Abstract
Bacterial binding protein-dependent ATP binding cassette (ABC) transporters facilitate uptake of essential nutrients. The crystal structure of Escherichia coli BtuF, the protein that binds vitamin B12 and delivers it to the periplasmic surface of the ABC transporter BtuCD, reveals a bi-lobed fold resembling that of the ferrichrome binding protein FhuD. B12 is bound in the “base-on” conformation in a deep cleft formed at the interface between the two lobes of BtuF. A stable complex between BtuF and BtuCD (with the stoichiometry BtuC2D2F) is demonstrated to form in vitro and was modeled using the individual crystal structures. Two surface glutamates from BtuF may interact with arginine residues on the periplasmic surface of the BtuCD transporter. These glutamate and arginine residues are conserved among binding proteins and ABC transporters mediating iron and B12 uptake, suggesting that they may have a role in docking and the transmission of conformational changes.
ATP binding cassette (ABC) transporters are a ubiquitous family of importer and exporter proteins that invariably consist of two membrane-spanning domains, which form a translocation pathway, and two cytoplasmic ABC domains, which power the transport reaction through binding and hydrolysis of ATP (1). Although most eukaryotic ABC transporters export hydrophobic molecules from the cytoplasm (2), the majority of bacterial ABC transporters import essential nutrients that are delivered to them by specific binding proteins (1, 3, 4). These proteins bind their substrates selectively and with high affinity, which is thought to ensure the specificity of the transport reaction (3). The association of a substrate-loaded binding protein with its cognate transporter has been shown to stimulate ATP hydrolysis by the cytoplasmic ABC domains (5). The binding protein remains docked to the cognate transporter until one or both of the hydrolysis products are released, as shown by experiments that used vanadate to trap an intermediate close to the transition state (6). This finding suggested that the binding protein, associated with the transporter during substrate translocation, may prevent the escape of substrate into the periplasmic space.
The structures of many different binding proteins have been solved, revealing a common architecture: two domains, each consisting of a central β-sheet and surrounding α-helices, with the substrate binding site located in a cleft between them (7). Recently, the crystal structure of the binding protein-dependent ABC transporter, BtuCD, which facilitates import of vitamin B12 into Escherichia coli, was determined at 3.2-Å resolution (8). We have now solved the crystal structure of E. coli BtuF, the cognate periplasmic binding protein for BtuCD (9, 10) at 2.0-Å resolution. In addition, we could form a stable complex between BtuF and BtuCD in vitro. These results provide general insights into the interaction of binding proteins with their cognate ABC transporters.
Materials and Methods
Purification and Crystallization.
The btuf gene (previously yadt) was amplified from E. coli genomic DNA. A construct with the OmpA signal sequence and an N-terminal Strep-tag II preceding the coding sequence of the mature form of BtuF was subcloned into pET22b (Novagen). The protein was expressed in BL21 DE3 cells (Novagen) grown at 30°C in Terrific Broth supplemented with 100 μg/ml ampicillin. The periplasmic extract from ≈4 g of BtuF-expressing cells was concentrated to 4 ml by using Centriprep 10 concentrator units. The sample was dialyzed overnight against 2 liters of 10 mM Tris (pH 7.5)/25 mM NaCl at room temperature and subsequently centrifuged to remove precipitated protein. The supernatant was applied to a 1-ml DEAE Sepharose column equilibrated in dialysis buffer and the flow through, containing 90–95% pure BtuF, was collected. Although present in the final construct, the Strep-tag II was not used for purification. Vitamin B12 (cyanocobalamin, Sigma) was added to a final concentration of ≈1 mM, and the sample was concentrated to 15–20 mg⋅ml−1. The ABC transporter BtuCD was expressed and purified as described (8).
BtuF with bound vitamin B12 was crystallized at 4°C by vapor diffusion in hanging drops containing 2 μl of protein and 2 μl of reservoir solution [30% PEG 400/0.1 M sodium acetate (pH 4.6)/0.1 M cadmium chloride/Hampton Crystal Screen 2, #12]. Ruby-red hexagonal bipyramidal crystals appeared within 3 days. Crystals were transferred to a 20-μl drop of reservoir solution containing ≈5 mM vitamin B12, incubated for 5–15 min, and frozen in liquid nitrogen. The crystals were in the space group P65 (a = b = 133.07 Å, c = 67.69 Å) with two molecules per asymmetric unit.
