A-ring and Pro164, a flexible loop in front of the active site becomes partly ordered. Interestingly, flavonols bound to 2,3QD are bent at the C2 atom, which is pyramidalized. The increased local sp3 character at this atom may stabilize a carbon-centered radical activated for dioxygen attack. Glu73 coordinates the copper through its Oɛ1 atom. The short distance of about 2.55 Å between its Oɛ2 atom and the flavonol O3 atom suggests that a hydrogen bond exists between the two atoms, indicating that Glu73 can act as a base in flavonol deprotonation and that it retains the proton. Structure-based geometric considerations indicate O2 binding to the flavonol C2 atom as the preferred route for flavonol dioxygenation.
In several aerobic metabolic pathways, O2 is incorporated into organic compounds through monooxygenase- or dioxygenase-catalyzed reactions (1). For instance, oxygenation is often used to make lipids and particularly aromatic molecules amenable to further biochemical transformations. However, owing to its triplet ground state, O2 cannot directly react with singlet ground state substrates to produce singlet state products because that would imply a violation of the conservation of the total angular momentum (2). Some form of activation, that is, some way to overcome the spin forbiddenness of the process, is therefore required to make dioxygen react with organic molecules. Often, metal cofactors are used for this purpose, with iron almost invariably being the element of choice for dioxygenases (3). Detailed crystallographic and spectroscopic studies have elucidated how the mononuclear non-heme iron center in these enzymes is exploited to accomplish catalysis (for reviews, see refs. 4 and 5). It seemed that the extradiol-type dioxygenases use an Fe2+ ion to directly ligate and activate O2, whereas the intradiol-type enzymes employ Fe3+ to activate the substrate before dioxygen attack.
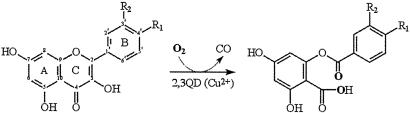
Quercetin 2,3-dioxygenase (2,3QD) belongs to the cupin superfamily (6, 7) and is the only dioxygenase unambiguously known to rely on a mononuclear copper center for activity (8–10). It is a type 2 copper-dependent enzyme expressed by Aspergillus species when grown on complex aromatic compounds, such as rutin or quercetin. 2,3QD catalyzes the cleavage of the O-heteroaromatic ring of flavonols, yielding the corresponding depside (phenolic carboxylic acid ester) and carbon monoxide (Fig. 1). The general lines of a possible mechanism for the enzymatic process have been suggested by the early biochemical work on Aspergillus flavus 2,3QD (11) and by model studies (12–14). To overcome the singlet-triplet spin barrier, the reaction has been proposed to proceed through a Cu+-flavonoxy radical formed from tautomerization of the Cu2+-flavonolate adduct. Dioxygen attack could then occur either directly at the metal or at the radical center.
Fig. 1.
Scheme of 2,3QD-mediated dioxygenation of the flavonol substrates quercetin (5,7,3′,4′-tetrahydroxy flavonol, R1 = R2 = OH) and kaempferol (5,7,4′-trihydroxy flavonol, R1 = OH, R2 = H). Flavonol ring and atom nomenclature is given on the left.
The environment of the copper ion of Aspergillus japonicus 2,3QD has been probed by x-ray crystallography (6) and electron paramagnetic resonance (EPR) spectroscopy (10) and found to be heterogeneous. The crystal structure at 1.6 Å resolution shows the metal ion chiefly bound in a distorted tetrahedral geometry by three histidine residues (His66, His68, and His112) and a water molecule. However, a minor mixed trigonal bipyramidal/square pyramidal coordination is also present. In the latter geometry, the water molecule is positioned in the equatorial plane, and the side chain of Glu73 additionally coordinates the metal. Interestingly, x-ray absorption spectroscopy (15) and EPR (10) data indicate that, on anaerobically binding of flavonol substrates [enzyme⋅substrate (E⋅S) complexes] the site becomes penta-coordinate and ordered. The copper ion maintains a formal Cu2+ oxidation state.
In this report, we present the crystal structures of A. japonicus 2,3QD anaerobically complexed with the substrate kaempferol (2,3QD⋅KMP) and with the natural substrate quercetin (2,3QD⋅QUE) at 1.90 and 1.75 Å resolution, respectively. These structures provide detailed insights into the enzymatic mechanism of this intriguing copper-dependent dioxygenase.
Materials and Methods
Expression and Purification.
2,3QD from A. japonicus was overexpressed in Aspergillus awamori as described (10). 2,3QD used for obtaining the 2,3QD⋅KMP complex was purified according to Fusetti et al. (6) whereas the material used for producing the 2,3QD⋅QUE complex was purified as described by Kooter et al. (10). The essential difference between the two purification protocols is that, in the latter, the step through a Cu-chelating Sepharose column was omitted. As a result, the protein obtained from this purification had a lower copper content (0.8 vs. 1.0 atoms of Cu per monomer as indicated by atomic absorption measurements).
