Abstract
The development and maintenance of immune homeostasis indispensably depend on signals from the gut flora. Lactic acid bacteria (LAB), which are gram-positive (G+) organisms, are plausible significant players and have received much attention. Gram-negative (G−) commensals, such as members of the family Enterobacteriaceae, may, however, be immunomodulators that are as important as G+ organisms but tend to be overlooked. Dendritic cells (DCs) are crucial immune regulators, and therefore, the present study aimed at investigating differences among human gut flora-derived LAB and G− bacteria in their patterns of DC polarization. Human monocyte-derived DCs were exposed to UV-killed bacteria, and cytokine secretion and surface marker expression were analyzed. Profound differences in the DC polarization patterns were found among the strains. While strains of LAB varied greatly in their capacity to induce interleukin-12 (IL-12) and tumor necrosis factor alpha (TNF-α), G− strains were consistently weak IL-12 and TNF-α inducers. All strains induced significant amounts of IL-10, but G− bacteria were far more potent IL-10 inducers than LAB. Interestingly, we found that when weakly IL-12- and TNF-α-inducing LAB and strong IL-12- and TNF-α-inducing LAB were mixed, the weakly IL-12- and TNF-α-inducing LAB efficiently inhibited otherwise strong IL-12- and TNF-α-inducing LAB, yet when weakly IL-12- and TNF-α-inducing LAB were mixed with G− bacteria, they synergistically induced IL-12 and TNF-α. Furthermore, strong IL-12- and TNF-α-inducing LAB efficiently up-regulated surface markers (CD40, CD83, CD86, and HLA-DR), which were inhibited by weakly IL-12- and TNF-α-inducing LAB. All G− bacteria potently up-regulated surface markers; however, these markers were not inhibited by weakly IL-12- and TNF-α-inducing LAB. These much divergent DC stimulation patterns among intestinal bacteria, which encompass both antagonistic and synergistic relationships, support the growing evidence that the composition of the gut flora affects immune regulation and that compositional imbalances may be involved in disease etiology.
Mammals coexist with an estimated 300 to 500 different species of commensal bacteria that colonize the gastrointestinal tract in a symbiotic relationship (19). It is now recognized that the immune cells of the gut are indeed influenced by stimuli coming from the commensal microflora. Although bacteria-immune cell interactions are traditionally alleged to induce immune activation, in healthy individuals this interaction in fact leads to gut immune homeostasis accompanied by tolerance to the microflora (13). Intestinal homeostasis implies that the immune system has the capability to discriminate appropriately between pathogens toward which an active immune response should be elicited and harmless antigens which should be tolerated by the active down-regulation of the immune response. Imbalances in this dual role of the mucosal immune system have been linked to conditions like food allergies and Crohn's disease, which are characterized by inappropriate immune responses to food antigens and intestinal bacteria, respectively (2, 41). Understanding of the regulating forces of the various gut flora components in the control of gut immune homeostasis is of key importance to the elucidation of disease etiology and therapeutic approaches.
The intestinal microflora has been ascribed many beneficial properties: increased maturation of the gut (40), pathogen antagonism (8, 9, 29, 36), and immune modulation (1, 11, 14, 15). Modern revisions of the hygiene hypothesis suggest that altered bacterial exposure during the establishment of the intestinal flora may cause changes in the microbial composition and, in turn, altered immunomodulating signaling that affects important stages of immune development during infancy (23, 49). Dendritic cells (DCs) reside in an immature stage in peripheral tissues and are scattered throughout the gut mucosa. When they encounter an antigenic stimulus, DCs undergo a maturation process, in which up-regulation of costimulatory molecules and induction of cytokine and chemokine production take place. These actions dictate whether a Th1, Th2, or T-regulatory response is induced (21, 22). In the intestine, DCs play a pivotal role in generating regulatory T cells and immunoglobulin A (IgA)-producing B cells through the production of cytokines, such as transforming growth factor β and interleukin-10 (IL-10), which contribute strongly to immune homeostasis (48). Factors of the microenvironment modulate the function and impact of DCs on the outcome of an immune response (37). Consequently, the heavy load of microbial compounds in the intestine is likely sensed by DCs via pattern recognition receptors, including Toll-like receptors (TLRs). It has become apparent that subepithelial DCs can sample luminal bacterial antigens by passing their dendrites between epithelial tight junctions into the gut lumen (18, 32). Furthermore, DCs interact directly with bacteria that have gained access via M cells. DC stimulation through TLRs allows the DCs to be polarized differently in response to different microorganisms, which may be of particular importance for the immune-regulating role of the intestinal DCs. In the gut, a very diverse microflora coexists in close proximity to the DCs (24, 35, 45).
We therefore found it highly relevant to further investigate the effects of different gut flora-derived bacteria on the maturation of human monocyte-derived DCs and also to take into account the variations between donors. Lactic acid bacteria (LAB), which are important members of the gut flora, have become the focus of much research due to their prevalent use in traditional dairy products and as probiotics (16). In previous work, based on very few bacterial strains, we found that different strains of Lactobacillus can induce very distinct DC maturation patterns (7). In the present study, however, we included, in addition to more strains of both Lactobacillus and Bifidobacterium (gram-positive [G+] organisms), a number of commensal strains of the family Enterobacteriaceae as representatives of the gram-negative (G−) bacteria of the gut flora, which are just as important and which tend to be neglected. The Escherichia coli Nissle 1917 strain has been ascribed probiotic properties: it outcompetes pathogenic E. coli strains (5) and is used for the treatment of ulcerative colitis (27) and Crohn's disease (5, 30). Hence, the effects of gut flora-derived LAB, as well as E. coli Nissle 1917 and other commensal E. coli and Klebsiella pneumoniae strains, on DCs were addressed in the present study. While a few studies have focused on the immunomodulating capacities of different G+ and G− bacteria (10, 20, 24, 25, 42), the literature on the effects of combinations of strains with different immunomodulating effects is sparse. Here, the responses of DCs after exposure to combinations of LAB and G− strains were studied. We found a strongly divergent DC stimulation pattern among intestinal bacteria that encompassed both antagonistic and synergistic relationships, however, with clear relations linked to G+ versus G− bacteria and also certain genera and species. Understanding of the mechanisms of the effects that the microflora manifest on immune homeostasis may enable refinement of probiotic treatments for use for specific diseases.
MATERIALS AND METHODS
Preparation of UV-killed LAB and G− bacteria.
The bacterial strains used in this study were primarily gut flora derived and are listed in Table 1. Lactobacilli and bifidobacteria were grown anaerobically overnight at 37°C in de Man, Rogosa, and Sharpe broth (MRS; Merck, Darmstadt, Germany). The cultures were harvested by centrifugation at 2,000 × g for 15 min, then washed two times in phosphate-buffered saline (PBS; pH 7.4), and finally, resuspended in 1/10 the growth volume of PBS. The bacteria were killed by a 20-min exposure to UV light and were stored at −80°C. For concentration determination, the dry weight was determined by lyophilization (corrected for buffer salt content). The E. coli and K. pneumoniae strains were grown aerobically overnight at 37°C in Luria-Bertani broth (LB; Merck). They were killed by a 45-min exposure to UV light. Killing was verified by replating the UV-exposed bacteria on MRS and LB plates.