Data Collection and Structure Determination.
Data were collected from a single crystal at the Stanford Synchrotron Radiation Laboratory beamline 9-2 at 100 K with a Quantum-315 CCD (ADSC, Poway, CA) detector and processed using denzo and scalepack (11). Experimental phases were obtained from a three-wavelength multiwavelength anomalous diffraction (MAD) experiment at the cobalt edge. The cobalt sites were found using solve (12) and their parameters refined with sharp (13). Solvent-flattening, twofold noncrystallographic symmetry (NCS) averaging and phase extension to 2.0 Å were performed with dm (14), and the resulting maps, calculated using fft (14), were of excellent quality. Anomalous difference Fourier maps were used to visualize the B12-bound cobalts as well as ordered cadmium and chlorine ions bound to the surface of the protein. Cadmium versus chlorine ions were distinguished by the intensities of the anomalous difference Fourier peaks. Peaks in the anomalous difference Fourier map were also observed at positions corresponding to sulfur atoms in cysteine and methionine residues, and to the phosphorus atom in each B12 molecule. The protein model was built using o (15) and was refined against data combined from all wavelengths by using cns (16). NCS restraints were released after initial rounds of refinement and each molecule was refined separately. Model quality was verified using procheck (17). The final model contains two molecules of BtuF including residues 23–266 (the entire mature protein) plus one residue at the N terminus from the Strep-tag II linker (Ala-22). The model includes 377 water molecules, 22 cadmium and 6 chlorine ions, and 1 PEG400 molecule. The coordinates have been deposited in the Protein Data Bank (PDB ID code ). Figs. 1, 2, and 4A were prepared using dino (www.dino3d.org).
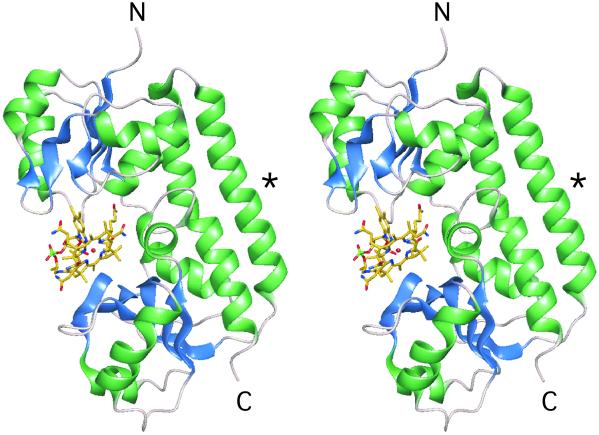
Fig. 1.
Structure of BtuF. A ribbon diagram of BtuF is shown in stereo with bound vitamin B12 in ball and stick. The N and C termini are labeled N and C, respectively. The backbone α-helix that bridges the two lobes of the protein is marked with an asterisk (see text).
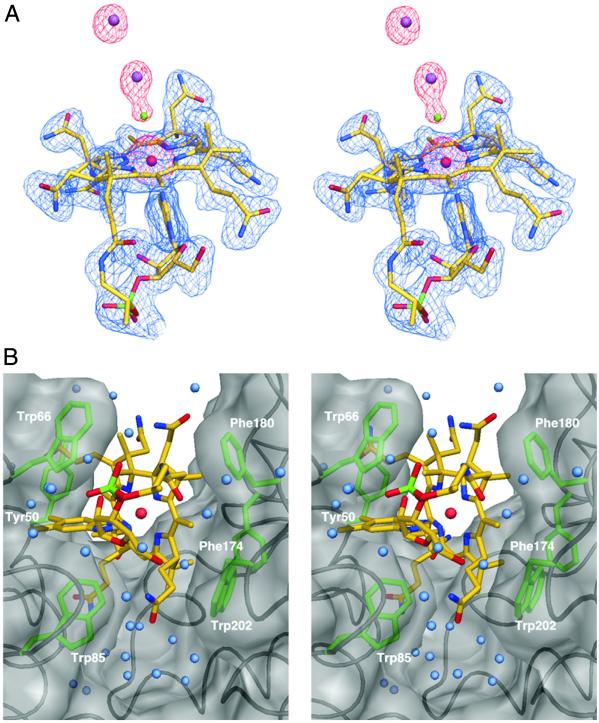
Fig. 2.