Crystallization and Soaking Experiments.
For crystallization purposes, the purified 2,3QD glycoenzyme (≈35% carbohydrate content, wt/wt) was treated with the deglycosylating enzyme Endoglycosidase-H (Boehringer-Mannheim) following the protocol indicated by the manufacturer. The released carbohydrates were removed by applying the protein solution to a Pharmacia Superdex G75 column. This procedure reduced the residual glycosylation to ≈3–5% (wt/wt). An activity assay, carried out according to the procedure of Oka et al. (16), indicated that deglycosylation had no adverse effect on the catalytic efficiency of 2,3QD.
Crystals of 2,3QD were grown at room temperature by using the hanging-drop method. The protein solution (15 mg/ml in 50 mM Mes buffer, pH 6.0, 2-(N-morpholino)-ethanesulfonic acid) was mixed in a 1:1 ratio with a reservoir solution containing PEG 8000 (21–23%, wt/vol), 200 mM ammonium sulfate, and 100 mM sodium citrate (pH 5.2). Crystals belonging to space group P21 grew in about 3 wk. 2,3QD⋅KMP and 2,3QD⋅QUE complexes were obtained by anaerobically soaking crystals of 2,3QD for about 18 h in a reservoir solution enriched by a 10-mM DMSO solution of the appropriate substrate. The DMSO content of the soaking solution was 10% (vol/vol). At this concentration, DMSO does not inhibit the enzyme (9, 11). All oxygen-free crystal manipulations were carried out in an anaerobic chamber (Belle Technology, Portesham, Dorset, U.K.) at an O2 pressure lower than 1–2 ppm. Before their use, all reagents were equilibrated in the chamber for at least 24 h.
Data Collections and Refinement of the Models.
Intensities from a crystal of 2,3QD⋅KMP were collected at beam line BM14 [European Molecular Biology Laboratory (EMBL)/Grenoble] by using a MAR345 image plate detector. Diffraction data from a 2,3QD⋅QUE crystal were collected by using synchrotron radiation at beam line ID14-1 (EMBL/Grenoble) employing a MAR charge-coupled device detector (MAR-Research, Hamburg, Germany). Both data collections were carried out at 100 K. MPD (2-methyl,2,4-pentanediol, 25% vol/vol) was used as a cryo-protectant. Integration, scaling, and merging of the data were done with the hkl suite (17). Data collection and processing results together with the final refinement statistics are summarized in Table 1. Restrained refinement of both models including TLS refinement (18, 19) was performed with the program refmac5 (20, 21). Initial phases were obtained from the model of native 2,3QD at 1.6 Å (PDB code ), from which all of the non-protein atoms and residue Glu73 were removed. No restraints on the metal-ligand distances were used. The molecular model of QUE was obtained from the Cambridge Structural Database (22) whereas that of KMP was derived from the QUE structure by removal of the 3′OH group.
Table 1.
Data collections and refinement statistics
| Data collection | ||
| Data set | 2,3QD⋅KMP | 2,3QD⋅QUE |
| Resolution range, Å | 50.0–1.90 | 20.0–1.75 |
| Space group | P21 | P21 |
| Unit cell | ||
| a, b, c, Å | 109.27, 55.38, 123.93 | 108.94, 55.65, 123.86 |
| β, ° | 98.33 | 98.26 |
| Unique reflections | 111,775 | 148,198 |
| Completeness, % | 96.7 (94.8) | 99.4 (69.4) |
| Rsymm, % | 8.5 (37.8) | 6.2 (25.0) |
| 〈I〉/〈σ(I)〉 | 12.0 (3.4) | 28.2 (3.6) |
| Refinement | ||
| Rfactor, %/Rfree, % | 16.6/20.0 | 15.8/18.9 |
| Non-H atoms | 12,618 | 12,784 |
| Average B all atoms, Å2 | 20.6 | 24.1 |
| Average B protein, Å2 | 18.9 | 22.3 |
| Average B Cu, Å2 | 18.6 | 30.3 |
| Average B ligand, Å2 | 24.2 | 27.7 |
| Average B solvent, Å2 | 29.0 | 33.2 |
| rms bond lengths, Å | 0.009 | 0.007 |
| rms bond angles, ° | 1.35 | 1.23 |
The number in parentheses refers to the highest resolution shell.
For the definitions of standard crystallographic quantities, the reader is referred to ref. 38.
Rfree (39) was calculated using 5% of the data.
Calculated with B values obtained after individual redistribution of the TLS parameters performed with the program tlsanl.
rms deviations from ideality (40).
Analysis of the Models.
Map inspections and rebuilding were carried out with the programs quanta (Accelrys, San Diego, CA) and xtalview (23). The stereochemical quality of the final structures was assessed with the program procheck (24). Programs of the ccp4 suite (25) were used to analyze the models throughout the refinement.
Accession Numbers.
