Abstract
Transfer RNAs are key components of the genetic code by virtue of aminoacylation reactions whereby each amino acid is linked to the tRNA that bears the anticodon for the attached amino acid. The L-shaped tRNA structure contains two domains connected at right angles through a corner formed from tertiary interactions involving loops of each domain. Some evidence suggests that the domains arose separately and eventually were fused into a single covalent structure. In this scenario, the present-day tRNA possibly developed through a noncovalently assembled heterodimeric intermediate. Trbp111 is an ancient structure-specific tRNA binding protein that interacts specifically with the outside corner of the L-shaped molecule. Plausibly, this protein could act as a chaperone to cover and protect the fragile corner and thereby have a historical role in the development of tRNA. Here we show that Trbp111 interacts with a noncovalently assembled tRNA-like structure, under conditions where it does not interact with individual tRNA domains. Trbp111 binding specifically requires formation of the tRNA-like corner. In a mixture of RNA domains, it selects those that can make the L-like structure. Thus, cofactors such as Trbp111 have the capacity to help assemble and stabilize RNA dimers that are tRNA-like.
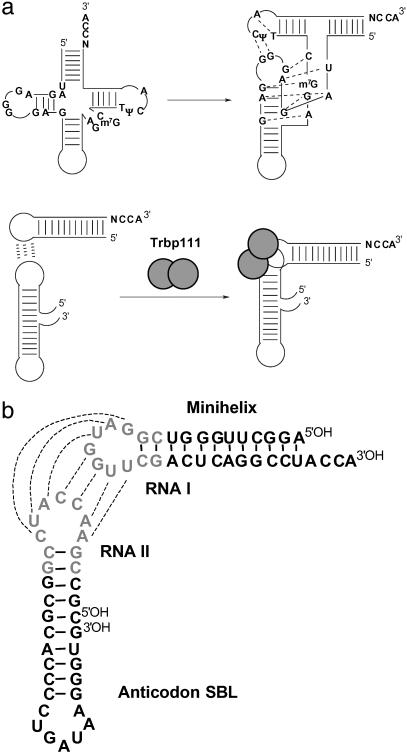
Transfer RNAs are central components of the genetic code used by all life forms, because they link amino acids to their cognate trinucleotides (1–3). They adopt an L-shaped structure composed of two domains, the acceptor-TΨC minihelix, which contains the amino acid attachment site at the 3′ end, and the anticodon stem-biloop (SBL), with one of the loops encoding the anticodon triplet (Fig. 1a). Several lines of evidence suggest that the acceptor-TΨC minihelix is the historical, earliest part of the two-domain structure, with the anticodon SBL being added later (4–8). With this scenario, in the transition from the putative RNA world to the theater of proteins, aminoacylation of the minihelix was established first. The anticodon SBL was then added as a template reading head for template-directed peptide synthesis (6). The two domains are connected by highly differentiated tertiary interactions at the corner of the resulting L-shaped structure (Fig. 1a).
Fig 1.
Establishment of L-shaped tRNA through noncovalent association of the two domains. (a Upper) The tRNA cloverleaf and its rearrangement into an L-shaped structure with tertiary interaction common to all tRNAs shown with dashed lines. (a Lower) Noncovalent assembly of RNA domains to make a tRNA-like structure. Trbp111 dimer is shown as gray spheres. Base pairing between loops is shown with dashed lines. (b) Sequence of the RNA minihelix and anticodon SBL used in the experiments. The 7-nt loop and the first two base pairs of the closing stem from RNA I and RNA II (12) are highlighted in pale gray.
A structure-specific tRNA binding protein such as Trbp111 may have played a role in stabilizing a dimeric tRNA-like intermediate (Fig. 1a; refs. 9–11). This protein, whose three-dimensional structure has been solved (10), achieves structure-specific binding by interacting with the outside corner of the L shape (11). To explore the possibility that Trbp111 might interact with a noncovalently assembled structure, we set out to construct a noncovalent dimeric RNA that was tRNA-like. For this purpose, we combined tRNA-like elements with a portion of a naturally occurring motif that generates “kissing” hairpins that have an L-like orientation (12–14). Although the corner is formed differently than that of a normal tRNA, we wanted to test whether Trbp111 would have the capacity to bind selectively and protect this kind of L-shaped structure, which is a plausible intermediate species in the development of a full tRNA. The “flexibility” of Trbp111 to recognize broadly the corners associated with L shapes would be significant (in a historical sense) for allowing it to make a wide selection of RNA dimeric complexes, some of which might proceed to develop into the full-length tRNA.