Stereo view of vitamin B12 bound to BtuF. (A) B12 is shown in ball and stick with carbon atoms in yellow, oxygens in red, nitrogens in blue, and phosphorus in green. The central cobalt atom is depicted as a red sphere. The experimental electron density at 2-Å resolution is shown as a blue mesh contoured at 1.2 σ, whereas the anomalous difference Fourier density is shown as a red mesh and contoured at 8 σ. Cadmium and chlorine ions are shown as purple and green spheres, respectively. (B) B12 binding site. The BtuF backbone is shown in black and the molecular surface of BtuF in transparent gray. Side chains of six aromatic residues in van der Waals contact with B12 are colored green. B12 is shown as in A, whereas water molecules are depicted as light blue spheres.
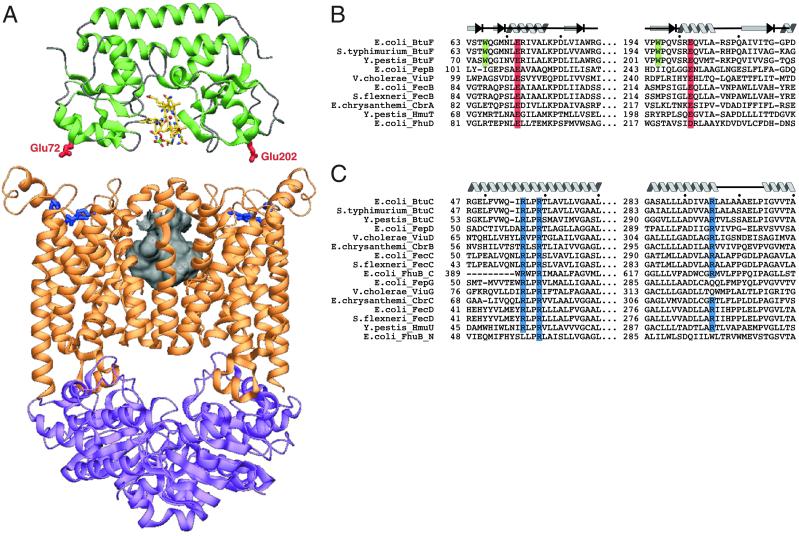
Fig. 4.
Proposed interaction between BtuF and BtuCD. (A) BtuF and BtuCD are depicted as ribbon diagrams in an orientation that places the BtuF surface glutamates adjacent to conserved BtuC arginines (see text for further explanation). Once docked, the molecules were separated along the vertical axis for clarity. BtuF is shown in green, with the critical glutamates shown in ball and stick and colored red. BtuC and BtuD are shown in orange and purple, respectively, with the critical BtuC arginines (Arg-56, -59, and -295) shown in ball and stick and colored blue. The size and location of the translocation pathway at the interface of the BtuC subunits (8) are represented by the molecular surface colored in gray. (B) Sequence alignment of iron siderophore/cobalamin binding proteins. The conserved surface glutamates are shown with a red background, with tryptophans contacting B12 on the green background. The secondary structure elements assigned for the E. coli BtuF structure are indicated above the sequence. (C) Alignment of the membrane-spanning subunits of the cognate ABC transporters with conserved arginines shown with a blue background. The secondary structure elements assigned for the structure of E. coli BtuC (8) are indicated above the sequence.
Results and Discussion
Structure of BtuF and the Vitamin B12 Binding Site.