TABLE 1.
Strains used in this study
| Name | Origin |
|---|---|
| Lactobacillus paracasei CRL431 | Child, fecesa |
| Lactobacillus acidophilus X37 | Adult, biopsy specimenb |
| Lactobacillus paracasei Z11 | Adult, biopsy specimenb |
| Lactobacillus reuteri DSM 12246:12002 | Pig, fecesb |
| Lactobacillus rhamnosus GG | Adult, fecesc |
| Bifidobacterium bifidum S131 | Child, fecesa |
| Bifidobacterium longum Q45 | Adult, biopsy specimenb |
| Bifidobacterium longum Q46 | Adult, biopsy specimenb |
| Bifidobacterium bifidum Z9 | Adult, biopsy specimenb |
| Bifidobacterium animalis subsp. lactis Bb12 | Dairy straina |
| Escherichia coli Nissle 1917 O6:K5:H1 | Adult, fecesd |
| Escherichia coli F18 OR:K1:H5 | Adult, fecesd |
| Escherichia coli BJ4 OR:K:H2 | Rat, fecesd |
| Escherichia coli MG1655 OR:K:H48 | Laboratoryd |
| Escherichia coli UTI | Urinary tractd |
| Klebsiella pneumoniae T74421-N | Adult, fecesd |
| Klebsiella pneumoniae H79129-N | Adult, fecesd |
C. Hansen A/S, Horsholm, Denmark.
The Royal Veterinary and Agricultural University, Frederiksberg, Denmark.
Valio, Helsinki, Finland.
Statens Serum Institut, Copenhagen, Denmark.
In vitro generation of dendritic cells from monocytes.
DCs were generated from peripheral blood mononuclear cells (PBMCs) as described by Zhou and Tedder, with minor modifications (6, 50). Briefly, PBMCs were isolated from buffy coats obtained from the local blood bank by Ficoll-Paque (Amersham Bioscience, Uppsala, Sweden) density gradient centrifugation. The PBMCs were washed twice with RPMI 1640, and then monocytes were isolated by magnetic sorting with MACS hCD14+ microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany). The yield of CD14+ cells from PBMCs was 14% ± 6%. The monocytes were cultured at a density of 6× 106 cells/3 ml/well in six-well tissue culture plates (Nunc, Roskilde, Denmark) for 6 days in culture medium (RPMI 1640 supplemented with 2 mM l-glutamine, 10% [vol/vol] heat-inactivated fetal calf serum [Cambrex BioWhittaker, Belgium], 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol) containing 30 ng/ml IL-4 (corresponding to 15% supernatant from a IL-4-transfected cell line) and 20 ng/ml recombinant human granulocyte-macrophage colony-stimulating factor from Biosource (Nivelles, Belgium). After 3 days of incubation, one-third of the volume was removed from each well, spun down, resuspended in half of the volume fresh medium containing full doses of IL-4 and granulocyte-macrophage colony-stimulating factor, and then returned to the respective wells and incubated for an additional 3 days. After 6 days, the CD14+ cells were differentiated into nonadherent immature DCs. The yield was 75% ± 10% of the seeded CD14+ cells. Ninety to 95% of the cells expressed the DC marker CD1a, measured by flow cytometry as described below.
Stimulation of DCs with bacteria.
On day 6, the immature DCs from each well were pooled, harvested (by centrifugation at 250 × g for 10 min), and reseeded in 48-well tissue culture plates (Nunc) at 6× 105 cells/500 μl/well. UV-killed bacteria were then added as 100 μl medium per well to a final concentration of 0.5, 5, or 50 μg/ml or as indicated. Lipopolysaccharide (LPS) O26:B6 (Sigma-Aldrich, St. Louis, MO) was used as a positive control at a final concentration of 0.5 μg/ml. As negative control, 100 μl/well of medium was added. The cells were incubated for 18 h at 37°C in 5% CO2.
Immunostaining and flow cytometry.
After stimulation, 600 μl ice-cold PBS containing 1% (vol/vol) fetal calf serum and 0.15% (vol/vol) sodium azide (PBS-Az) was added to each well to prevent the internalization of surface markers. During all work, the cells were kept at temperatures below 4°C and under low levels of light exposure. The cells were collected by gentle pipetting and were centrifuged at 250 × g for 10 min. The culture supernatant was collected and stored at −80°C until cytokine analysis. The cells were resuspended in PBS-Az, and trypan blue exclusion showed that the culture contained ≥95% viable cells. The cells were transferred to a round-bottom 96-well plate at 3× 105 cells/100 μl/well. One hundred microliters of PBS-Az containing fluorochrome-conjugated antibodies (20 μl/106 cells) was added to each well, and the plate was incubated for 45 min. Subsequently, the cells were washed twice with 200 μl/well PBS-Az and finally resuspended in 200 μl/well PBS-Az for flow cytometric analysis with a FACSarray flow cytometer (BD Biosciences, San Jose, CA). The analysis was based on the counting of 10,000 cells. The following antibodies used for staining were obtained from BD Biosciences: phycoerythrin (PE)-conjugated anti-human CD1a, allophycocyanin-conjugated anti-human CD83, PE-conjugated anti-human CD40, PE-conjugated mouse IgG1(κ), and allophycocyaninconjugated mouse IgG1(κ). The following antibodies from Southern Biotech (Birmingham, AL) were used: PE-conjugated anti-human HLA-DR and PE-conjugated anti-human CD86. The level of staining was expressed as the geometric mean of the mean fluorescence intensity.
Cytokine quantification in culture supernatants.
The production of IL-12(p70) and tumor necrosis factor (TNF-α) was analyzed by using commercially available enzyme-linked immunosorbent assay (ELISA) kits (R&D, Minneapolis, MN), according to the instructions of manufacturer. IL-10 was analyzed by using matched antibody pairs purchased from BD Biosciences.
RESULTS
LAB differentially induce IL-10, IL-12(p70), and TNF-α independently of the donor.
