Abstract
Site-specific DNA binding molecules offer the potential for genetic manipulation of mammalian cells. Peptide nucleic acids (PNAs) are a DNA mimic in which the purine and pyrimidine bases are attached to a polyamide backbone. PNAs bind with high affinity to single-stranded DNA via Watson–Crick base pairing and can form triple helices via Hoogsteen binding to DNA/PNA duplexes. Dimeric bis-PNAs capable of both strand invasion and triplex formation can form clamp structures on target DNAs. As a strategy to promote site-directed recombination, a bis-PNA was coupled to a 40-nt donor DNA fragment homologous to an adjacent region in the target gene. The PNA–DNA conjugate was found to mediate site-directed recombination with a plasmid substrate in human cell-free extracts, resulting in correction of a mutation in a reporter gene at a frequency at least 60-fold above background. Induced site-specific recombination was also seen when the bis-PNA and the donor DNA were co-mixed without covalent linkage. In addition, the bis-PNA and the bis-PNA–DNA conjugate were found to induce DNA repair specifically in the target plasmid. Both the PNA-induced recombination and the PNA-induced repair were found to be dependent on the nucleotide excision repair factor, XPA (xeroderma pigmentosum complementation group A protein). These results suggest that the formation of a PNA clamp on duplex DNA creates a helical distortion that strongly provokes DNA repair and thereby sensitizes the target site to recombination. The ability to promote recombination in a site-directed manner using PNA–DNA conjugates may provide a useful strategy to achieve targeted correction of defective genes.
Keywords: triple helix, DNA repair
Synthetic oligonucleotides offer the potential to rationally design therapeutic agents that selectively modulate gene expression (1). In the antisense approach, the oligonucleotide is targeted to bind to mRNA, leading to degradation or inhibition of translation (1). In the antigene strategy, triplex-forming oligonucleotides (TFOs) are designed to bind in a sequence-specific manner as third strands in the major groove of duplex DNA at polypurine/polypyrimidine stretches (reviewed in refs. 2 and 3). The specificity arises from the base triplets formed either by Hoogsteen or reverse Hoogsteen hydrogen bonding between the third strand and the purine strand of the duplex. TFOs have been used successfully to inhibit transcription (4), damage DNA through the delivery of a mutagen (5–8), and provoke mutagenesis through high-affinity binding (9, 10). Triple helix formation has also been shown to stimulate recombination in mammalian cells in both episomal and chromosomal targets (11–14).
Peptide nucleic acids (PNAs) represent another class of DNA-binding reagents that can be used for gene targeting (15). PNAs consist of purine and pyrimidine bases attached to a polyamide backbone, maintaining a spacing similar to DNA, but yielding an achiral, neutrally charged molecule. PNAs can bind to DNA via Watson–Crick hydrogen bonds, with binding affinities significantly higher than those of the corresponding DNA oligomers (16). Under conditions that promote opening of the duplex, either in vitro (e.g., low salt or high temperature) or in vivo (e.g., transcription), PNAs can mediate strand invasion in duplex DNA (17, 18). This results in the displacement of one DNA strand to form a D-loop (19).
Highly stable PNA:DNA:PNA triplexes can be formed by two PNA strands and a homopurine DNA strand. If connected by a linker of sufficient flexibility, the two PNA strands can be contained in a single molecule (bis-PNA), yielding a clamp structure on DNA binding (PNA clamp). In this structure, one strand forms Watson–Crick base pairs with the DNA strand in an antiparallel orientation, whereas the other strand forms Hoogsteen base pairs to the hompurine DNA strand in the DNA–PNA duplex (20, 21). Although, as with DNA triple helices, a homopurine DNA strand is needed to allow formation of a stable PNA/DNA/PNA triplex, PNA clamps can form at shorter homopurine sequences than DNA triplexes and do so with greater stability.
Several studies have shown that PNA clamps bind to DNA with high affinity and specificity (20, 21). In fact, PNA clamps are sufficiently stable to block transcription initiation and inhibit elongation (22). They can inhibit binding of proteins, such as restriction enzymes and transcription factors, to their target sites (23). As a result, PNAs are currently being developed as antigene drugs to inhibit gene activity on the transcriptional level. In addition, our group has shown that a high-affinity PNA clamp transfected into mouse cells can generate mutations at a chromosomal target site (24).