Atomic coordinates and structure factors have been deposited through the European Bioinformatics Institute with the Protein Data Bank (RCSB Rutgers University) with entry codes and .
Results
Overall Structure and Linker Region.
The overall structures of 2,3QD⋅KMP and 2,3QD⋅QUE (collectively E⋅S complexes) are virtually identical. The average rms deviation for the independent superposition of the Cα atoms of the four molecules present in the asymmetric unit (344 Cα atoms in chains A and B, and 343 Cα atoms in chains C and D) is 0.10 Å only. However, the structures of the E⋅S complexes show some clear differences with the structure of the native enzyme. Least-squares structural alignments of the Cα atoms of the most complete native atomic model (chain B encompassing 339 residues) with the 2,3QD⋅KMP and 2,3QD⋅QUE models (chain B) give an equal rms deviation of 0.86 Å, with a maximum positional difference of 7.75 Å for the Cα atoms of Ser166. Ser166 is part of the linker region (residues 146–205), which connects the N-terminal and C-terminal domains). The different conformation of residues 164–176 in this linker region accounts for the large rms differences between native 2,3QD and the E⋅S complexes.
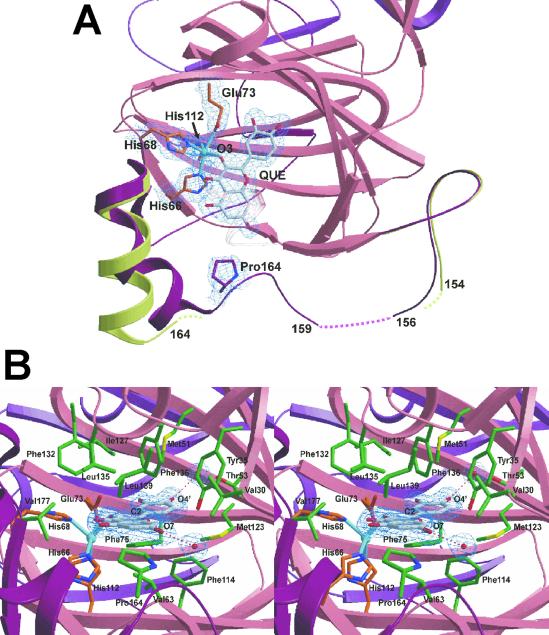
Residues 155–169 of the linker region are located in front of the active site entrance. In native 2,3QD, these residues are generally not defined in the electron density maps. Only in molecule B was electron density visible starting from residue 164 (Fig. 2A), with residues 165–174 forming a straight α-helix. In the E⋅S complexes, the flexibility of this part of the linker region is largely removed. Clear electron density is present in all four molecules of the asymmetric unit, which allowed the modeling of the amino acid chain starting from residue 159 (Fig. 2A). The 165–176 region now comprises two helices (165–170 and 171–176) angled by ≈150°, which form part of the lid that covers the active site entrance. The curved conformation is probably due to a van der Waals interaction between the Pro164 ring and the A-ring of the flavonol substrate, which blocks Pro164 more or less in the middle of the active site entrance. Its position is displaced by ≈6.0 Å compared with that occupied in the native enzyme. Residues 159–164 do not possess any defined secondary structure, and residues 157 and 158 remain disordered also in the substrate-bound form.
Fig. 2.
(A) Top view of the active site of 2,3QD⋅QUE, with a superposition of the linker regions of 2,3QD and 2,3QD⋅QUE. The N-terminal domain (residues 1–145), part of the C-terminal domain (206–350), and the linker region (146–205) of 2,3QD⋅QUE are represented in pink, violet, and dark red, respectively. The linker region of native 2,3QD is in light green. Pro164 and QUE make a van der Waals contact. This contact induces a conformational change in the linker region, stabilizing a large part of it. 2Fo-Fc electron density, for the copper site, bound quercetin, and Pro164, is contoured at the 1σ level. (B) View from the solvent into the active site of 2,3QD⋅QUE. Protein copper ligands are represented by orange sticks, QUE is in gray, and the residues in van der Waals contact with the substrate are in green. 2Fo-Fc electron density, for the substrate and the water molecules hydrogen bonded to it, is contoured at the 1σ level. Figs. 2 and 3 were generated with the program bobscript (41) and rendered with the program raster3d (42).
Substrate Binding and Copper Coordination.
KMP and QUE bind to the copper ion in a monodentate fashion through their O3 atom (Fig. 2A). They displace the water molecule observed in the native enzyme. Substrate chelation (i.e., bidentate coordination), generally observed in flavonol complexes (26, 27), does not occur, owing to the shape of the active site cavity. The substrates' B-ring binds in the active site in a region lined by the side chains of residues Tyr35, Met51, Thr53, Glu73, Phe75, Phe114, Met123, and Ile127, and by the backbone atoms of Gly125 mainly through van der Waals interactions (Fig. 2B). In this geometry, the carbonyl O4 of the flavonol molecule cannot approach the metal being blocked at a noncoordinating distance of about 3.5 Å. The A and C rings of the flavonol are also stabilized by van der Waals contacts, with the side chains of several residues that form the shallow access to the catalytic center. They are more numerous on the flavonol side trans to the position of the copper. In line with the hydrophobic character of the active site cavity, only two hydrogen bonds are present. They connect the O7 and O4′ oxygen atoms of the flavonol hydroxyl groups at the A- and B-ring, respectively, to two well defined solvent molecules.