Materials and Methods
Preparation of RNAs.
The minihelix 35-mer and the anticodon SBL 34-mer were synthesized on an Expedite 8909 synthesizer (PE Biosystems, Foster City, CA) and purified as described (15). 5′-end 32P labeling was carried out with T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP and purified using a NAP-10 column (Amersham Pharmacia Biosciences).
Protein Preparation.
Aquifex aeolicus Trbp111 and mutant S82A were expressed and purified as described (9).
Gel Retardation Assay.
The reaction mixture (total volume, 10 μl) was incubated for 30 min at room temperature. A 20% sucrose solution (10 μl) containing trace dye was added and immediately loaded onto a 0.8-mm-thick, 8% 29:1 acrylamide:bis-acrylamide gel containing 45 mM Tris-borate (pH 7.0), 5 mM MgCl2, and 0.001% Triton X-100. Gels were run at 4°C at 130 V (10–13 mA) in 45 mM Tris-borate (pH 7.0), 5 mM MgCl2, and 0.001% Triton X-100 for 2.5 h. The gels were dried on a Whatman type 3 filter sheet in a vacuum drier at 80°C for 60 min and analyzed using a PhosphorImager (Molecular Dynamics). The intensity of the band was quantified and analyzed using ORIGIN 6.0 (Microcal Software, Northampton, MA). For a nonlinear curve fit, the following equation was used:
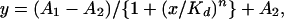 |
where A1 and A2 are the maximum and minimum value for y, Kd is the equilibrium dissociation constant, and n is the Hill constant (16).
Results
RNA Dimer Formation.
To noncovalently associate the two domains of a tRNA, we chose complementary sequences in the two loops that join together the corner of the L-shaped structure. These loops were based on RNA I and RNA II encoded by the colEI plasmid (12–14). The loops of these RNAs hybridize together to give an L-shaped structure with an angle between the domains of ≈80° (Fig. 1b; refs. 12 and 14). In our construction, the 7-nt loop and first two base pairs of the closing stem from RNA I replaced the TΨC loop and the first two closing pairs of minihelixIle. The single-stranded loop and first two base pairs of RNA II replaced the D-loop and its closing pairs in the anticodon SBL of tRNAIle.
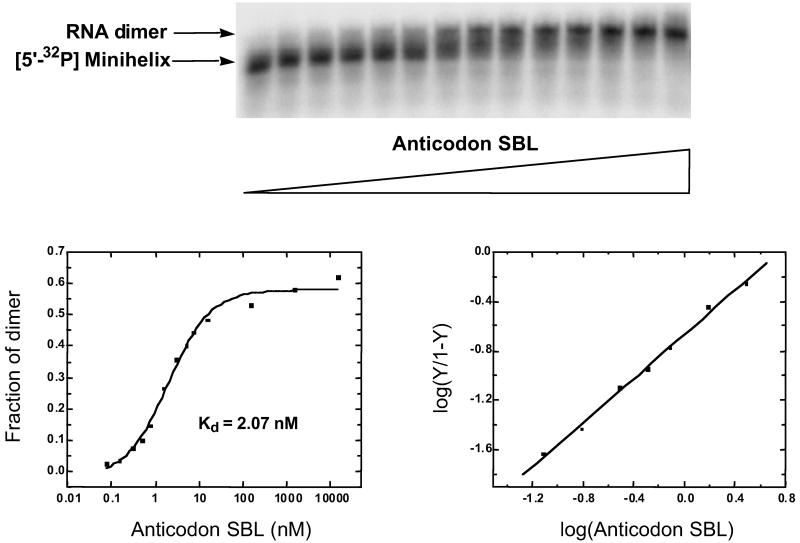
The two tRNAIle-like domains were mixed together to examine potential dimer formation by gel retardation electrophoresis on an 8% polyacrylamide gel. One domain was radioactively labeled and mixed with the unlabeled, opposite domain. When trace amounts of the 32P minihelix (≈1 nM) were added to increasing concentrations of the unlabeled anticodon SBL, a clear shift in the labeled band was observed (Fig. 2). The intensity of the shifted band was quantified by PhosphorImager (Molecular Dynamics) to determine the fraction of labeled RNA in the complex. This fraction was then plotted on a logarithmic scale to obtain the binding profile. A theoretical curve based on a simple bimolecular binding equilibrium (assuming 1:1 complex formation) gave an apparent dissociation constant Kd = 2.07 nM. The data were further analyzed by plotting log[Y/1 − Y] (Y = fraction of complex) against log concentration of unlabeled RNA. The plot thus obtained gave a Hill constant n of 0.89 and an apparent Kd of 5.60 nM.