In previous work studying the ability of DNA TFOs to mediate targeted recombination, we designed a bifunctional oligonucleotide composed of a TFO coupled to a short donor DNA fragment homologous to the target gene (TFO–DNA). In this tethered-donor strategy, the TFO is intended to bind to the gene and thereby position the donor fragment for recombination and information transfer. In addition, the formation of the triple helix can stimulate DNA repair (9), producing recombinagenic intermediates such as strand breaks, and thereby sensitizing the target site to recombination (13). Such a bifunctional molecule was found to mediate recombination with an episomal target on transfection into monkey COS cells in culture (11) and with a plasmid substrate in vitro in human cell-free extracts (25).
Because of the favorable binding characteristics of bis-PNAs, we sought to investigate the ability of a bis-PNA to substitute for the TFO in the tethered-donor strategy. In the work reported here, we have synthesized a PNA–DNA hybrid molecule by using a post-synthetic conjugation method, and we show that a bis-PNA coupled to a short donor DNA fragment can mediate specific sequence changes within the supFG1 reporter gene in vitro in human cell-free extracts. Furthermore, we show that the PNA–DNA conjugate is more active than its TFO–DNA counterpart and that the bis-PNA can stimulate recombination even without covalent linkage to the donor DNA. Both the bis-PNA alone and the PNA–DNA hybrid molecule were able to stimulate DNA repair within the target pSupFG1 plasmid at levels even higher than that induced by high-affinity DNA triple helix formation. The nucleotide excision repair (NER) damage recognition factor, XPA (xeroderma pigmentosum complementation group A protein), was found to be required for repair of the PNA clamp and for the ability of the PNA to induce recombination. Overall, the bis-PNA was found to effectively mediate target site recognition and induced recombination, thereby providing a potentially useful tool for directed gene modification.
Materials and Methods
Oligonucleotides.
Oligonucleotides were synthesized by the Midland Certified Reagent Company (Midland, TX), using cyanoethyl phosphoramidite chemistry. After removal of the protecting groups by hydrolysis with concentrated ammonium hydroxide, the product was purified by reversed-phase HPLC. The ammonium salt form of the oligonucleotide was dissolved in distilled water and further purified using a NAP 5 Sephadex G25 filtration column (Amersham Pharmacia Biotech). To resist 3′-exonuclease activity, the donor DNA–TFO oligonucleotide, which consisted primarily of phosphodiester linkages, was modified at the 3′ end (of the TFO domain) with three terminal phosphorothioate linkages. The donor fragment and the TFO domain were synthesized on a single column, with the following sequence: 5′-AGG GAG CAG ACT CTA AAT CTG CCG TCA TCG ACT TCG AAG G-linker-AGG AAG GGG GGG GTG GTG GGG GAG GGG GAG-3′, where the 40 nt at the 5′ end represent donor A and the 30 nt at the 3′ end represent the TFO domain. The linker segment, which allows flexibility for binding of the individual domains, consisted of the sequence 9TT9TT9, where “9” represents a 9-carbon polyethylene glycol linker, O-methoxytrityl-triethylene glycol, 1-[(2-cyanoethyl)-(N,N-diisopropyl)]phosphoramidite (Spacer 9, Glen Research, Sterling, VA). In the case of PNA–DNA, the donor fragment used in the formation of the hybrid molecule was synthesized in continuity with the same linker segment but otherwise was made as a distinct molecule from the bis-PNA. It did not contain any phosphorothioate linkages, because the 3′-TFO domain was substituted by the PNA, which is, by itself, nuclease resistant. To allow coupling to the PNA, the combined donor DNA fragment and linker segment were synthesized with a 3′-thiol modifier, introduced as a C3 S-S CPG. The sequence, donor A plus linker, was as follows: 5′-AGG GAG CAG ACT CTA AAT CTG CCG TCA TCG ACT TCG AAG G 9TT9TT9 S-S (C3)-3′.
PNA.
The PNA was synthesized at Applied Biosystems according to standard procedure and purified by reverse-phase HPLC with trifluoroacetic acid present. The PNA was designed to bind as a clamp and synthesized to include two distinct segments connected by two 8-amino-3,6-dioxaoctanoic acid (O-linker) units to allow flexibility in binding. The PNA clamp was synthesized with the sequence JJJ-JJT-TJJ-T-O-Lys-(SMCC)-O-TCC-TCC-CCC-C, with the heterofunctional crosslinker succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC) covalently attached to the lysine residue. Pseudoisocytosine (J) that mimics N-3-protonated cytosine is routinely used in PNA clamps for recognition of double-stranded DNA. This modified base is important for pH-independent binding to G in the Hoogsteen mode of triplex formation in a PNA:DNA:PNA triplex.
Synthesis of PNA–DNA Conjugate.