The crystal structure of E. coli BtuF with bound vitamin B12 was solved using phases obtained from a MAD experiment at the cobalt edge. Data collection, phasing, and refinement statistics are presented in Table 1. BtuF has two structurally similar domains (lobes), each consisting of a central five-stranded β-sheet surrounded by helices (Rossmann-like fold). The domains are connected by a single “backbone” α-helix spanning the length of the protein (Fig. 1). This pattern of interdomain connectivity has also been observed in the Mn2+-binding protein PsaA from Streptococcus pneumoniae (18), the Zn2+-binding protein TroA from Treponema pallidum (19), and the ferrichrome binding protein FhuD from E. coli (20). None of these binding proteins fall into the previously defined Group I or Group II, characterized by three or two interdomain connections, respectively (7), but instead form a third Group, characterized by a single α-helical segment bridging the two lobes.
Table 1.
Statistics of data collection, phasing, and structure refinement
| Data collection | Peak | Inflection | Remote |
|---|---|---|---|
| Wavelength, Å | 1.60268 | 1.60518 | 1.03317 |
| Resolution, Å | 20–2.0 | 20–2.0 | 20–2.0 |
| Unique reflections | 46,209 | 46,201 | 46,346 |
| Redundancy | 19.5 | 19.5 | 10.1 |
| Completeness | 99.9 (98.7) | 99.9 (98.5) | 100 (100) |
| I/σ(I) | 35.8 (7.7) | 35.8 (7.4) | 34.6 (11.1) |
| Rsym | 9.3 (28.8) | 9.0 (29.0) | 5.8 (18.2) |
| Phasing from Co MAD dataset | |||
| Resolution, Å | 20–2.5 | ||
| Isomorphous phasing power† from peak (centric/acentric) | 7.61/11.93 | ||
| Anomalous phasing power from peak (acentric) | 2.22 | ||
| Isomorphous phasing power from inflection (centric/acentric) | 7.69/12.98 | ||
| Anomalous phasing power from inflection (acentric) | 2.06 | ||
| Figure of merit (centric/acentric) | 0.82/0.82 | ||
| Refinement | |||
| Resolution, Å | 20–2.0 | ||
| Reflections | 41,592 | ||
| Test reflections | 4,651 | ||
| Number non-H atoms | 4,416 | ||
| Rwork, % | 0.189 | ||
| Rfree, % | 0.210 | ||
| Average B factors, Å2 | |||
| Protein | 28.1 | ||
| B12 | 25.5 | ||
| Water molecules | 38.5 | ||
| Cadmium/chlorine/PEG | 38.8 | ||
| rms deviation (rmsd) bond length, Å | 0.0067 | ||
| rmsd bond angles, ° | 1.32 | ||
| Ramachandran analysis | |||
| Most favored, % | 90.7 | ||
| Allowed, % | 9.3 | ||
Numbers in parentheses refer to the highest-resolution shell.
Phasing power as calculated by sharp (13).
Α single, well ordered molecule of vitamin B12 is bound in a deep cleft between the two lobes of BtuF (Figs. 1 and 2). Similar to B12 free in solution, BtuF-bound B12 exists in the “base-on” conformation, i.e., with the N3B nitrogen of the dimethylbenzimidazole (DMB) base serving as an axial ligand to the central cobalt atom. A similar conformation of bound B12 has been observed in diol dehydratase (21) and class II ribonucleotide reductase (22). In contrast, B12 is bound in the base-off conformation by methionine synthase (23), methylmalonyl-coA mutase (24), and glutamate mutase (25). In these enzymes, the pseudonucleotide tail of B12 is extended and the nitrogen of a nearby histidine side chain acts as an axial ligand to the cobalt. Although B12 is bound by BtuF in the base-on conformation, it is not known whether passage through the BtuCD transporter requires unfolding.