The copper ion is penta-coordinated, with a square pyramidal geometry. HisNɛ266 (the Nɛ2 atom of His66), HisNɛ2112, GluOɛ173, and the O3 atom of the substrate form the equatorial base. HisNɛ268 is the apical ligand (Fig. 2B). Some distortion toward a trigonal bipyramidal geometry is present, as evidenced by a τ index (28) of 0.38 and 0.41 for 2,3QD⋅KMP and 2,3QD⋅QUE, respectively. Histidine-copper bond lengths do not differ much between the complexes (see Table 2), but variable metal-ligand distances are observed for the oxygen ligands. In 2,3QD⋅QUE the two Cu-O interactions are longer by 0.2–0.3 Å (see Table 2). These distances result in an average coordination distance of 2.17 Å in 2,3QD⋅QUE and of 2.04 Å in 2,3QD⋅KMP. The latter value, given the positional crystallographic uncertainty (≈0.1 Å), compares well with the average distance of 2.00 Å obtained previously for the complexes with QUE and myricetin (5′-hydroxy quercetin) from x-ray absorption spectroscopy data (15). The longer distances for the Cu-O bonds in the 2,3QD⋅QUE complex most likely reflect a partial occupation of the copper site (see Materials and Methods). We believe that the histidine-copper distances are less sensitive to a low copper content due to the fixed positioning of the imidazole rings.
Table 2.
Coordination geometries
| Atoms | 2,3QD⋅KMP | 2,3QD⋅QUE |
|---|---|---|
| Distances, Å | ||
| Cu-HisNɛ266 | 2.00 (0.09) | 2.11 (0.03) |
| Cu-HisNɛ268 | 2.09 (0.06) | 2.12 (0.05) |
| Cu-GluOɛ173 | 1.97 (0.13) | 2.28 (0.09) |
| Cu-HisNɛ2112 | 2.07 (0.04) | 2.05 (0.03) |
| Cu-FLAO3 | 2.08 (0.08) | 2.29 (0.06) |
| Angles, ° | ||
| HisNɛ266-Cu-HisNɛ268 | 103 (4) | 104 (3) |
| HisNɛ266-Cu-GluOɛ173 | 172 (4) | 169 (3) |
| HisNɛ266-Cu-HisNɛ2112 | 93 (1) | 100 (2) |
| HisNɛ266-Cu-FLAO3 | 105 (2) | 98 (3) |
| HisNɛ268-Cu-GluOɛ173 | 80 (5) | 86 (1) |
| HisNɛ268-Cu-HisNɛ2112 | 104 (2) | 115 (4) |
| HisNɛ268-Cu-FLAO3 | 96 (3) | 90 (3) |
| GluOɛ173-Cu-HisNɛ2112 | 82 (4) | 77 (2) |
| GluOɛ173-Cu-FLAO3 | 78 (4) | 80 (4) |
| HisNɛ2112-Cu-FLAO3 | 149 (4) | 144 (3) |
Distances and angles reported here are the average of the values found for the four molecules contained in the asymmetric unit. In parentheses, the SD is given.
Similarly to what was observed in the presence of the inhibitor kojic acid (29), the carboxylate group of the side chain of Glu73 coordinates the copper (Fig. 2 A and B). In both complexes, it binds with a syn-monodentate geometry through its Oɛ1 atom. The χ3 angle is ≈90° rotated compared with that observed in the coordinating conformation of native 2,3QD. In this geometry, the GluOɛ273 atom is located at ≈3.23 Å from the metal (in 2,3QD⋅KMP; 3.37 Å in 2,3QD⋅QUE) and at the short distance of 2.43/2.66 Å from the O3 atom of the flavonol substrate (KMP/QUE, respectively).
Substrate Bending and QUE Geometry.