The 3′-thiol-modified donor fragment A (1; 78.87 μg, 569 μM) was dissolved in 150 μl of 0.1 M DTT and the reduction of the thiol group was allowed to proceed overnight at 37°C. The solution was then applied to a NAP 5 filtration column preequilibrated in 0.1 M sodium phosphate buffer (pH 7.0). The reduced oligonucleotide (2) was eluted under gravity with 1 ml of phosphate buffer (pH 7.0) directly into a siliconized Eppendorf tube containing a 20-μl aliquot of maleimide-derivatized PNA (3). The reaction was allowed to proceed for 4 h at room temperature, followed by the removal of excess unreacted PNA through application to a NAP 10 filtration column and elution with water. The amount of product (4) eluted was quantified by A260 before concentration in vacuo and analysis by denaturing 12% PAGE (7 M urea). The PNA–DNA conjugate, A–PNA, was purified by means of preparative gel electrophoresis.
Plasmid Vector and Cells.
The shuttle vector plasmid pSupFG1/G144C, containing a supFG1 gene with an inactivating G:C to C:G point mutation at position 144, has been described (11). Construction of Escherichia coli SY302 lacZ125 (Am) recA56 hsdR2:Tn10 trp-49 has been described (26). HeLa cells were maintained and grown by the National Cell Culture Center (Minneapolis) and were obtained as frozen cell pellets for extract preparation.
Proteins and Antibodies.
XPA protein was produced using an E. coli expression vector [obtained from R. Wood (27)], as described (25, 28). Rabbits were immunized with the purified XPA protein (100 μg/injection) to produce high-affinity antibodies specific to human XPA, as described (25, 28).
In Vitro Recombination Assay.
Cell-free extracts were prepared as described (25, 26). Recombination reactions consisting of 3 μg of pSupFG1/G144C plasmid DNA and 3 μg each of selected oligonucleotide or PNA were carried out as described (25). Plasmid DNA recovered from the reactions was used to transform E. coli by electroporation followed by genetic analysis of supFG1 gene function as described (25).
In Vitro DNA Repair Assay.
The plasmid pSupFG1 (300 ng), which contains the triplex binding site, was incubated overnight at 37°C with the TFO or PNA clamp (at a concentration of 1 μM), in triplex binding buffer or water, respectively. As an internal control, the plasmid pIND/lacZ, which lacks the triplex binding site, was included in each sample. In some samples, as a positive control, the pSupFG1 DNA was damaged by exposure to short-wavelength (254 nm) UV light at a dose of 5,000 J/m2, as described (29). Cell-free extracts were prepared, and the reactions were carried out and analyzed as described (9). Quantification was performed using a phosphorimager. Each assay was conducted a minimum of three times.
Immunodepletion of HeLa Extracts.
Anti-XPA sera (10 μl) was incubated with pre-swollen protein A-agarose beads for 45 min at 4°C. The beads were washed three times with 1× TBS buffer [10 mM Tris⋅HCl (pH 7.5)/100 mM NaCl] and then incubated with 50 μl of HeLa extract for 2 h on ice with gentle rotation. The resulting supernatant (XPA-depleted extract) was recovered by means of centrifugation and subsequently examined by Western blot analysis before use in the DNA repair or recombination assays. Reactions containing XPA-depleted extracts (−XPA extracts) were in some cases supplemented with 500 ng of purified, recombinant XPA protein (+XPA extracts), prepared as described above.
Results
Design and Synthesis of PNA–DNA Hybrid Molecule.
Previous work to study the ability of DNA triple helix formation to promote targeted recombination focused on the DNA–TFO molecule, A-AG30 (11). This molecule consists of the TFO, AG30, tethered via a linker segment to a single-stranded donor DNA fragment, A, homologous to bp 121–160 of the supFG1 reporter gene. The linkage of the donor DNA to the TFO exploits triplex formation both as a means of target site recognition and as a tool to stimulate repair, thereby inducing recombination. In the prior work, it was found that A-AG30 could mediate directed recombination in the supFG1-144 gene in the reporter vector both in vitro in cell-free extracts and in vivo in cells in culture (11, 25).