Six aromatic residues, three from each lobe of BtuF, contact B12 in the binding site (Fig. 2B). Two of these, Trp-66 and -85, are situated on either side of the DMB group. In this respect, the binding site of BtuF resembles that of the ferrichrome binding protein FhuD (20). A network of ordered waters is observed between several corrin ring side chains and BtuF side and main chain atoms at the bottom of the binding site. There are also direct hydrogen bonds involving corrin ring side chains with the main chain nitrogen of Ala-32 and the side chain atoms of Asp-242 and Arg-246. Although cyanocobalamin was used in the crystallization, no cyano group is apparent in the electron density at the upper axial ligand position. Instead, strong peaks in the anomalous difference Fourier map led us to model a chlorine ion (from the crystallization solution) with associated cadmium ions and water molecules (Fig. 2A). Although these ions occupy this space in the crystal structure, there appears to be room for other axial ligands to the cobalt in vivo.
Conformational Dynamics in BtuF Structural Homologs.
The release of substrate into the translocation pathway of an ABC transporter is undoubtedly coupled to conformational changes in the binding protein (5, 7). In the absence of transporter, binding proteins such as the maltose binding protein (MBP) exhibit large hinge and twist movements of one lobe relative to the other between the liganded and unliganded states (7). In contrast, binding proteins such as BtuF and FhuD with a backbone α-helix are thought to be less likely to undergo such motions (18–20). Recently, the structure of one such binding protein, T. pallidum TroA, was solved with (19) and without (26) bound Zn2+. The difference between liganded and unliganded TroA was indeed found to be a mere 4° tilting of the C-terminal domain about the long axis of the protein without bending or unwinding of the backbone helix. This movement is very different from that observed for MBP and yet the result is a partial collapse of the binding site and the loss of the proper coordination geometry for Zn2+.
A search using the Dali server (27) revealed that there is a striking structural similarity between the two lobes of BtuF and the αII and αIII domains of the molybdenum iron (MoFe) protein of nitrogenase (Z = 9.9 with 203 structurally equivalent residues; ref. 28). Analogous to vitamin B12 in BtuF, the iron-molybdenum cofactor (FeMoco) is situated at the interface between the two domains. In the absence of cofactor (29), one of the two protein ligands to the FeMoco is shifted by ≈5 Å, and other segments of the αIII domain are displaced or disordered, without a notable hinge movement or bending of the helix that corresponds to the backbone helix in BtuF. The differences in the structures of the cofactor-free MoFe protein compared with the MoFe holoprotein may be instructive for the potential conformational changes in binding proteins during the transport cycle, because, analogous to the rest of the MoFe protein, the membrane-spanning domains of the ABC transporter may constrain the movements of the two lobes of the attached binding protein.
BtuCD and BtuF Interact in Vitro.
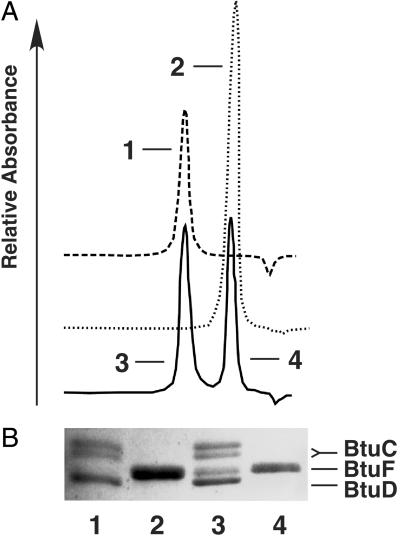
To show that the purified proteins used to determine the crystal structures can interact, BtuF and BtuCD were mixed at a molar ratio of ≈5:1 (binding protein:transporter). A complex forms that is stable at 4°C for at least 72 h and can be isolated by size exclusion chromatography. The presence of BtuF, BtuC, and BtuD was confirmed by gel electrophoresis (Fig. 3) and the stoichiometry of the complex (1 BtuF:2 BtuC:2 BtuD) was determined by densitometry of the Coomassie-stained gel.
Fig. 3.
In vitro interaction of BtuF and BtuCD. (A) BtuF and BtuCD were mixed in detergent solution (0.1% lauryldimethylamine-N-oxide) at a molar ratio of ≈5:1 in the presence of vitamin B12. The mixture was loaded onto an Superdex 200 10/30 column (Amersham Pharmacia) with pure BtuF (dotted line) and BtuCD (dashed line) as controls. Note that because of the presence of a large detergent micelle, pure BtuCD elutes at essentially the same volume as the complex of BtuCD and BtuF. (B) Peak fractions from the gel filtration chromatography shown in A were analyzed by SDS/PAGE. Lane numbers correspond to peaks as labeled in A. Note that BtuC runs as a doublet.