Fig 2.
Formation of the RNA dimer complex through complementary loop–loop interactions. A gel retardation assay was carried out at pH 7, 4°C, with 5′-32P-labeled minihelix 35-mer (≈1 nM) in which unlabeled anticodon SBL was added in increasing concentrations from 0 to 15.5 μM. The shifted band was quantified and plotted on a logarithmic scale to obtain the binding profile shown on the left. The corresponding Hill plot is shown on the right.
A similar experiment was carried out in the opposite way, with the 32P anticodon SBL and the unlabeled minihelix. The same band shift was observed. A quantitative analysis of the data gave Kd = 2.24 nM and n = 0.78. These results collectively are consistent with the two domains forming a 1:1 dimer.
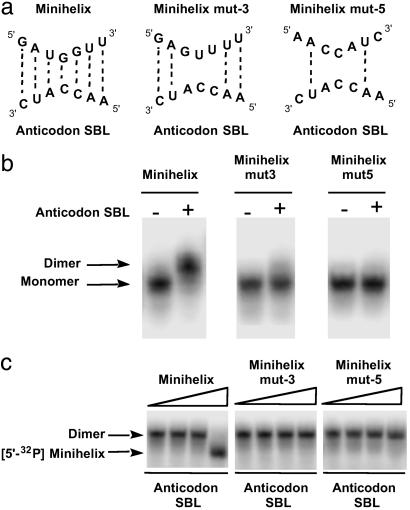
To further confirm that the RNA dimer complex formation is through complementary interaction at the loops, mutations were introduced in the loop sequence. Two mutant RNAs were synthesized and designated minihelix (mut-3) and minihelix (mut-5). These mutant RNAs disrupt 3 and 5 bp, respectively, between the minihelix and the anticodon SBL (Fig. 3a). Both (mut-3) and (mut-5) abolished dimer formation (Fig. 3b). Thus, dimer stability requires complementary base pairing interactions between the loops. Further characterizations were done by competition assays. The unlabeled “wild-type” (mut-3) or (mut-5) was added in increasing concentrations to the labeled minihelix mixed with the unlabeled anticodon SBL. The amount of labeled RNA dimer was lowered on addition of the unlabeled minihelix. In contrast, this competition was not seen when (mut-3) or (mut-5) was used (Fig. 3c). This result further demonstrates the importance of correct base pairing at the loop region.
Fig 3.
Mutation in the loop sequence disrupts complex formation. (a) Sequence of the loop used for minihelices (mut-3) and (mut-5). The potential base pairing interactions are shown with dotted lines. (b) Gel retardation assay using mutant forms of minihelix. Each 5′-32P-labeled minihelix (≈900 nM) was incubated with unlabeled anticodon SBL for 30 min at room temperature before loading on the gel. (c) Competition assay using unlabeled minihelices. Each unlabeled minihelix was added in increasing concentrations from 10 nM to 10 μM into the mixture of 5′-32P minihelix (≈1 nM) and unlabeled anticodon SBL (1.6 μM).
Ternary Complex Formation with Trbp111.