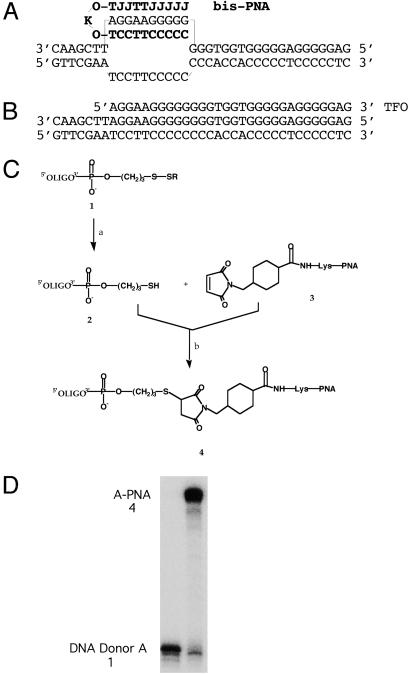
Because of the special DNA binding properties of PNAs, we hypothesized that substitution of the TFO domain with a bis-PNA might improve on this bifunctional oligonucleotide strategy. Whereas the TFO, AG30, was designed to bind as a third strand in the major groove of bp 167–196 of the supFG1 gene [and has been shown to bind there with high affinity in the antiparallel triplex motif (7)], the bis-PNA was designed to bind as a clamp to the homopurine DNA strand at positions 167–176 in the gene (Fig. 1 A and B). The bis-PNA was synthesized to include two distinct PNA segments connected by a flexible linker composed of two 8-amino-3,6-dioxaoctanoic acid (O-linker) units flanking a lysine. The distal (C-terminal or 3′) stretch of the PNA was designed to strand-invade the target site and form Watson–Crick hydrogen bonds to the complementary homopurine DNA strand. The proximal (N-terminal or 5′) PNA segment can fold over to form a PNA/DNA/PNA triplex by Hoogsteen hydrogen bonds consistent with the parallel pyrimidine motif for triple helix formation (Fig. 1A). The cytosines in this triplex-forming portion of the PNAs were replaced by pseudoisocytosine (J), which is a substitute for protonated cytosine and forms stable triplex structures at neutral pH (20). Previously, we demonstrated that a bis-PNA of the composition used here binds to a duplex target of this sequence (24), and similar PNA/DNA/PNA clamps have been found to be highly stable structures with melting temperatures of >70°C (20).
Fig 1.
Design and synthesis of a bifunctional PNA–DNA conjugate. (A) Sequence of and predicted clamp formation by the bis-PNA designed to bind to the homopurine strand at positions 167–176 of the supFG1 gene. (B) Orientation of triple helix formation by the TFO, AG30, which binds to bp 167–196 of the supFG1 gene. (C) Preparation of the PNA–DNA bifunctional molecule. The DNA oligonucleotide-bis-PNA conjugation was carried out by means of reaction between the 3′-thiol-modified oligonucleotide and the maleimido-modified bis-PNA. a, 0.1 M DTT; b, 0.1 M phosphate buffer (pH 7.0). (D) Analysis of PNA–DNA conjugate by denaturing 12% PAGE (7 M urea). Lane 1, DNA donor fragment A; lane 2, A–PNA hybrid molecule after purification by preparative gel electrophoresis.
To produce the bifunctional PNA–DNA molecule, our strategy was to synthesize the DNA chain by using standard phosphoramidite chemistry and then couple it to the PNA clamp by means of post-synthetic conjugation. The PNA clamp was converted to the maleimide derivative by covalently attaching the heterofunctional crosslinker, SMCC, to a lysine residue during synthesis (Fig. 1C). The DNA fragment, A, was synthesized to include a 3′ linker segment with a thiol modifier at the 3′ end. The synthesis of the PNA–DNA hybrid molecule is depicted in Fig. 1C. After reduction of the thiol group to the free sulfhydryl and removal of excess reducing agent, the oligonucleotide was eluted directly into a solution containing the derivatized bis-PNA. Following a 4-h reaction period, unreacted material was removed using a gel filtration column, and the product was eluted with water. Typically, 90% of the conjugate was found to elute under these conditions, as judged by absorbance of the solution at 260 nm. The purity of the conjugate at this stage was determined by denaturing polyacrylamide gel electrophoresis and found to be >95% (Fig. 1D). The PNA–DNA conjugate was purified by means of preparative gel electrophoresis.
PNA Clamp-Induced Recombination in HeLa Cell-Free Extracts.
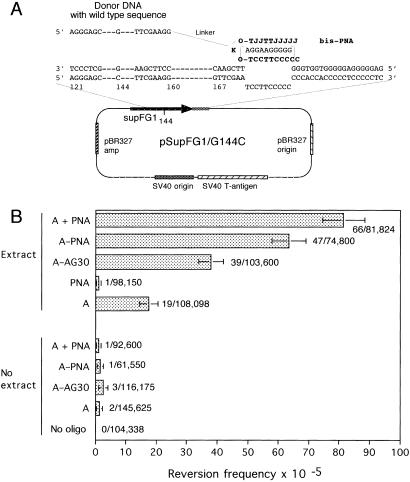
To test the ability of a bis-PNA to induce recombination, the plasmid vector pSupFG1/G144C was used as the substrate for targeted recombination (Fig. 2A). Recombination of the donor fragment with the vector corrects the inactivating G:C to C:G mutation at bp 144 in the supFG1 reporter gene. Because supFG1 encodes an amber suppressor tRNA, functional correction of the gene can be assayed by transformation of the plasmid into bacteria carrying an amber stop codon in the lacZ gene, with consequent β-galactosidase activity determined using a chromogenic substrate. The PNA–DNA conjugate and various related molecules were each incubated with the target vector in HeLa cell-free extracts (Fig. 2B). Following a 2-h incubation, the plasmid DNA was isolated and used to transform indicator bacteria to score for supFG1 gene function. As in previous work, the host bacteria were recA deficient (11, 25).