The production of IL-10, IL-12(p70), and TNF-α by DCs upon exposure to a range of gut flora-derived lactobacilli and bifidobacteria (Table 1) at different concentrations was measured (Fig. 1). No cytokine production was found in unexposed immature DCs. In general, the synthesis of IL-12 correlated well with the synthesis of TNF-α; however, the levels of induction of IL-12 and TNF-α varied remarkably among the bacterial strains. The most striking result was that, independently of the donor, strains of Bifidobacterium induced low levels of IL-12 and TNF-α production, while the various strains of Lactobacillus differed substantially in their capacities to induce IL-12 and TNF-α, with Lactobacillus acidophilus X37 and Lactobacillus paracasei Z11 consistently exhibiting the strongest capacities. All of the bacteria tested induced the production of IL-10, but to various degrees. All bifidobacteria and Lactobacillus reuteri DSM12246 were potent IL-10 inducers, while the remaining strains induced relatively small amounts of IL-10. When higher bacterial concentrations were used (e.g., 100 μg/ml or higher), the cytokine production in the DCs was not further increased; rather, for some bacteria, very high bacterial concentrations led to weak cytokine production. The threshold concentration causing this “overstimulation” was donor dependent (data not shown). The maximum production of IL-12 and TNF-α was already reached at approximately 5 μg/ml, whereas the maximum production of IL-10 demanded higher bacterial concentrations (approximately 50 μg/ml). These data revealed a dose-dependent response that varied among the bacterial strains and the cytokine in question. A study was conducted in order to elucidate the variation between the responses of the different donors (data not shown). The levels of cytokine produced varied between donors, but the overall cytokine pattern for each bacterial strain seemed to be donor independent.
FIG.1.
LAB-induced cytokine production in DC cultures. IL-12(p70), IL-10, and TNF-α in culture supernatants of human monocyte-derived DCs stimulated with 0.5, 5, or 50 μg/ml UV-killed bacteria or no stimulus for 18 h in 48-well plates with 6× 105 cells/600 μl/well were analyzed by ELISA. The data represent the means ± standard deviations derived from one representative donor of eight donors tested in triplicate.
Commensal G− bacteria induce large amounts of IL-10 and small amounts of IL-12 and TNF-α.
Five commensal E. coli strains as well as two strains of Klebsiella pneumoniae were also included in the study as representatives of G− bacteria. These bacteria included E. coli Nissle, which is a strain known to outcompete pathogenic E. coli in vivo (5) and which, accordingly, is categorized as a probiotic. In contrast to the LAB, the different strains of G− bacteria induced similar cytokine production patterns in the DCs, characterized by large amounts of IL-10 but only modest amounts of IL-12 and TNF-α (Fig. 2). However, E. coli MG1655 diverged somewhat from that pattern by inducing larger amounts of IL-12 and TNF-α, although the amounts were still much smaller than the amounts induced by the strongest IL-12- and TNF-α-inducing LAB. It is noteworthy that the G− bacteria possessed an extremely potent capacity to induce IL-10, with an almost maximal response induced at a concentration as low as 0.5 μg/ml. Quite the opposite applied to the LAB, where maximum IL-10 production was reached only with very high bacterial concentrations. LPS at 0.5 μg/ml was included as a reference and induced the same response pattern as the G− bacteria, suggesting that LPS is the major stimulating component of the G− bacteria. As was observed for LAB, high bacterial concentrations (100 μg/ml or higher, depending on donor) caused “overstimulation” of the DCs.
FIG. 2.
G− bacteria-induced cytokine production in DC cultures. IL-12(p70), IL-10, and TNF-α in culture supernatants of human monocyte-derived DCs stimulated with 0.5, 5, or 50 μg/ml UV-killed bacteria, 0.5 μg/ml LPS, or no stimulus for 18 h in 48-well plates with 6× 105 cells/600 μl/well were analyzed by ELISA. The data represent the means ± standard deviations derived from one representative donor of eight donors tested in triplicate.
LAB differentially up-regulated surface maturation markers.
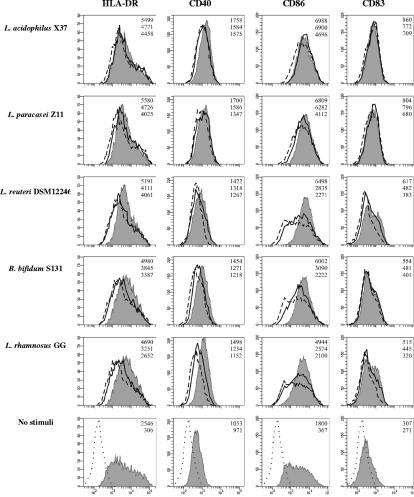
In order to further analyze the maturation patterns of DCs exposed to the different bacterial strains, the expression of HLA-DR, CD40, CD83, and CD86 was determined by flow cytometry upon stimulation with selected representative strains (L. acidophilus X37, L. paracasei Z11, L. reuteri DSM12246, Bifidobacterium bifidum S131, and Lactobacillus rhamnosus GG) (Fig. 3). The differences among the strains were most evident at low bacterial concentrations (0.3 and 3 μg/ml). At low concentrations, L. acidophilus X37 and L. paracasei Z11 were significantly more effective than the remaining strains in inducing the up-regulation of all the surface markers tested. This applied in particular to CD86 expression. All strains of Bifidobacterium included in the study (Table 1) up-regulated maturation markers at levels similar to that by B. bifidum S131 (data not shown). The strain-dependent capability to up-regulate surface markers correlated well with the cytokine induction profile (Fig. 1): those strains of Bifidobacterium and Lactobacillus which were weak IL-12 and TNF-α inducers were also relatively weak up-regulators of surface markers, whereas L. acidophilus X37 and L. paracasei Z11, which induced high levels of IL-12 and TNF-α, were also potent up-regulators of surface markers.
FIG. 3.
Up-regulation of HLA-DR, CD40, CD83, and CD86 on DCs upon exposure to L. acidophilus X37, L. paracasei Z11, L. reuteri DSM12246, B. bifidum S131, and L. rhamnosus GG at concentrations of 30 μg/ml (filled histograms), 3 μg/ml (solid histograms), and 0.3 μg/ml (hatched histograms) of UV-killed bacteria. The results for unstimulated immature DCs are also depicted (filled histogram), with the dotted histogram indicating the unspecific binding of isotype antibodies. The numbers in the upper right corner of each histogram represent the geometric mean fluorescences obtained with 30, 3, and 0.3 μg/ml, from top to bottom, respectively.
Commensal G− bacteria effectively up-regulated maturation markers.
When DC surface marker up-regulation upon exposure to G− bacteria was analyzed, no significant differences were found among the strains tested. The results showed that the G− bacteria were extremely potent in up-regulating the maturation markers on the DCs (Fig. 4). Even at doses as low as 3 ng/ml, the G− bacteria induced substantial expression of HLA-DR, CD40, CD83, and CD86. Thus, while the LAB exhibited various capacities to up-regulate DC surface markers, the G− bacteria induced a consistent pattern among different strains and were, at 100 times lower doses, remarkably more effective in up-regulating the maturation markers than the LAB.
FIG. 4.
Up-regulation of HLA-DR, CD40, CD83, and CD86 on DCs upon exposure to E. coli Nissle. The result for E. coli Nissle is representative of the results for the G− bacteria included in this study (Table 1). The concentrations tested included 300 ng/ml (filled histograms), 30 ng/ml (solid histograms), and 3 ng/ml (hatched histograms). The dotted line shows the expression on immature DCs. The numbers in the upper right corner of each histogram represent the geometric mean fluorescences obtained with 300 ng/ml, 30 ng/ml, 3 ng/ml, and immature DCs, from top to bottom, respectively.