A Model for BtuF Docking to BtuCD.
The availability of the crystal structures of BtuF and BtuCD, combined with the evidence for their docking in vitro, provides an opportunity to model the interaction of an ABC transporter with its cognate binding protein. Electrostatic surface potential maps reveal two negatively charged “knobs” near the apex of each BtuF lobe and two positively charged “pockets” on the periplasmic surface of BtuC. The knobs on the surface of BtuF, corresponding to Glu-72 and -202, are ≈46 Å apart, with the vitamin B12 binding site located halfway between them. The pockets on BtuC are ≈48 Å apart and are lined by three arginine residues: Arg-56 and -59 from transmembrane helix TM2 and Arg-295 from TM9. Both glutamates and all three arginines are conserved in iron siderophore/cobalamin transport systems from various organisms (Fig. 4 B and C), suggesting that these residues may form interprotein salt bridges that are critical for proper interaction of the binding protein with the transporter. When BtuF is manually docked onto the periplasmic face of BtuCD, aligning the conserved glutamates and arginines, bound B12 is positioned over the entrance to the translocation pathway (Fig. 4A).
A competitive peptide mapping study of FhuB, an ABC transporter for iron siderophores that is homologous to BtuCD, provides independent biochemical support for the proposed docking model of BtuF to BtuCD (30). It demonstrated that several peptides derived from FhuB bind to its cognate binding protein FhuD and inhibit transport of ferrichrome when introduced into the periplasm. One of these peptides corresponds to a loop on the periplasmic surface of BtuC located between the conserved arginine pockets and the translocation pathway where it could interact with BtuF.
Despite significant differences in architecture, salt bridges may also play a crucial role for the interaction of the binding proteins for maltose and histidine, MBP and HisJ, with their cognate ABC transporters. When Glu-214, located near the binding cleft of MBP, is mutated to lysine, maltose import is significantly impaired in an in vivo transport assay, even though binding of maltose to MBP appears unaffected (31). Similarly, mutations in HisJ at residues Asp-144, Asp-149, and Arg-154 on lobe II of HisJ, have been shown to inhibit histidine transport with no significant effect on histidine binding (32). Mutation of Asp-149 and Arg-154 also inhibits ATP hydrolysis by the HisQMP2 transporter. In addition, chemical crosslinking studies revealed that binding of these HisJ mutants to the membrane-spanning subunit of the transporter, HisQ, was impaired. Although there is no high-resolution structure of either the maltose or histidine transporter, these mutation studies suggest that salt bridges between surface-exposed residues may play an important role in the attachment of binding proteins.
Conclusions
The structure of B12-bound BtuF reveals that vitamin B12 is presented to the BtuCD transporter in the “base-on” conformation. Conserved acidic residues on the surface of BtuF may be important for interaction with BtuCD. Our observations provide a model for the docking of a periplasmic binding protein to its cognate ABC transporter. The ability to form a stable complex between BtuF and BtuCD should allow their docking to be studied structurally, which may reveal crucial conformational changes and provide a more detailed understanding of the B12 transport reaction.
Abbreviations
ABC, ATP binding cassette
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ).