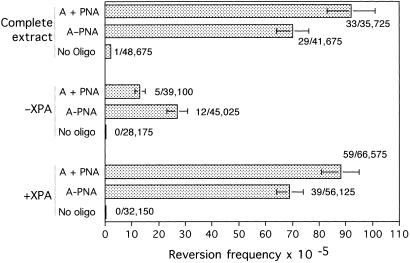
Fig 2.
Targeted recombination induced by PNA clamp formation. (A) Design of the plasmid vector assay to detect targeted recombination in the supFG1 reporter gene by PNA–DNA conjugates and related molecules. (B) PNA-induced recombination in vitro in HeLa cell-free extracts. The pSupFG1/G144C plasmid DNA was incubated in vitro with the indicated PNAs or oligonucleotides in the presence or absence of human cell-free extracts, as indicated. After 2 h, the plasmid DNA was isolated and used to transform indicator bacteria for genetic analysis of the supFG1 gene. The bars indicate the frequency of blue colonies (representing recombinants) out of total colonies, with the actual count given to the right of each bar. The samples A–AG30 and A–PNA indicate molecules in which the donor DNA fragment, A, was covalently linked either to the TFO, AG30, or to the bis-PNA, respectively. The sample A + PNA represents the donor DNA fragment and the bis-PNA mixed together as separate, unlinked molecules. Error bars indicate standard errors.
The results show that the hybrid molecule, A–PNA, mediated correction of the supFG1-144 mutation at a frequency of 62 × 10−5 (Fig. 2B). Note that no recombinant products above background were observed on transformation of the A–PNA sample directly into bacteria without prior incubation in extract. Hence, it can be concluded that the induced recombination occurred in the human cell-free extracts and not in the host bacteria. As reported (25), we again found that the corresponding bifunctional DNA–TFO oligonucleotide, A-AG30, can mediate recombination at a level substantially above background (38 × 10−5). However, the DNA–PNA molecule appeared to be more active in inducing recombination than the DNA–TFO molecule.
As would be expected, the donor fragment A, by itself, was somewhat active: addition of A to the plasmid led to a low level of supFG1-144 reversion (Fig. 2B). This is consistent with the ability of short fragments of DNA to mediate recombination (30, 31). However, the effect of A–PNA was 3-fold higher than the donor A alone, demonstrating the influence of the PNA clamp and providing direct evidence for PNA clamp-induced recombination in vitro. By itself, the PNA domain produced minimal reversion over background, indicating the need for the sequence information provided by the donor fragment A.
Interestingly, the sample in which the bis-PNA and the donor oligonucleotide were not synthetically linked, but were simply co-mixed as separate molecules together with the plasmid substrate (indicated as A + PNA), produced a level of recombination even higher than that produced by the linked A–PNA conjugate (frequency of 81 × 10−5; Fig. 2B). This frequency is 5-fold higher than that produced by the donor alone, providing further evidence that a PNA clamp can stimulate recombination between a fragment and a target gene, even if the fragment is not covalently linked to the PNA. This result specifically demonstrates that a PNA clamp can stimulate recombination in a manner distinct from its ability to deliver a tethered donor fragment to the target site. In fact, it is possible that the linkage of the bis-PNA to the DNA may provide some steric hindrance that could somewhat inhibit recombination of the donor fragment with the target gene.
In Vitro DNA Repair Assay.