Weakly IL-12-inducing LAB inhibited DC maturation when coexposed with strongly IL-12-inducing LAB.
Since it was previously shown for L. reuteri DSM12246 that this strain can inhibit IL-12 and TNF-α production in murine DCs (7), we wanted to investigate whether this phenomenon also applied to other weakly IL-12-inducing gut flora-derived LAB. To this end, we coexposed L. acidophilus X37, the strain that induced the highest levels of IL-12 and TNF-α, to three bacteria that induce only low levels of IL-12 and TNF-α, namely, B. bifidum S131, L. reuteri DSM12246, and L. rhamnosus GG. All three strains efficiently inhibited the IL-12- and TNF-α-inducing capacity of L. acidophilus X37 in a dose-dependent manner, while IL-10 production was unaltered (Fig. 5). Since B. bifidum S131 and L. reuteri DSM12246 alone induced high levels of IL-10, while L. rhamnosus GG induced only modest levels of IL-10 (Fig. 1), the IL-12 and TNF-α inhibitory effect appeared to be IL-10 independent. Likewise, the three LAB strains also inhibited the IL-12 and TNF-α responses induced by L. paracasei Z11 (data not shown). Furthermore, all the strains of Bifidobacterium included in the study (Table 1) demonstrated an IL-12 and a TNF-α inhibitory capacity (data not shown). Hence, it seems to be a general phenomenon that weakly IL-12- and TNF-α-inducing strains of lactobacilli and bifidobacteria possess a capability to inhibit the IL-12 and TNF-α response induced by otherwise strongly IL-12- and TNF-α-inducing LAB strains.
FIG. 5.
Capabilities of weakly IL-12- and TNF-α-inducing LAB strains (B. bifidum S131, L. reuteri DSM12246, and L. rhamnosus GG) to inhibit the otherwise strong IL-12 and TNF-α response induced by L. acidophilus X37. Monocyte-derived DCs were stimulated with L. acidophilus X37 together with either B. bifidum S131, L. reuteri DSM12246, or L. rhamnosus GG for 18 h in 48-well plates with 6× 105 cells/600 μl/well. Culture supernatants were collected and analyzed for IL-12(p70), TNF-α, and IL-10 by ELISA. The data represent the means ± standard deviations derived from one representative donor of eight donors tested in triplicate.
We furthermore examined whether the weakly IL-12- and TNF-α-inducing strains, which are also weak in up-regulating surface maturation markers, were also capable of inhibiting the induction of maturation markers in DCs. We found that when L. reuteri DSM12246 or B. bifidum S131 was coexposed to L. acidophilus X37, the up-regulation of CD40 and CD86 was reduced in a dose-dependent manner (Fig. 6). Inhibition occurred even at a concentration as low as 0.3 μg/ml of L. reuteri DSM12246 or B. bifidum S131; yet, on the contrary, very high concentrations did not lead to complete inhibition. The same trend observed for CD40 and CD86 expression was observed for HLA-DR and CD83 expression (data not shown). L. rhamnosus GG was also capable of inhibiting surface marker expression similarly to B. bifidum S131 (data not shown). When the responses from DCs derived from six different donors were compared, the inhibitory effects were found to be completely donor independent.
FIG. 6.
Histograms representing the expression of CD40 and CD86 on DCs exposed to 1 μg/ml of L. acidophilus X37 mixed with either L. reuteri DSM12246 (left) or B. bifidum S131 (right) at concentrations of 0 μg/ml (filled histograms), 0.3 μg/ml (hatched histograms), 1 μg/ml (dotted histograms), or 3 μg/ml (solid histograms). The numbers in the upper right corner of each histogram represent the geometric mean fluorescences at each of the four concentrations, from top to bottom, respectively.
Coexposure of G− bacteria and LAB to DCs synergistically induces cytokine expression and surface marker expression.
G− bacteria and LAB coexist in the gut, and thus, the combinatory effects of these groups of bacteria on DC maturation are of relevance. The effect of the simultaneous presence of E. coli Nissle and those LAB with inhibitory capacities (L. rhamnosus GG, B. bifidum S131, or L. reuteri DSM12246) was investigated. Surprisingly, this combination of bacteria exerted a strong synergistic capacity to induce IL-12 and TNF-α in the DCs, reaching levels several times higher than the levels induced by the bacteria per se (Fig. 7). An additive-like pattern of IL-10 production was observed. These effects were likewise observed when the LAB mixed with the other E. coli and K. pneumoniae strains were examined, although the effects were not as great for the K. pneumoniae strains (data not shown). While the synergistic effect on the induction of IL-12 and TNF-α was observed for G− bacteria in combination with weakly IL-12- and TNF-α-inducing LAB strains, any combination of two weakly IL-12- and TNF-α-inducing LAB did not act in this manner (data not shown), indicating the necessity of specific molecular signaling originating uniquely from each of these groups of bacteria.
FIG. 7.
Weakly IL-12- and TNF-α-inducing LAB strains (B. bifidum S131, L. reuteri DSM12246, and L. rhamnosus GG) act synergistically with E. coli Nissle on IL-12 and TNF-α production by DCs. DCs were stimulated simultaneously with E. coli Nissle and either B. bifidum S131, L. reuteri DSM12246, or L. rhamnosus GG at different concentrations for 18 h in 48-well plates with 6× 105 cells/600 μl/well. Culture supernatants were collected and analyzed for IL-12(p70), TNF-α, and IL-10 by ELISA. The data represent the means ± standard deviations derived from one representative donor of eight donors tested in triplicate.
We then investigated whether a similar synergistic effect pertained to the expression of surface markers. The expression of CD40 and CD86 on DCs stimulated with E. coli Nissle (3 ng/ml) in combination with L. reuteri DSM12246 or B. bifidum S131 at different concentrations (0.3 and 1 μg/ml) was analyzed (Fig. 8). In contrast to the inhibitory effect of L. reuteri DSM12246 and B. bifidum S131 on L. acidophilus X37-stimulated DC surface marker expression, L. reuteri DSM12246 and B. bifidum S131 did not inhibit the surface marker expression induced by E. coli Nissle; rather, an enhancement of the expression was seen. This is in line with the synergistic effect on cytokine production induced by the G− bacteria that was observed. Other combinations of inhibitory LAB strains and G− strains showed the same tendency. The same trend for CD40 and CD86 expression was observed for HLA-DR and CD83 expression (data not shown).
FIG. 8.