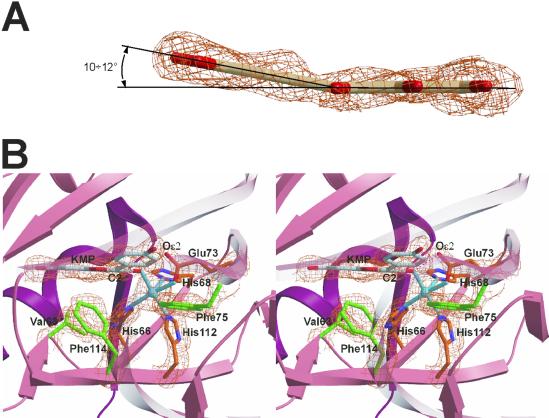
Flavonols are flat molecules, owing to their highly conjugated aromatic rings (Fig. 1). The crystal structure of quercetin [Cambridge Structural Database (CSD) (22), code FEFBEX01 (30)] shows only a 7° twist about the C2—C1′ bond, which connects the exocyclic phenyl B-ring to the rest of the molecule (see Fig. 1). Quercetin bound to 2,3QD shows a similar 6° rotation. However, differently from what is observed in the quercetin crystal structure and other Cu-flavonol complexes [CSD codes KIRCIX (31), ROKFAY (32), and XAJZUD (33)], QUE and KMP display a pyramidalization of the C2 flavonol atom when bound to 2,3QD. This result shows that the C2 atom has obtained some sp3 character (Fig. 3A). As a result of this pyramidalization, their B-rings are bent out of the plane defined by the rest of the molecule by ≈10–12°. This bending is such that the pyramidal vertex at C2 faces an empty region of the active site cavity that is lined with the side chains of Val63, His66, Phe75, His112, and Phe114 (Fig. 3B). This void surrounds the open sixth coordination site of the copper coordination sphere.
Fig. 3.
(A) Lateral view of the substrate QUE in 2,3QD⋅QUE (molecule A). Simulated annealing omit Fo-Fc electron density (43), calculated with the program cnx (Accelrys), is contoured at the 3.0σ level. In both crystallographically determined E⋅S complexes, the substrates evidence a pyramidalization of the C2 atom. (B) Side view of the catalytic center of 2,3QD⋅KMP. The copper coordination is best defined as square pyramidal, with His68 as apical ligand. Phe75, Phe114, Val63, His66, and His112 define a cavity available for the dioxygen molecule. Simulated annealing omit Fo-Fc electron density is contoured at the 3.0σ level.
The main reason for the bending seems to be the conformation of the side chains of residues Phe75 and Phe114 (Fig. 3B). Their positions preclude the B-ring to be coplanar with the C-ring because that would cause steric clashes, particularly between the PheCɛ275 and flavonolC4′, PheCξ75 and flavonolC3′, and PheCɛ2114 and flavonolC6′ atoms. Also contributing to the stabilization of the bent geometry is possibly an attractive CH/π interaction (34) between the terminal methyl group of Met51 and the flavonol B-ring. In the E⋅S complexes, this methyl group is rotated such that it points toward the aromatic B-ring (Fig. 2B). A density functional theory (DFT) calculation performed with the program adf (35) indicates the bent geometry to be about 1 kcal/mol higher in energy than the flat one.
Compared with KMP, the natural substrate QUE possesses an additional hydroxyl group at the 3′ position of the B-ring. Two conformations are therefore possible for QUE while preserving an extended electronic delocalization: cis-QUE, in which the 3′-OH group is cis to the O-heteroatom of the C-ring, and trans-QUE, in which the 3′-OH group is on the opposite side. In the x-ray structure of quercetin (30), crystal contacts induce QUE to adopt a cis conformation. In the 2,3QD⋅QUE complex, QUE is instead observed in the trans conformation only (Fig. 2A). In this geometry, the 3′OH group fills a void formed by the backbone atoms of Asn74 and the side chain of Glu73. It is not hydrogen bonded to any atom. Unfavorable interactions between the 3′OH group and the side chains of Met123 and Thr53 would exist if the cis conformation was adopted.
Discussion
Formation of the E⋅S Complex and Role of Glu73.
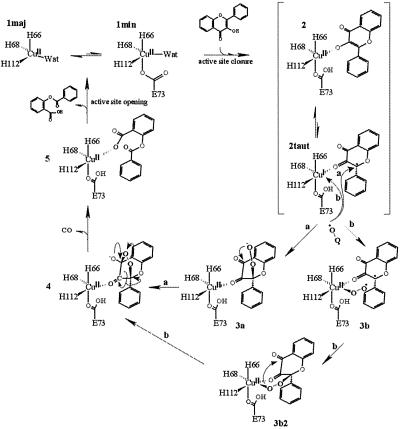
A possible mechanism for 2,3QD-mediated dioxygenation of flavonols is shown in Fig. 4. It combines the information from the present and previous crystallographic studies (6, 29), EPR investigations (10), and bio-mimetic studies (12–14). The initial step of the catalytic cycle requires complexation of the flavonol substrate to the copper ion. On substrate binding, the heterogeneous distorted tetrahedral (major)/distorted trigonal bipyramidal (minor) native copper (structures 1maj and 1min in Fig. 4, respectively) gains structural order, changing to a square pyramidal geometry represented by structure 2. Concomitantly, the portion of the linker peptide located in front of the active site entrance becomes ordered and assists, via the van der Waals interaction between Pro164 and the substrate molecule, the stabilization of the loaded substrate.
Fig. 4.