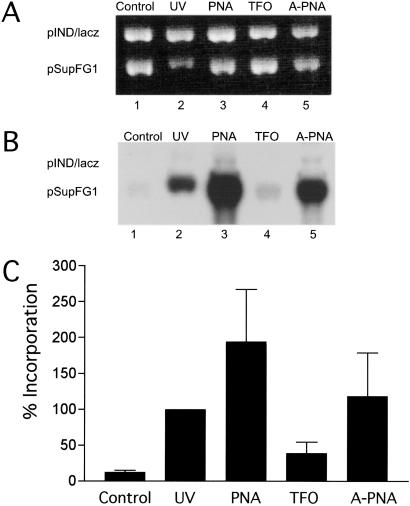
The PNA clamp constitutes an altered helical structure consisting of a PNA:DNA:PNA triplex in conjunction with a D-loop. To determine whether such a structure can provoke DNA repair, we measured the ability of the bis-PNA to induce repair synthesis in the target plasmid in HeLa cell-free extracts. Selected molecules, including either the bis-PNA, the PNA–DNA conjugate, or the DNA TFO, AG30, were incubated with supercoiled pSupFG1 plasmid in extracts supplemented with [α-32P]dCTP. As an internal control, the plasmid, pIND/lacZ, which lacks the PNA and TFO target site, was included with all samples. After incubation in the extracts, the plasmids were linearized by EcoRI digestion and analyzed by agarose gel electrophoresis. Incorporation of the labeled nucleotide into the plasmid was quantified by phosphor storage analysis and was taken as a measure of DNA repair synthesis (9).
The bis-PNA and to a slightly lesser extent the PNA–DNA conjugate were both found to stimulate labeling of the target pSupFG1 plasmid but not of the control pIND/lacZ plasmid (Fig. 3). The bis-PNA induced repair synthesis at an intensity 16-fold above background, and the PNA–DNA conjugate did so at an intensity of 7-fold above background (Fig. 3). The level of repair induced by the bis-PNA was as high as that seen in UV-damaged plasmid DNA, suggesting that the PNA clamp structure can strongly induce repair. The extent of repair synthesis induced by the PNA was higher than that produced by the TFO, AG30, which had been previously found to stimulate DNA repair synthesis in several sets of experiments (9, 32). This greater effect of the bis-PNA suggests that the PNA clamp may create a helical distortion even greater than that produced by a DNA triple helix. The difference in the extent of repair induction may also be due to the nonnatural chemical composition of the PNA as opposed to the DNA TFO.
Fig 3.
DNA repair synthesis stimulated by the formation of a PNA clamp in HeLa cell-free extracts. Supercoiled plasmids pSupFG1 and pIND/lacZ were preincubated at 37°C with PNA and TFO oligomers to allow binding via either clamp or triplex formation. The DNA was then added to HeLa extracts supplemented with [α-32P]dCTP. After 3 h at 30°C, the reactions were terminated and the plasmid DNAs were isolated, linearized by EcoRI digestion, and analyzed by agarose gel electrophoresis. (A) Visualization of the plasmid DNA by ethidium bromide staining. (B) Autoradiogram showing labeled nucleotide incorporation indicative of DNA repair synthesis. (C) Quantification of incorporation of [α-32P]dCTP into plasmid DNA. Each bar represents the ratio of intensity of the pSupFG1 band over the intensity of the control pIND/lacZ band representing background. The absolute incorporation was then normalized by setting the value for the UV-induced damage at 100%. The data are the average obtained from three individual experiments, with error bars calculated as the standard error.
The Role of Nucleotide Excision Repair Factor, XPA.
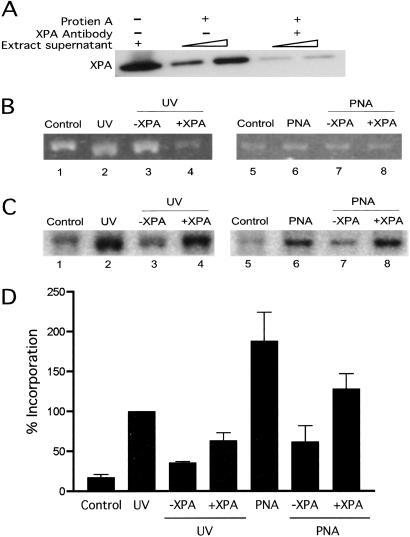
The results above establish that a PNA clamp can stimulate DNA repair synthesis in HeLa cell-free extracts. Because the ability of triplex formation to stimulate DNA metabolism depends on the NER pathway (9) and because the NER damage recognition factor, XPA, is important for recognition and repair of a triple helix (28), we tested the requirement for XPA in the PNA-induced DNA repair synthesis. A rabbit polyclonal antibody directed against recombinant human XPA protein was used to deplete XPA from the extracts by immunoprecipitation (25, 28). Depletion of XPA was confirmed by Western blot analysis of the residual extracts (Fig. 4). Because repair of UV damage depends on XPA (33), control samples included UV-irradiated plasmid DNAs. Depletion of XPA was found to substantially reduce the amount of induced DNA repair synthesis in both the PNA and UV samples (Fig. 4).