Expression of CD40 and CD86 on DCs exposed to 30 ng/ml of E. coli Nissle mixed with either L. reuteri DSM12246 (left) or B. bifidum S131 (right) at concentrations of 1 μg/ml (solid histograms), 0.3 μg/ml (hatched histograms), or 0 μg/ml (filled histograms). The dotted histograms represent 1 μg/ml of L. reuteri DSM12246 or 1 μg/ml of B. bifidum S131 alone. The numbers in the upper right corner of each histogram represent the geometric mean fluorescences for E. coli in combination with L. reuteri DSM12246 or B. bifidum S131, E. coli alone, L. reuteri DSM 12246, and B. bifidum alone, from top to bottom, respectively.
LPS was responsible for the synergistic effect observed with G− bacteria.
An obvious component that may account for the stimulatory capacity of G− bacteria is LPS. Thus, we tested the response in DCs when L. reuteri DSM12246 or B. bifidum S131 was coexposed to LPS (Fig. 9). LPS in combination with L. reuteri DSM12246 or B. bifidum S131 induced a synergistic IL-12 and TNF-α response, while an additive effect was seen on IL-10 production (data not shown), resembling the synergistic effect of whole G− bacteria (Fig. 7). These data indicate that LPS is the major component of the G− bacteria responsible for the synergistic effect observed in the DCs.
FIG. 9.
Weakly IL-12- and TNF-α-inducing LAB strains (B. bifidum S131 and L. reuteri DSM12246) act synergistically with LPS on the IL-12 and TNF-α induction in DCs. DCs were stimulated with 0.5 μg/ml LPS and either B. bifidum S131 or L. reuteri DSM12246 simultaneously for 18 h in 48-well plates with 6× 105 cells/600 μl/well. Culture supernatants were collected and analyzed for IL-12(p70) and TNF-α by ELISA. The data represent the means ± standard deviations derived from one representative donor of six donors tested in triplicate.
DISCUSSION
In the present work, we studied the maturation pattern of human monocyte-derived DCs stimulated with a collection of different bacteria mainly derived from the human gut flora. The bacteria included both LAB, including strains of Lactobacillus and Bifidobacterium, which are G+ bacteria, and member of the family Enterobacteriaceae, including strains of E. coli and K. pneumoniae, which are G− bacteria and which are phenotypically very different from the lactobacilli and bifidobacteria. Interesting data for the DC responses were found both when the DCs were stimulated with the various bacteria per se and when they were costimulated with mixtures of bacteria with very distinct individual stimulation patterns.
When the bacterial capacities to induce the production of the proinflammatory cytokines IL-12 and TNF-α, the anti-inflammatory cytokine IL-10, and surface maturation markers in DCs were compared, vast dissimilarities were found among the different genera of bacteria (lactobacilli, bifidobacteria, and members of the family Enterobacteriaceae), whereas differences also occurred at the species and strain levels only for the genus Lactobacillus (Table 2). The Lactobacillus strains can be divided into two groups: strongly and weakly IL-12- and TNF-α-inducing strains. All of the Lactobacillus strains augmented IL-10 production, but to different degrees. Both L. rhamnosus GG and L. reuteri DSM12246 induced low IL-12 and TNF-α responses, but only L. reuteri DSM12246 induced a strong IL-10 response, indicating marked differences among the bacteria in the IL-12/IL-10 ratio induced.
TABLE 2.
Relative patterns of DC response to groups of intestinal bacteria
| Intestinal bacteria | Response pattern
|
||
|---|---|---|---|
| IL-12/TNF-αa | IL-10 | Costimulatory moleculesb | |
| Lactobacillic | Strong or weak | Moderate or weak | Moderate or weak |
| Bifidobacteria | Weak | Moderate | Weak |
| Enterobacteriaceae | Moderate | Strong | Strong |
The IL-12 and TNF-α responses were coordinately induced.
CD40 and CD86.
The response strengths varied greatly among strains.
For the bifidobacteria, all strains tested induced significant levels of production of IL-10, while only very low levels of production, if any, of IL-12 and TNF-α were induced. Furthermore, the bifidobacteria were generally weak in inducing the up-regulation of costimulatory molecules. In contrast to both the lactobacilli and the bifidobacteria, the Enterobacteriaceae consistently induced remarkably strong IL-10 responses in the DCs, with substantial IL-10 production at bacterial concentrations at which no IL-10 production was induced by any of the LAB. The Enterobacteriaceae stimulated the DCs to produce only moderate amounts of IL-12 and TNF-α, with the levels produced being much less than those induced by the most potent lactobacilli. Furthermore, it was found that whereas the ability of LAB to up-regulate maturation markers, including costimulatory molecules, on the DCs correlated well with the production of IL-12 and TNF-α, the Enterobacteriaceae, despite their weakly IL-12- and TNF-α-inducing capacities, were extremely potent inducers of expression of surface markers, with the maximum level of surface marker expression induced at doses of Enterobacteriaceae 100 times lower than the lowest dose of LAB necessary for incipient surface marker up-regulation. These data, summarized in Table 2, point toward a highly diverse capacity of the various bacteria present in the human gut to induce different maturation patterns in DCs and that some generalities exist among different genus of bacteria.
It is known that DCs sense and respond to multiple signals present in their environment, which are then conveyed to T cells to direct the appropriate immune responses (37). It is well evidenced that, depending on the nature of the stimuli received, myeloid DCs can develop into different subsets that possess unique biological functions, determined by the combination of surface molecule expression and cytokine secretion (22). The phenotypes of the different DC subsets that function to induce a Th1, Th2, or T-regulatory response, however, seem to vary depending, among other things, on the tissue in which the DCs reside (22). Whereas consensus exists that IL-12-producing DCs induce a Th1 response, no strict phenotype for the DCs that induce a Th2 or T-regulatory response is defined. Even the DC subset found to induce T-regulatory cells is a very heterogeneous population of cells encompassing phenotypes with different degrees and patterns of maturation, in particular, at the level of costimulatory molecule expression (43). On basis of the existing diverse literature on DCs, it would not be reasonable to try to define a link between the different ways in which the DCs are activated by the different bacteria and the resulting immune response induced by the DCs. However, as more in vivo data accumulate, they may allow more firm conclusions to be drawn. Yet, it is indeed plausible that the diversity of the stimulatory capacities of the gut flora-derived bacteria affects the DC phenotype and, consequently, the T-cell response and immune deviation in the gut.