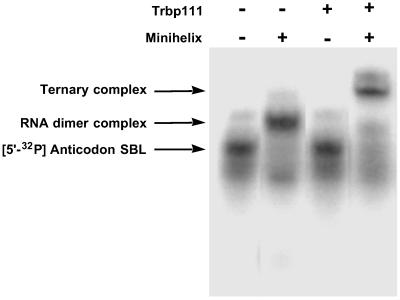
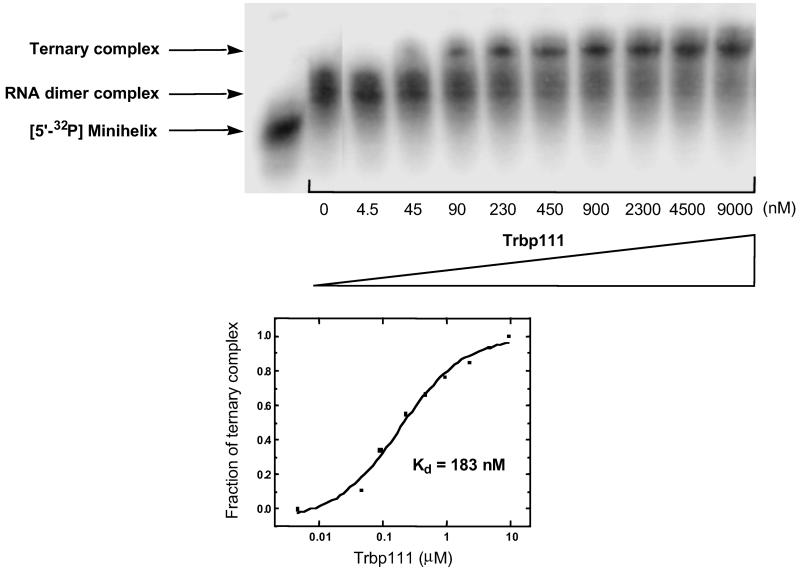
Having established noncovalent complex formation between the two domains, we set out to see whether Trbp111 (a homodimer of 111 aa) could bind to this noncovalent complex. Trbp111 was incubated with labeled anticodon SBL and unlabeled minihelix. The mixture was then subjected to gel electrophoresis. A new, more retarded band (compared with the dimer complex) was seen in the presence of Trbp111 (Fig. 4). The position of the new shifted band roughly corresponded to that of a Trbp111–tRNA complex (9). The new band was not observed when the labeled anticodon SBL was mixed alone with Trbp111. This result suggested that Trbp111 formed a ternary complex with the dimer, but not with monomeric RNA. The fraction of the ternary complex was quantified and plotted on a logarithmic scale to obtain a binding profile (Fig. 5). The resulting Hill plot gave Kd = 183 nM and n = 0.96 (plot not shown). Thus, Trbp111 binds to the tRNA-like structure formed by noncovalent association of the canonical domains of a tRNA.
Fig 4.
Gel retardation assay with Trbp111. Anticodon SBL was 32P-labeled and added with indicated RNA species. The identity of each shifted band is denoted on the left.
Fig 5.
Binding profile of Trbp111 to the RNA dimer complex. Trbp111 was added in increasing concentrations to the preformed RNA dimer with 5′-32P-labeled minihelix (≈900 nM) and unlabeled anticodon SBL (16 μM). The band corresponding to the ternary complex was quantified and plotted on a logarithmic scale to obtain the binding profile shown below.
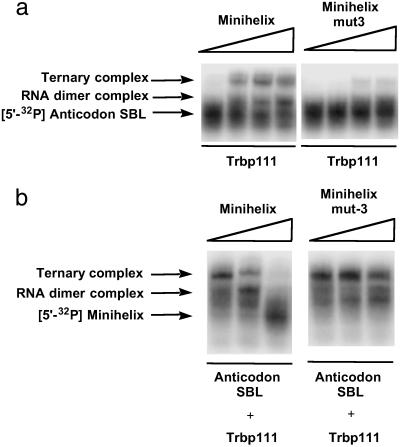
Further experiments demonstrated that binding of Trbp111 to the RNA dimer requires complementary loop–loop interactions. For example, as shown in Fig. 6a, when Trbp111 was mixed with labeled anticodon SBL and unlabeled (mut-3), ternary complex formation was not observed. In the competition assay, the amount of labeled band corresponding to the ternary complex was lowered on addition of unlabeled wild-type minihelix (Fig. 6b). However, this competition was not seen when (mut-3) was used. Thus, in a mixture of RNA domains (minihelix, mut-3 minihelix, and anticodon SBL), Trbp111 selects the combination that makes the L-shaped domain.
Fig 6.
(a) Mutation in the loop sequence disrupts ternary complex formation. 5′-32P-labeled anticodon SBL (≈850 nM) was incubated with Trbp111 (37 μM) and unlabeled minihelix and minihelix (mut-3) in increasing concentrations from 100 nM to 100 μM. (b) Competition assay using unlabeled minihelices. 5′-32P minihelix was incubated with unlabeled anticodon SBL (20 μM) and Trbp111 (1.9 μM) to which unlabeled minihelix or minihelix (mut-3) was added in increasing concentrations from 100 nM to 100 μM.