Proposed reaction mechanism for 2,3QD-mediated dioxygenation of flavonols.
The formation of the E⋅S complex likely entails the loss of the 3OH flavonol proton. Although deprotonation of the 3OH group may result from the lowering of its pKa on its complexation to the copper ion or simply from exchange to water, the 1,000-fold decrease in activity on a Glu73→Gln mutation (I. M. Kooter, personal communication) and the proper positioning of the side chain of Glu73 strongly support a role for this residue in substrate deprotonation. The observation of a close interaction between the GluOɛ273 atom and the O3 atom of the flavonol (2.43 Å in 2,3QD⋅KMP and 2.66 Å in 2,3QD⋅QUE) suggests in addition that Glu73 might be bound to the copper ion retaining the 3OH proton. In mechanistic terms, the retention of the 3OH proton in the vicinity of the copper center could be advantageous for protonating the leaving depside product (step 5 → 1). Additionally, a Glu residue bound to the metal is an effective way to allow, if needed, small adjustments in coordination geometries.
Dioxygen Attack.
The reaction catalyzed by 2,3QD is a spin-forbidden process. The triplet ground state O2 reacts with a singlet ground state flavonol to form singlet ground state products (depside and CO). Such a reaction requires some form of activation. Similarly to what is observed in the case of the Fe3+-dependent intradiol-type catechol dioxygenases, EPR (10) and x-ray absorption spectroscopy (15) experiments provide evidence that the copper center of 2,3QD in complex with its flavonol substrates retains a formal high oxidation state. Normally, complexes of Cu2+, even when coordinatively unsaturated, do not bind O2 (36). This result is not surprising because good ligands for high oxidation state metal ions are usually relatively strong bases, and O2 is not. The spin-forbiddeness of the reaction can, nonetheless, be circumvented by allowing the existence of a flavonoxy radical-Cu+ valence tautomer (2taut), arising from the flow of one electron from the flavonol into the copper. The radical center of this complex can react with the diradical O2 (route a in Fig. 4). The pyramidalization of the flavonol C2 atom, independently observed in both E⋅S complexes, is consistent with the stabilization of the unpaired electron on this atom. Analysis of the shape of the active site cavity shows that room is available for a dioxygen molecule to directly attack the flavonol C2 atom from the copper side. O2 binding would then produce the peroxide 3a, which, after nucleophilic attack on the C4 atom, would generate the endoperoxide 4. In these peroxidic intermediates, the C2 atom is sp3 hybridized. The pyramidalization observed in the bound substrates indicates that, apart from likely radical stabilization, the protein structure also preorganizes a conformation of the E⋅S complex favorable for subsequent intermediates.
The existence of an active flavonoxy radical-Cu+ complex 2taut allows, in principle, also to hypothesize a reaction mechanism in which O2 is activated by coordination to the Cu+ ion (route b) (14). An a priori prediction of whether this complex possesses the reducing equivalents to do so is difficult. However, from a geometrical point of view a binding site for O2 is available in the E⋅S complex. Differently from what was generally assumed from the chelating behavior of flavonols, the protein matrix constrains the flavonol to bind to the metal with only its O3 atom. As a consequence, the coordination position trans to His68 might be used by the dioxygen molecule to directly bind to the metal (3b). The crystal structure of 2,3QD complexed with kojic acid (29) shows that an oxygen atom can indeed occupy this sixth coordination position, producing a pseudooctahedral copper site. Owing to this result, we have tried to model the relevant structures along the reaction path b: 3b and 3b2. We find that, although formation of 3b2 from 3b is geometrically feasible, intramolecular nucleophilic attack of the oxygen atom, bound to the copper on the C4 flavonol atom required to generate the endoperoxide 4 from 3b2, seems more difficult. The model indicates that the attacking oxygen atom would be at about 3.5–4.0 Å from the C4 atom and not above the carbonyl plane, which represents the most likely direction of attack. In contrast, such an arrangement seems favored in 3a.
On the basis of geometrical considerations, we therefore conclude that the mechanism of substrate activation (route a), which assumes the formation of the endoperoxide 4 from 3a, is favored. Experiments of radical-initiated oxygenation of flavonols support a mechanism of substrate activation (37). In the presence of free radicals such as TEMPO (2,2,6,6-tetramethyl-1-piperidinyloxyl) or galvinoxyl (2,6-di-tert-butyl-a-(3,5-di-tert-butyl-4-oxo-2,5-cyclohexadien-1-ylidene)-p-tolyloxyl) flavonols undergo catalytic dioxygenation, giving the same products as the enzymatic reaction. Owing to the absence of copper, a mechanism such as that of route b is, for this reaction, impossible. O2 attack has been proposed to occur on the C2 atom of the substrate as in route a. The only difference between the enzymatic reaction and this biomimetic process seems to be the way in which the flavonoxy radical subjected to O2 attack is generated. In the former, the redox properties of the metal center are exploited whereas, in the latter, the hydrogen atom in position 3 is abstracted by the radical initiator.