In agreement with the present data, Smits et al. (42) showed in a recent study that four different G− bacteria, including a strain of E. coli, induced no detectable IL-12 but large amounts of IL-10 in monocyte-derived DCs; yet, they all primed for Th1 development, which nevertheless could be inhibited by blocking IL-12, as well as IL-23 and IL-27. This indicates that a lack of significant IL-12 production in DCs upon stimulation of bacteria cannot stand alone as a predictor of an anti-inflammatory capacity. Our data showed that in comparison with LAB, G− bacteria exerted an extremely potent capacity to up-regulate DC surface marker expression. This may explain the Th1-driving capacity even with a low or undetectable level of IL-12 production. Furthermore, Smits et al. (42) found that DCs stimulated with G+ bacteria primed for neither Th1 nor Th2 development. Unfortunately, the reported data were based on studies with four different bacteria (including a strain of Lactobacillus and a strain of Bifidobacterium) that were all low-level IL-12 inducers, and data for high-level IL-12-inducing bacteria were not included. However, in a later study by Mohamadzadeh et al. (31), it was concluded that lactobacillus-treated human monocyte-derived DCs lead to Th1 polarization. In that study, the three strains of lactobacilli included induced the DCs to produce IL-12 but not IL-10. In the present and previous studies, we have found that those strains of Lactobacillus that were strong IL-12 inducers also induced significant up-regulation of surface markers (7). These data together indicate that the bacterial capacity to induce up-regulation of costimulatory molecules determines whether the outcome is induction of priming DCs or tolerogenic DCs. Furthermore, the contradictory outcomes of these similar in vitro studies demonstrate that different members of the microflora can give very diverse responses and that this response diversity cannot be related strictly to a G+ bacterium or G− bacterium dichotomy. However, the responses to G− bacteria seem somewhat consistent, indicating a strong capacity to induce IL-10 and surface marker expression, which most likely leads to a Th1 response, despite low-level IL-12 induction. In support of this generalization, Hessle et al. have shown in monocytes that G− bacteria preferentially stimulate IL-10 production (20). Moreover, Cross et al. (10) have reported that different strains of E. coli induce similar immune responses: an enteropathogenic E. coli strain induced the same levels of cytokine production by the murine monocyte cell line J774A.1 as E. coli Nissle, with high levels of IL-10 production and low levels of IL-12 production.
Another key finding of the present study was that DCs responded differently to combinations of distinct bacteria than they responded to either bacterium alone. Strains of both Lactobacillus and Bifidobacterium that were weak inducers of IL-12 and TNF-α showed an ability to inhibit the production of these cytokines and to diminish the up-regulation of surface markers induced by other strains of Lactobacillus. In a murine DC model, we have previously shown such an inhibitory effect of L. reuteri DSM12246 (7). The inhibitory effect is seemingly independent of IL-10, since strains that induce small amounts of IL-10, like L. rhamnosus GG, were still capable of inhibiting the IL-12 and TNF-α production induced by other lactobacilli. Another study in our laboratory with murine dendritic cells has shown that the inhibitory ability is a general property of weakly IL-12-inducing LAB and that members of the Bifidobacterium genus in general are weak IL-12 inducers (unpublished data). Data on the inhibitory potential of gut flora-derived LAB are sparse, although inhibition is an important property in vivo where bacteria coexist. However, live L. reuteri has been reported to inhibit IL-8 production in TNF-α-activated epithelial cell lines (28).
In contrast to the IL-12-inhibiting potential of the weakly IL-12-inducing LAB, the G− bacteria did not inhibit IL-12 production in DCs costimulated with strongly IL-12-inducing LAB. Instead, an interesting finding was that when the weakly IL-12- and TNF-α-inducing LAB were combined with G− bacteria or simply LPS, the strong synergistic induction of IL-12 and TNF-α and at least an additive effect on the up-regulation of surface markers were observed. To our knowledge there have been no previous reports on such a synergistic effect in DCs of G− bacteria when they are present in combination with lactobacilli or bifidobacteria. It should be mentioned that studies on the response of intestinal DCs toward commensal bacteria are limited, and it remains unknown whether monocyte-derived DCs resemble intestinal DCs in their responses. However, it has been shown that murine colonic DCs are able to respond to both LPS and Bifidobacterium longum in vitro but only B. longum induced IL-10 production, whereas both stimuli led to some IL-12 production (38).
The mechanisms behind the inhibitory and the synergistic capacities among commensal G+ and G− bacteria observed in the present study are unknown. DCs recognize and respond to microbial stimuli through pattern recognition receptors, including the TLRs. These receptors recognize microbial motifs and activate a set of genes that lead to cytokine production (45). Traditionally, TLRs have been regarded as sensors of microbial infections, and their role is to induce an inflammatory response. However, the motifs recognized by TLRs are not unique to pathogens but are general motifs shared by entire classes of microorganisms, and it is not fully understood how the immune system differentiates between commensal and pathogenic bacteria via the TLRs. Recently, data have shown that TLRs, despite their role in the induction of the inflammatory response, also play a role in maintaining intestinal homeostasis by recognizing the commensal microflora (34). This dual role of TLRs (protection from infection and maintenance of homeostasis) demands that the TLR signaling system be complex. The mechanism of the inhibitory effect observed in the present study could involve TLRs and/or other receptors with suppressive properties, e.g., C-type lectin receptors (CLRs). CLRs (e.g., DC-SIGN, a member of the CLR family) are implicated in the abrogation of activation via TLRs (46). Pathogens that use DC-SIGN for immune surveillance have been identified; e.g., Mycobacterium tuberculosis interacts with DCs by binding to DC-SIGN, which thereby blocks TLR-induced maturation and induces IL-10 production (17, 26). Recently, Smits et al. (44) have elegantly demonstrated that certain LAB prime for regulatory T-cell development via monocyte-derived DCs, and this priming was shown to be abrogated by blocking DC-SIGN. The inhibitory effect of some strains of Lactobacillus and Bifidobacterium observed in the present study could be explained by the fact that commensal bacteria are recognized by a complex combination and different amounts of pro- and anti-inflammatory receptors, which, by acting together, determine the outcome of the response. The synergistic effect that G− bacteria in combination with low-level IL-12- and TNF-α-inducing LAB manifest may likewise involve two or more distinct receptors. The component responsible for the synergistic effect was identified simply as LPS; therefore, the synergistic response could be a product of simultaneous signaling through TLR 4 and a receptor that interacts with an as yet unidentified component of the LAB.
Denaturing gradient gel electrophoresis analyses have revealed that the composition of the microflora differs between individuals but that it is remarkably stable over time in adults. On the contrary, it is less complex and less stable in children and highly unstable in infants (39, 47). Breast-fed infants are mainly colonized by bifidobacteria, whereas formula-fed infants have a more adult-like microflora (12). Studies have also shown that infants with allergies have less bifidobacteria and lactobacilli than nonallergic infants (3, 4). Furthermore, it appears that the levels of bifidobacteria are lower in patients with Crohn's disease and ulcerative colitis (33). Taken together, these data indicate that the treatment of individuals prone to the development of allergies or individuals with inflammatory bowel diseases with probiotics that possess certain immunomodulating properties may have a positive effect. However, the data presented in this study emphasize not only that the immunomodulating effect is very strain dependent but also that the individual response to a probiotic treatment may be strongly influenced by the individual's gut flora. In the intestinal environment, DCs constantly sample and monitor a complex mixture of both G+ and G− bacteria by protruding their dendrites across the intestinal wall (18). This in vitro study has revealed that DCs in contact with two different microbes respond differently than they respond to either microbe alone. We have observed both immunosuppressive effects of certain strains of Lactobaccillus and Bifidobacterium and synergistic effects when these Lactobaccillus and Bifidobacterium strains are combined with E. coli and K. pneumoniae strains. These findings therefore might contribute to our increasing understanding of the immune homeostasis maintained in healthy individuals.