Trbp111 Protects the L-Shaped Structure.
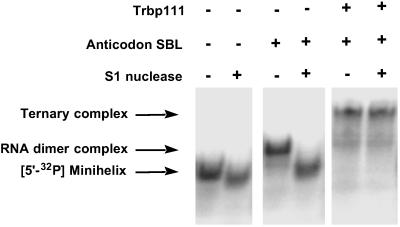
To see whether Trbp111 can protect the RNA dimer against nuclease cleavage, both the dimer and the ternary complex were treated with the single-strand-specific S1 nuclease and subjected to gel electrophoreses. Previous RNA footprint analysis with native tRNA showed that Trbp111 protects the outside corner of the L-shaped tRNA structure (11). As shown in Fig. 7, the band corresponding to the dimer disappeared on S1 nuclease addition, whereas the ternary complex stayed intact. Thus, Trbp111 protects the outside corner of the noncovalently assembled tRNA-like structure, just as it does with native tRNA (11).
Fig 7.
Trbp111 protection from S1 nuclease cleavage. 5′-32P-labeled minihelix (≈1 nM) was incubated with indicated RNA species. Anticodon SBL (1.6 μM), Trbp111 (43 μM), and S1 nuclease (≈1 unit) were used.
S82A Mutation Disrupts Complex Formation.
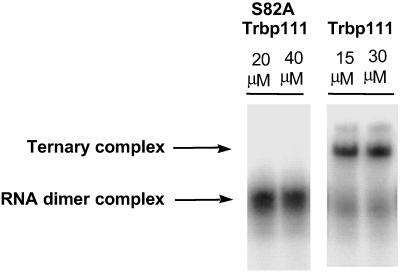
To demonstrate whether the same protein determinants for tRNA binding were also required for interaction with the noncovalently associated RNA dimer, a mutant protein was studied. The S82A substitution was shown previously to disrupt binding of Trbp111 to tRNA (10). S82 is at the center of the RNA binding cleft of the Trbp111 dimer. S82A Trbp111 was incubated with labeled anticodon SBL and mixed with unlabeled minihelix. The S82A mutant protein did not bind to the RNA dimer (Fig. 8) even at the highest concentration tested. This observation is consistent with Trbp111 binding to the noncovalently assembled tRNA-like structure through the same RNA binding cleft used for its interaction with tRNA (10).
Fig 8.
S82A mutant Trbp111 disrupts binding to the RNA dimer. 5′-32P-labeled anticodon SBL (≈300 nM) was added with unlabeled minihelix (1.6 μM) and Trbp111 or S82A Trbp111 at indicated concentrations.
Discussion
An L shape is the distinguishing feature of the tRNA structure and that shape, in turn, depends on base-base interactions through loops that form a corner. The results collectively show that Trpb111 binds selectively to that combination of RNA domains that can noncovalently associate by loop–loop interactions to form a tRNA-like structure. Domains that bear mutations preventing stable loop–loop associations, such as (mut-3) minihelix combined with anticodon SBL, were not brought together by Trbp111. Thus, in a scenario where the domains of tRNA arose separately, ancient proteins like Trbp111 [such as Arc1p (17), EMAPII (18), the C-terminal domain of mammalian tyrosyl-tRNA synthetase (19, 20), and the C-terminal domain of methionyl-tRNA synthetases (21, 22)] would exert strong pressure for selecting L-shaped dimeric RNA complexes that required protection and stabilization of a fragile corner.
The RNA I–RNA II complex used as a model to develop the loop–loop interactions used in this study differs in details from the corner of the L-shaped tRNA. In contrast to the 7-bp loop–loop interactions of the RNA I–RNA II complex (12, 14), the tRNA corner is stabilized by interactions that include non-Watson–Crick pairing and not just loop-to-loop interactions (23–25). However, both corners lead to an angle of ≈90° between the two domains. Thus, Trbp111 appears to be designed to recognize corners associated with L shapes, even though the details of corners are not the same. This feature would be important for allowing Trbp111 to give selective advantage to RNA complexes that could develop into full tRNAs.
Acknowledgments
We thank Professor Susan Martinis for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant GM15539 and a fellowship from the National Foundation for Cancer Research.
Abbreviations
SBL, stem-biloop
References
- 1.Lapointe J. & Giegé, R. (1991) in Translation in Eukaryotes, ed. Trachsel, H. (CRC, Boca Raton, FL), pp. 35–69.