Fig 4.
Role of the NER pathway in PNA clamp-induced repair synthesis. DNA repair synthesis in HeLa cell-free extracts was measured as in Fig. 3. pSupFG1 plasmid DNA was untreated (control), treated with UV light, or incubated with the bis-PNA, as indicated. Extract samples were unmodified, XPA-depleted (−XPA), or XPA-depleted followed by supplementation with recombinant XPA protein (+XPA). (A) Immunodepletion of the NER factor, XPA, from HeLa cell-free extracts. Extract samples were immunodepleted using a rabbit polyclonal XPA antibody premixed with protein A agarose beads (25, 28). The immunoprecipitate was removed by centrifugation, and the remaining supernatant was examined by Western blot analysis. (B) Visualization by ethidium bromide staining of plasmid DNAs isolated from the extract reactions. (C) Autoradiogram showing labeled nucleotide incorporation indicative of repair synthesis. (D) Quantification of incorporation of [α-32P]dCTP into plasmid DNA. The amount of incorporation in each case was normalized by setting the value for the UV-induced damage at 100%. The data are the average obtained from three individual experiments, with error bars calculated as the standard error.
Following XPA immunodepletion, we tested the ability of XPA protein to restore the PNA-induced repair activity. The results show that the addition of XPA provides functional complementation in the depleted extracts (Fig. 4). These findings suggest that the ability of the bis-PNA to induce DNA repair depends on XPA, and they support the hypothesis that the NER pathway can recognize a PNA clamp as a lesion, thereby provoking DNA metabolism that can produce recombinagenic intermediates.
Requirement for XPA in PNA Clamp-Induced Recombination.
To directly determine whether the NER pathway participates in PNA clamp-induced recombination, we performed the recombination assay in extracts in which the levels of XPA had been manipulated by either immunodepletion or immunodepletion plus supplementation with purified protein, as above (Fig. 5). We found that XPA depletion substantially decreases recombination induced by the bis-PNA, whether the PNA is linked or not linked to the donor DNA. Interestingly, the activity of the unlinked mixture of A + PNA appeared to be more dependent on XPA than did that of the linked A–PNA conjugate. The activity of the A + PNA sample in the XPA-depleted extracts was reduced to 14% of the activity of that sample seen in the complete extracts, whereas the activity of the linked A–PNA sample in the depleted extracts was 39% of that in the complete extract. It is possible that the ability of the bis-PNA (i) to deliver the donor to the target region, (ii) to mediate strand invasion, and (iii) to thereby create an open, D-loop-containing structure may promote recombination to some extent, even when the NER pathway is experimentally disrupted.
Fig 5.
Role of the NER pathway in PNA clamp-induced recombination. Targeted recombination induced by PNA clamp formation was assayed in vitro in HeLa cell-free extracts as in Fig. 2. The pSupFG1/G144C plasmid DNA was incubated with the indicated PNAs and oligonucleotides in extract samples that were complete, XPA-depleted (−XPA), or XPA-depleted followed by supplementation with recombinant XPA protein (+XPA). After 2 h, the plasmid DNA was isolated and used to transform indicator bacteria for genetic analysis of the supFG1 gene. The bars indicate the frequency of blue colonies (representing recombinants) out of total colonies, with the actual count given to the right of each bar. A–PNA indicates molecules in which the donor DNA fragment, A, was covalently linked either to the bis-PNA, whereas the sample A + PNA represents the donor DNA fragment and the bis-PNA mixed together as separate, unlinked molecules. Error bars indicate standard errors.
Discussion
In the work described here, we have explored the use of PNAs to induce recombination at a specific site. Our results show that a bis-PNA designed to bind to a 10-bp region in the supFG1 gene was capable of stimulating recombination between the target vector and a 40-nt donor fragment in human cell-free extracts. Recombination was observed not only when the PNA and the donor DNA were covalently linked but also when the two were unlinked. Interestingly, a higher frequency was observed in the case where the molecules were not covalently joined, suggesting that such a linkage may create some steric hindrance to the recombination reaction.
The ability of a PNA clamp to induce recombination between its binding site and a separate donor DNA molecule is important because it may obviate the need for the synthesis of a PNA–DNA conjugate. Such a synthesis imposes some constraints on reagent design, such as limiting the length of the donor DNA to what can be made synthetically (to incorporate a functional group for conjugation) with reasonable purity and yield (typically <100 nt). As a result, much longer donor DNA molecules could be used, potentially increasing the efficiency of recombination (34). However, the ability of an unlinked mixture of a bis-PNA and a donor DNA to mediate recombination in a cell remains to be determined, and it may be influenced by the need for efficient cotransfection and simultaneous intranuclear colocalization of the two molecules. As a result, the PNA–DNA conjugate may still prove superior for intracellular use because of the inherent colocalization of the molecules. Also, the PNA domain in the bifunctional molecule can mediate an enzyme-independent homology search to thereby position the linked DNA at the cognate site, a property distinct from the ability of the PNA clamp to provoke repair and recombination.