Acknowledgments
We thank A. Lanzavecchia, Institute for Research in Biomedicine, Bellinzona, Switzerland, for providing the IL-4-producing cell-line and K. Krogfelt, Statens Serum Institut, Copenhagen, Denmark, and P. L. Møller, The Royal Veterinary and Agricultural University, Frederiksberg, Denmark, for providing the bacterial strains.
This work was supported by the Centre for Advanced Food Studies, The Dairy Research Foundation, and the Danish Research and Development Program for Food Technology (FØTEK).
REFERENCES
- 1.Arunachalam, K., H. S. Gill, and R. K. Chandra. 2000. Enhancement of natural immune function by dietary consumption of Bifidobacterium lactis (HN019). Eur. J. Clin. Nutr. 54:263-267. [DOI] [PubMed] [Google Scholar]
- 2.Bischoff, S., and S. E. Crowe. 2004. Food allergy and the gastrointestinal tract. Curr. Opin. Gastroenterol. 20:156-161. [DOI] [PubMed] [Google Scholar]
- 3.Bjorksten, B., P. Naaber, E. Sepp, and M. Mikelsaar. 1999. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin. Exp. Allergy 29:342-346. [DOI] [PubMed] [Google Scholar]
- 4.Bjorksten, B., E. Sepp, K. Julge, T. Voor, and M. Mikelsaar. 2001. Allergy development and the intestinal microflora during the first year of life. J. Allergy Clin. Immunol. 108:516-520. [DOI] [PubMed] [Google Scholar]
- 5.Boudeau, J., A. L. Glasser, S. Julien, J. F. Colombel, and A. Darfeuille-Michaud. 2003. Inhibitory effect of probiotic Escherichia coli strain Nissle 1917 on adhesion to and invasion of intestinal epithelial cells by adherent-invasive E. coli strains isolated from patients with Crohn's disease. Aliment. Pharm. Ther. 18:45-56. [DOI] [PubMed] [Google Scholar]
- 6.Chiang, B. L., Y. H. Sheih, L. H. Wang, C. K. Liao, and H. S. Gill. 2000. Enhancing immunity by dietary consumption of a probiotic lactic acid bacterium (Bifidobacterium lactis HN019): optimization and definition of cellular immune responses. Eur. J. Clin. Nutr. 54:849-855. [DOI] [PubMed] [Google Scholar]
- 7.Christensen, H. R., H. Frokiaer, and J. J. Pestka. 2002. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J. Immunol. 168:171-178. [DOI] [PubMed] [Google Scholar]
- 8.Coconnier, M. H., M. F. Bernet, G. Chauviere, and A. L. Servin. 1993. Adhering heat-killed human Lactobacillus acidophilus, strain LB, inhibits the process of pathogenicity of diarrhoeagenic bacteria in cultured human intestinal cells. J. Diarrhoeal. Dis. Res. 11:235-242. [PubMed] [Google Scholar]
- 9.Cross, M. L. 2002. Microbes versus microbes: immune signals generated by probiotic lactobacilli and their role in protection against microbial pathogens. FEMS Immunol. Med. Microbiol. 1442:1-9. [DOI] [PubMed] [Google Scholar]
- 10.Cross, M. L., A. Ganner, D. Teilab, and L. M. Fray. 2004. Patterns of cytokine induction by gram-positive and gram-negative probiotic bacteria. FEMS Immunol. Med. Microbiol. 42:173-180. [DOI] [PubMed] [Google Scholar]
- 11.Cross, M. L., R. R. Mortensen, J. Kudsk, and H. S. Gill. 2002. Dietary intake of Lactobacillus rhamnosus HN001 enhances production of both Th1 and Th2 cytokines in antigen-primed mice. Med. Microbiol. Immunol. 191:49-53. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, C. A., and A. M. Parrett. 2002. Intestinal flora during the first months of life: new perspectives. Br. J. Nutr. 88:S11-S18. [DOI] [PubMed] [Google Scholar]
- 13.Elson, C. O., Y. Z. Cong, N. Iqbal, and C. T. Weaver. 2001. Immune-bacterial homeostasis in the gut: new insights into an old enigma. Semin. Immunol. 13:187-194. [DOI] [PubMed] [Google Scholar]
- 14.Erickson, K. L., and N. E. Hubbard. 2000. Probiotic immunomodulation in health and disease. J. Nutr. 130:403S-409S. [DOI] [PubMed] [Google Scholar]
- 15.Fang, H., T. Elina, A. Heikki, and S. Seppo. 2000. Modulation of humoral immune response through probiotic intake. FEMS Immunol. Med. Microbiol. 29:47-52. [DOI] [PubMed] [Google Scholar]
- 16.Fuller, R. 1989. Probiotics in man and animal. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 17.Geijtenbeek, T. B., S. J. van Vliet, E. A. Koppel, M. Sanchez Hernandez, C. M. J. E. Vandenbroucke-Grauls, B. J. Appelmelk, and Y. van Kooyk. 2005. Mycobacteria target DC-SIGN to suppress dendritic cell function. J. Exp. Med. 197:7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granucci, F., C. Vizzardelli, N. Pavelka, S. Feau, M. Persico, E. Virzi, M. Rescigno, G. Moro, and P. Ricciardi-Castagnoli. 2001. Inducible IL-2 production by dendritic cells revealed by global gene expression analysis. Nat. Immunol. 2:882-888. [DOI] [PubMed] [Google Scholar]
- 19.Guarner, F., and J. R. Malagelada. 2003. Gut flora in health and disease. Lancet 361:512-519. [DOI] [PubMed] [Google Scholar]
- 20.Hessle, C., B. Andersson, and A. E. Wold. 2000. Gram-positive bacteria are potent inducers of monocytic interleukin-12 (IL-12) while gram-negative bacteria preferentially stimulate IL-10 production. Infect. Immun. 68:3581-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwasaki, A., and B. L. Kelsall. 1999. Mucosal immunity and inflammation. I. Mucosal dendritic cells: their specialized role in initiating T cell responses. Am. J. Physiol. 276:G1074-G1078. [DOI] [PubMed] [Google Scholar]
- 22.Kalinski, P., C. M. U. Hilkens, E. A. Wierenga, and M. L. Kapsenberg. 1999. T-cell priming by type-1 and type-2 polarized dendritic cells: the concept of a third signal. Immunol. Today 20:561-567. [DOI] [PubMed] [Google Scholar]
- 23.Kalliomaki, M., P. Kirjavainen, E. Eerola, P. Kero, S. Salminen, and E. Isolauri. 2001. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J. Allergy Clin. Immunol. 107:129-134. [DOI] [PubMed] [Google Scholar]