Product Release.
Endoperoxide intermediate 4 represents the most convenient structure to rationalize first the release of carbon monoxide and then the formation of the depside-bound form 5 of the enzyme, for which a putative signal corresponding to a trigonal bipyramidal geometry has been observed by EPR spectroscopy (10). Carbon monoxide liberation would occur with the breakage of one O—O and two C—C bonds. Depside release and regeneration of the native enzyme would occur by proton transfer from Glu73 to the bound product, concomitant with the reopening of the active site mouth.
Conclusions
The crystal structures of 2,3QD in complex with KMP and QUE have revealed how substrates are bound in the active site of this copper-dependent dioxygenase and how the E⋅S complex is ready to be attacked by molecular oxygen. Most likely, O2 binding occurs at the activated flavonol C2 atom. This mechanism of action shares commonalities with that advanced for the intradiol-type, iron-containing catechol dioxygenases (5). These enzymes deprotonate the catechol moieties, which asymmetrically chelate the iron center. In the E⋅S state, they maintain a formal ferric oxidation state. Intradiol-type catechol dioxygenases have been proposed to activate the substrate molecules through the formation of an Fe2+-semiquinone radical complex arising from valence tautomerization of the Fe3+-catecholate complex. Because of the involvement of the former, some distortion from planarity was expected at the activated carbon atom, but such a deformation has never been observed, owing to the unfavorable shape of the substrate. In contrast, the presence of the accessory B-ring in flavonol molecules allows the bending to be unambiguously discerned in the case of 2,3QD⋅flavonol complexes. Although copper-dependent 2,3QD and Fe-containing intradiol-type catechol dioxygenases are not evolutionarily related, their apparent mechanistic similarities, particularly when considered along with genetic and structural evidences obtained by comparing 2,3QD to other metalloproteins sharing the cupin fold (6), support the view that the copper site of 2,3QD has evolved from a preexisting, possibly iron-containing, metal center. Metal swap, likely due to more adequate redox properties of copper for the particular process to be catalyzed, would have then occurred later when this metal became biologically available.
Acknowledgments
The staff scientists operating the beam lines ID14-1 and BM14 (European Molecular Biology Laboratory/Grenoble) are acknowledged for their help during the experiments. Dr. Marcel Swart is thanked for his help with the density functional theory (DFT) calculation. This research was supported by the Netherlands Foundation for Chemical Research (CW), with financial aid from the Netherlands Foundation for Scientific Research (NWO). We thank the European Union for supporting the work at the synchrotron sites through the Access to Large Installations, Human Capital and Mobility, Program.
Abbreviations
2,3QD, quercetin 2,3-dioxygenase (also known as quercetinase)
KMP, kaempferol (3,5,7,4′-tetrahydroxy flavone)
QUE, quercetin (3,5,7,3′,4′-pentahydroxy flavone)
E⋅S, enzyme⋅substrate
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes and ).
References
- 1.Hayaishi O. (1974) in Molecular Mechanisms of Oxygen Activation, ed. Hayaishi, O. (Academic, New York), pp. 1–28.
- 2.Hamilton G. A. (1974) in Molecular Mechanisms of Oxygen Activation, ed. Hayaishi, O. (Academic, New York), pp. 405–451.
- 3.Bairoch A. (1993) Nucleic Acids Res. 21, 3155-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broderick J. B. (1999) Essays Biochem. 34, 173-189. [DOI] [PubMed] [Google Scholar]
- 5.Que L. (1999) in Bioinorganic Catalysis, eds. Reedijk, J. & Bouwman, E. (Dekker, New York), pp. 269–321.
- 6.Fusetti F., Schröter, K. H., Steiner, R. A., van Noort, P. I., Pijning, T., Rozeboom, H. J., Kalk, K. H., Egmond, M. R. & Dijkstra, B. W. (2002) Structure 10, 259-268. [DOI] [PubMed] [Google Scholar]
- 7.Dunwell J. M., Khuri, S. & Gane, P. J. (2000) Microbiol. Mol. Biol. Rev. 64, 153-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oka T. & Simpson, F. J. (1971) Biochem. Biophys. Res. Commun. 43, 1-5. [DOI] [PubMed] [Google Scholar]
- 9.Hund H. K., Breuer, J., Lingens, F., Huttermann, J., Kappl, R. & Fetzner, S. (1999) Eur. J. Biochem. 263, 871-878. [DOI] [PubMed] [Google Scholar]
- 10.Kooter I. M., Steiner, R. A., Dijkstra, B. W., van Noort, P. I., Egmond, M. R. & Huber, M. (2002) Eur. J. Biochem. 12, 2971-2979. [DOI] [PubMed] [Google Scholar]
- 11.Oka T., Simpson, F. J. & Krishnamurty, H. G. (1972) Can. J. Microbiol. 18, 493-508. [DOI] [PubMed] [Google Scholar]
- 12.Speier G. (1991) in Dioxygen Activation and Homogeneous Catalytic Oxidation, ed. Simándi, L. I. (Elsevier Science, Amsterdam), pp. 269–278.