References
- 1.Higgins C. F. (2001) Res. Microbiol. 152, 205-210. [DOI] [PubMed] [Google Scholar]
- 2.Dean M., Rzhetsky, A. & Allikmets, R. (2001) Gen. Res. 11, 1156-1166. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H. & Hall, J. A. (1998) Methods Enzymol. 292, 3-20. [DOI] [PubMed] [Google Scholar]
- 4.Boos W. & Lucht, J. M. (1996) in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. 1, pp. 1175–1209. [Google Scholar]
- 5.Davidson A. L. (2002) J. Bacteriol. 184, 1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J., Sharma, S., Quiocho, F. A. & Davidson, A. L. (2001) Proc. Natl. Acad. Sci. USA 98, 1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quiocho F. A. & Ledvina, P. (1996) Mol. Microbiol. 20, 17-25. [DOI] [PubMed] [Google Scholar]
- 8.Locher K. P., Lee, A. T. & Rees, D. C. (2002) Science 296, 1091-1098. [DOI] [PubMed] [Google Scholar]
- 9.Van Bibber M., Bradbeer, C., Clark, N. & Roth, J. R. (1999) J. Bacteriol. 181, 5539-5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cadieux N., Bradbeer, C., Reeger-Schneider, E., Köster, W., Mohanty, A. K., Wiener, M. & Kadner, R. J. (2002) J. Bacteriol. 184, 706-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otwinowski Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 12.Terwilliger T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de La Fortelle E. & Bricogne, G. (1997) Methods Enzymol. 276, 472-494. [DOI] [PubMed] [Google Scholar]
- 14.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 15.Jones T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 16.Brünger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 17.Laskowski R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 18.Lawrence M. C., Pilling, P. A., Epa, V. C., Berry, A. M., Ogunniyi, A. D. & Paton, J. C. (1998) Structure (London) 6, 1553-1561. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y.-H., Deka, R. K., Norgard, M. V., Radolf, J. D. & Hasemann, C. A. (1999) Nat. Struct. Biol. 6, 628-633. [DOI] [PubMed] [Google Scholar]
- 20.Clarke T. E., Ku, S.-Y., Dougan, D. R., Vogel, H. J. & Tari, L. W. (2000) Nat. Struct. Biol. 7, 287-291. [DOI] [PubMed] [Google Scholar]
- 21.Shibata N., Masuda, J., Tobimatsu, T., Toraya, T., Suto, K., Morimoto, Y. & Yasuoka, N. (1999) Structure (London) 7, 997-1008. [DOI] [PubMed] [Google Scholar]
- 22.Sintchak M. D., Arjara, G., Kellogg, B. A., Stubbe, J. & Drennan, C. L. (2002) Nat. Struct. Biol. 9, 293-300. [DOI] [PubMed] [Google Scholar]
- 23.Drennan C. L., Huang, S., Drummond, J. T., Matthews, R. G. & Ludwig, M. L. (1994) Science 266, 1669-1674. [DOI] [PubMed] [Google Scholar]
- 24.Mancia F., Keep, N. H., Nakagawa, A., Leadlay, P. F., McSweeney, S., Rasmussen, B., Bösecke, P., Diat, O. & Evans, P. R. (1996) Structure (London) 4, 339-350. [DOI] [PubMed] [Google Scholar]
- 25.Reitzer R., Gruber, K., Jogl, G., Wagner, U. G., Bothe, H., Buckel, W. & Kratky, C. (1999) Structure (London) 7, 891-902. [DOI] [PubMed] [Google Scholar]
- 26.Lee Y.-H., Dorwart, M. R., Hazlett, K. R. O., Deka, R. K., Norgard, M. V., Radolf, J. D. & Hasemann, C. A. (2002) J. Bacteriol. 184, 2300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holm L. & Sander, C. (1998) Nucleic Acids Res. 26, 316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J. & Rees, D. C. (1992) Science 257, 1677-1682. [DOI] [PubMed] [Google Scholar]
- 29.Schmid B., Ribbe, M. W., Einsle, O., Yoshida, M., Thomas, L. M., Dean, D. R., Rees, D. C. & Burgess, B. K. (2002) Science 296, 352-356. [DOI] [PubMed] [Google Scholar]
- 30.Mademidis A., Killmann, H., Kraas, W., Flechsler, I., Jung, G. & Braun, V. (1997) Mol. Microbiol. 26, 1109-1123. [DOI] [PubMed] [Google Scholar]
- 31.Szmelcman S., Sassoon, N. & Hofnung, M. (1997) Protein Sci. 6, 628-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C. E., Liu, P.-Q., Wolf, A., Lin, E. & Ames, G. F.-L. (1999) J. Biol. Chem. 274, 739-747. [DOI] [PubMed] [Google Scholar]