- 24.Karlsson, H., C. Hessle, and A. Rudin. 2002. Innate immune responses of human neonatal cells to bacteria from the normal gastrointestinal flora. Infect. Immun. 70:6688-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlsson, H., P. Larsson, A. E. Wold, and A. Rudin. 2004. Pattern of cytokine responses to gram-positive and gram-negative commensal bacteria is profoundly changed when monocytes differentiate into dendritic cells. Infect. Immun. 72:2671-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koppel, E. A., I. S. Ludwig, M. Sanchez Hernandez, T. L. Lowary, R. R. Gadikota, A. B. Tuzikov, C. M. J. E. Vandenbroucke-Grauls, Y. van Kooyk, B. J. Appelmelk, and T. B. H. Geijtenbeek. 2004. Identification of the mycobacterial carbohydrate structure that binds the C-type lectins DC-SIGN, L-SIGN and SIGNR1. Immunobiology 209:117-127. [DOI] [PubMed] [Google Scholar]
- 27.Kruis, W., P. Fric, and M. S. Stolte. 2001. Maintenance of remission in ulcerative colitis is equally effective with Escherichia coli Nissle 1917 and with standard mesalamine. Gastroenterology 120:A127. [Google Scholar]
- 28.Ma, D., P. Forsythe, and J. Bienenstock. 2004. Live Lactobacillus reuteri is essential for the inhibitory effect on tumor necrosis factor alpha-induced interleukin-8 expression. Infect. Immun. 72:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mack, D. R., S. Michail, S. Wei, L. McDougall, and M. A. Hollingsworth. 1999. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am. J. Physiol. 276:G941- G950. [DOI] [PubMed] [Google Scholar]
- 30.Malchow, H. A. 1997. Chrohn's disease and Escherichia coli. A new approach in therapy to maintain remission of colonic Crohn's disease? J. Clin. Gastroenterol. 25:653-658. [DOI] [PubMed] [Google Scholar]
- 31.Mohamadzadeh, M., S. Olson, W. V. Kalina, G. Ruthel, G. L. Demmin, K. L. Warfield, S. Bavari, and T. R. Klaenhammer. 2005. Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc. Natl. Acad. Sci. USA 102:2880-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niess, J. H., S. Brand, X. B. Gu, L. Landsman, S. Jung, B. A. McCormick, J. M. Vyas, M. Boes, H. L. Ploegh, J. G. Fox, D. R. Littman, and H. C. Reinecker. 2005. CX(3)CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307:254-258. [DOI] [PubMed] [Google Scholar]
- 33.Ott, S. J., M. Musfeldt, D. F. Wenderoth, J. Hampe, O. Brant, U. R. Folsch, K. N. Timmis, and S. Schreiber. 2004. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118:229-241. [DOI] [PubMed] [Google Scholar]
- 35.Re, F., and J. L. Strominger. 2001. Toll-like receptor 2 (TLR2) and TLR4 differentially activate human dendritic cells. J. Biol. Chem. 276:37692-37699. [DOI] [PubMed] [Google Scholar]
- 36.Reid, G., and J. Burton. 2002. Use of Lactobacillus to prevent infection by pathogenic bacteria. Microbes Infect. 4:319-324. [DOI] [PubMed] [Google Scholar]
- 37.Rescigno, M. 2002. Dendritic cells and the complexity of microbial infection. Trends Microbiol. 10:425-431. [DOI] [PubMed] [Google Scholar]
- 38.Rigby, R. J., S. C. Knight, M. A. Kamm, and A. J. Stagg. 2005. Production of interleukin (IL)-10 and IL-12 by murine colonic dendritic cells in response to microbial stimuli. Clin. Exp. Immunol. 139:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satokari, R. M., E. E. Vaughan, H. Smidt, M. Saarela, J. Matto, and W. M. de Vos. 2003. Molecular approaches for the detection and identification of bifidobacteria and lactobacilli in the human gastrointestinal tract. Syst. Appl. Microbiol. 26:572-584. [DOI] [PubMed] [Google Scholar]
- 40.Schiffrin, E. J., and S. Blum. 2002. Interactions between the microbiota and the intestinal mucosa. Eur. J. Clin. Nutr. 56:S60-S64. [DOI] [PubMed] [Google Scholar]
- 41.Shanahan, F. 2002. Crohn's disease. Lancet 359:62-69. [DOI] [PubMed] [Google Scholar]
- 42.Smits, H. H., A. J. van Beelen, C. Hessle, R. Westland, E. de Jong, E. Soeteman, A. Wold, E. A. Wierenga, and M. L. Kapsenberg. 2004. Commensal gram-negative bacteria prime human dendritic cells for enhanced IL-23 and IL-27 expression and enhanced Th1 development. Eur. J. Immunol. 34:1371-1380. [DOI] [PubMed] [Google Scholar]
- 43.Smits, H. H., E. C. de Jong, E. A. Wierenga, and M. L. Kapsenberg. 2005. Different faces of regulatory DCs in homeostasis and immunity. Trends Immunol. 26:123-129. [DOI] [PubMed] [Google Scholar]
- 44.Smits, H. H., A. Engering, D. van der Kleij, E. C. de Jong, K. Schipper, T. M. M. van Capel, B. A. J. Zaat, M. Yazdanbakhsh, E. A. Wierenga, Y. van Kooyk, and M. L. Kapsenberg. 2005. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J. Allergy Clin. Immunol. 115:1260-1267. [DOI] [PubMed] [Google Scholar]
- 45.Underhill, D. M., and A. Ozinsky. 2002. Toll-like receptors: key mediators of microbe detection. Curr. Opin. Immunol. 14:103-110. [DOI] [PubMed] [Google Scholar]
- 46.van Kooyk, Y., and T. B. H. Geijtenbeek. 2003. DC-sign: escape mechanism for pathogens. Nat. Rev. Immunol. 3:697-709. [DOI] [PubMed] [Google Scholar]
- 47.Vaughan, E. E., M. C. de Vries, E. G. Zoetendal, K. Ben Amor, A. D. L. Akkermans, and W. M. de Vos. 2002. The intestinal LABs. Antonie Leeuwenhoek 82:341-352. [PubMed] [Google Scholar]
- 48.Viney, J. L., A. M. Mowat, J. M. O'Malley, E. Williamson, and N. A. Fanger. 1998. Expanding dendritic cells in vivo enhances the induction of oral tolerance. J. Immunol. 160:5815-5825. [PubMed] [Google Scholar]
- 49.Wills-Karp, M., J. Santeliz, and C. L. Karp. 2001. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat. Rev. Immunol. 1:69-75. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, L. J., and T. F. Tedder. 1996. CD14+ blood monocytes can differentiate into functionally mature CD83+ dendritic cells. Proc. Natl. Acad. Sci. USA 93:2588-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]