- 13.Barhács L., Kaizer, J. & Speier, G. (2001) J. Mol. Catal. 172, 117-125. [Google Scholar]
- 14.Barhács L., Kaizer, J., Pap, J. & Speier, G. (2001) Inorg. Chim. Acta 320, 83-91. [Google Scholar]
- 15.Steiner R. A., Meyer-Klaucke, W. & Dijkstra, B. W. (2002) Biochemistry 41, 7963-7968. [DOI] [PubMed] [Google Scholar]
- 16.Oka T., Simpson, F. J., Child, J. J. & Mills, C. (1971) Can. J. Microbiol. 17, 111-118. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 18.Schomaker V. & Trueblood, K. N. (1968) Acta Crystallogr. B 24, 63-76. [Google Scholar]
- 19.Winn M. D., Isupov, M. N. & Murshudov, G. N. (2001) Acta Crystallogr. D 57, 122-133. [DOI] [PubMed] [Google Scholar]
- 20.Murshudov G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. (1999) Acta Crystallogr. D 55, 247-255. [DOI] [PubMed] [Google Scholar]
- 22.Allen F. H., Bellard, S., Brice, M. D., Cartwright, B. A., Doubleday, A., Higgs, H., Hummelink, T., Hummelink-Peters, B. G., Kennard, O., Motherwell, W. D. S., Rodgers, J. R. & Watson, D. G. (1979) Acta Crystallogr. B 35, 2331-2339. [Google Scholar]
- 23.McRee D. E. (1999) J. Struct. Biol. 125, 156-165. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski R. A., Moss, D. S. & Thornton, J. M. (1993) J. Mol. Biol. 231, 1049-1067. [DOI] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 26.Balogh-Hergovich E., Kaizer, J., Speier, G., Huttner, G. & Zsolnai, L. (2000) Inorg. Chim. Acta 304, 72-77. [Google Scholar]
- 27.Speier, G., Fülöp, V. & Parkanyi, L. (1990) J. Chem. Soc. Chem. Commun., 512–513.
- 28.Addison A. W., Hendriks, H. M. J., Reedijk, J. & Thompson, L. K. (1981) Inorg. Chem. 20, 103-110. [Google Scholar]
- 29.Steiner R. A., Kooter, I. M. & Dijkstra, B. W. (2002) Biochemistry 41, 7955-7962. [DOI] [PubMed] [Google Scholar]
- 30.Jin G.-Z., Yamagata, Y. & Tomita, K. (1990) Acta Crystallogr. C 46, 310-313. [Google Scholar]
- 31.Balogh-Hergovich, E., Speier, G. & Argay, G. (1991) J. Chem. Soc. Chem. Commun., 551–552.
- 32.Lippai I., Speier, G., Huttner, G. & Zsolnai, L. (1997) Acta Crystallogr. C 53, 1547-1549. [Google Scholar]
- 33.Balogh-Hergovich E., Kaizer, J., Speier, G., Huttner, G. & Jacobi, A. (2000) Inorg. Chem. 39, 4224-4229. [DOI] [PubMed] [Google Scholar]
- 34.Nishio M., Umezawa, Y., Hirota, M. & Takeguchi, Y. (1995) Tetrahedron 51, 8665-8701. [Google Scholar]
- 35.te Velde G., Bickelhaupt, F. M., Baerends, E. J., Fonseca Guerra, C., van Gisbergen, S. J. A., Snijders, J. G. & Ziegler, T. (2001) J. Comput. Chem. 22, 931-967. [Google Scholar]
- 36.Ho R. Y. N., Liebman, J. F. & Valentine, J. S. (1995) in Active Oxygen in Biochemistry, eds. Valentine, J. S., Foote, C. S., Greenberg, A. & Liebman, J. F. (Blackie A & P, Glasgow, U.K.), pp. 1–36.
- 37.Kaizer J. & Speier, G. (2001) J. Mol. Cat. 171, 33-36. [Google Scholar]
- 38.Drenth J., (1999) Principles of Protein X-ray Crystallography (Springer, New York).
- 39.Brünger A. T. (1992) Nature 355, 472-475. [DOI] [PubMed] [Google Scholar]
- 40.Engh R. A. & Huber, R. (1991) Acta Crystallogr. A 47, 392-400. [Google Scholar]
- 41.Esnouf R. M. (1997) J. Mol. Graphics 15, 133-138. [DOI] [PubMed] [Google Scholar]
- 42.Merritt E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505-524. [DOI] [PubMed] [Google Scholar]
- 43.Brünger A. T., Adams, P. D. & Rice, L. M. (1997) Structure 5, 325-336. [DOI] [PubMed] [Google Scholar]