- 2.Giegé R., Puglisi, J. D. & Florentz, C. (1993) Prog. Nucleic Acid Res. Mol. Biol. 45, 129-206. [DOI] [PubMed] [Google Scholar]
- 3.Carter C. W. (1993) Annu. Rev. Biochem. 62, 715-748. [DOI] [PubMed] [Google Scholar]
- 4.Weiner A. M. & Maizels, N. (1987) Proc. Natl. Acad. Sci. USA 84, 7383-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noller H. F. (1993) in The RNA World, eds. Gesteland, R. F. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 137–156.
- 6.Schimmel P., Giegé, R., Moras, D. & Yokoyama, S. (1993) Proc. Natl. Acad. Sci. USA 90, 8763-8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maizels N. & Weiner, A. M. (1994) Proc. Natl. Acad. Sci. USA 91, 6729-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schimmel P. & Ribas de Pouplana, L. (1995) Cell 81, 983-986. [DOI] [PubMed] [Google Scholar]
- 9.Morales A. J., Swairjo, M. A. & Schimmel, P. (1999) EMBO J. 18, 3475-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swairjo M. A., Morales, A. J., Wang, C. C., Ortiz, A. R. & Schimmel, P. (2000) EMBO J. 19, 6287-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nomanbhoy T., Morales, A. J., Abraham, A. T., Vörtler, C. S., Giegé, R. & Schimmel, P. (2001) Nat. Struct. Biol. 8, 344-348. [DOI] [PubMed] [Google Scholar]
- 12.Marino J. P., Gregorian, R. S., Jr., Csankovszki, G. & Crothers, D. M. (1995) Science 268, 1448-1454. [DOI] [PubMed] [Google Scholar]
- 13.Predkil P. F., Nayak, L. M., Gottlieb, M. B. & Regan, L. (1995) Cell 80, 41-50. [DOI] [PubMed] [Google Scholar]
- 14.Lee A. J. & Crothers, D. M. (1998) Structure (London) 6, 993-1005. [DOI] [PubMed] [Google Scholar]
- 15.Wincott F., DiRenzo, A., Shaffer, C., Grimm, S., Tracz, D., Workman, C., Sweedler, D., Gonzalez, C., Scaringe, S. & Usman, N. (1995) Nucleic Acids Res. 23, 2677-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantor C. R. & Schimmel, P. R., (1980) Biophysical Chemistry (Freeman, New York), pp. 849–886.
- 17.Simos G., Segref, A., Fasiolo, F., Hellmuth, K., Shevchenko, A., Mann, M. & Hurt, E. C. (1996) EMBO J. 15, 5437-5448. [PMC free article] [PubMed] [Google Scholar]
- 18.Kao J., Ryan, J., Brett, G., Chen, J., Shen, H., Fan, Y. G., Godman, G., Familletti, P. C., Wang, F., Pan, Y. C., Stern, D. & Clauss, M. (1992) J. Biol. Chem. 267, 20239-20247. [PubMed] [Google Scholar]
- 19.Kleeman T. A., Wei, D., Simpson, K. L. & First, E. A. (1997) J. Biol. Chem. 272, 14420-14425. [DOI] [PubMed] [Google Scholar]
- 20.Wakasugi K. & Schimmel, P. (1999) Science 284, 147-151. [DOI] [PubMed] [Google Scholar]
- 21.Barker D. G., Ebel, J. P., Jakes, R. & Bruton, C. J. (1982) Eur. J. Biochem. 127, 449-457. [PubMed] [Google Scholar]
- 22.Nureki O., Muramatsu, T., Suzuki, K., Kohda, D., Matsuzawa, H., Ohta, T., Miyazawa, T. & Yokoyama, S. (1991) J. Biol. Chem. 266, 3268-3277. [PubMed] [Google Scholar]
- 23.Ladner J. E., Jack, A., Robertus, J. D., Brown, R. S., Rhodes, D., Clark, B. F. & Klug, A. (1975) Proc. Natl. Acad. Sci. USA 72, 4414-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rich A. & RajBhandary, U. L. (1976) Annu. Rev. Biochem. 45, 805-860. [DOI] [PubMed] [Google Scholar]
- 25.Kim S. (1979) in Transfer RNA: Structure, Properties and Recognition, eds. Schimmel, P., Söll, D. & Abelson, J. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 83–100.