With regard to repair, in experiments to measure DNA repair synthesis in the cell-free extracts, the PNA clamp was found to be a potent substrate for induction of repair. Unlike DNA triplex formation, in which the underlying duplex remains essentially intact and the third strand binds in the major groove, PNA clamp formation occurs by strand invasion of the duplex and binding of one DNA strand by two PNA oligomers to form a PNA/DNA/PNA triplex, with consequent displacement of the other DNA strand and formation of a D-loop. Our results indicate that such a structure is particularly effective at inducing DNA repair.
Detection of PNA clamp-induced repair synthesis prompted investigation of the importance of the NER pathway in the recognition and repair of this type of lesion. NER is a versatile DNA damage-removal pathway that protects DNA integrity from a multitude of DNA lesions (35). Within the divergent spectrum of NER-recognized lesions, the common denominator appears to be significant chemical or physical distortion of the DNA helix (36). XPA is a key NER factor responsible for verifying the altered DNA conformation, and it is crucial for correct assembly of the remaining repair machinery around the lesion (36). Because a PNA clamp significantly alters DNA helical structure and chemistry, we hypothesized that XPA would play a role in the repair of such a structure. We found that immunodepletion of XPA from the extracts resulted in a substantial reduction in the ability of the bis-PNA to induce repair synthesis. In addition, supplementation of depleted extracts with recombinant XPA protein restored PNA-induced repair activity. These results support a model in which repair of PNA-clamp structures is XPA dependent.
We also directly tested the role of XPA in the PNA clamp-induced recombination reaction. We found that depletion of XPA from the extracts causes a substantial reduction in the PNA-induced recombination, both when the bis-PNA is linked to the donor DNA and when it is unlinked. This finding is consistent with our model in which PNA clamp formation creates a helical distortion that can provoke DNA repair and thereby lead to recombinagenic intermediates. However, the bis-PNA appears to have some ability to promote recombination of a linked donor fragment even when XPA is depleted. This observation suggests that the capacity of the PNA to carry out site-directed DNA binding in an enzyme-independent manner combined with its ability to mediate strand invasion and displacement may stimulate recombination even in the absence of NER.
Currently, most gene targeting strategies are based on selection of cells that have undergone homologous recombination of a vector into the matching chromosomal locus (37). However, such selection techniques do not increase the absolute frequency of homologous recombination; rather, they serve to eliminate cells in which a targeting event has not occurred. Methods for selection have been needed because the frequency of homologous integration of transfected DNA is low in mammalian cells in comparison to that of nonhomologous integration (34). Because of this, investigators have recognized that techniques to enhance the frequency of homologous recombination in mammalian cells are needed (34).
DNA oligonucleotide-mediated triple helix formation represents one potential tool for stimulation of target-site recombination, and substantial efforts are underway to explore the utility of triplex formation for this purpose (11–13, 25, 34). However, in the work presented here, we have demonstrated that a bis-PNA can also stimulate repair and induce recombination in a site-directed manner by means of a mechanism dependent, in part, on the NER factor, XPA. Compared with a TFO-mediated triplex, the PNA clamp was able to induce higher levels of repair and recombination, suggesting that the clamp structure creates an even more severe helical distortion that is particularly effective at provoking DNA repair and promoting recombination. The ability of PNAs to sensitize a target site to homologous recombination and of PNA–DNA conjugates to direct recombination to a selected gene may provide an important research tool and may eventually enable correction of defective genes associated with hereditary diseases.
Acknowledgments
We thank L. Narayanan, D. Campisi, H. Datta, M. Macris, R. Franklin, S. J. Baserga, and L. Cabral for their help. This work was supported by a Scholar Award from the Leukemia and Lymphoma Society (to P.M.G.) and by National Institutes of Health Grants CA64186 and GM54731. F.A.R. was supported by an institutional National Research Service Award from the National Institutes of Health (T32 CA09259) and by a fellowship from the Lucille B. Markey Charitable Trust.
Abbreviations
NER, nucleotide excision repair
PNA, peptide nucleic acid
TFO, triplex-forming oligonucleotide
XPA, xeroderma pigmentosum complementation group A protein
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Opalinska J. B. & Gewirtz, A. M. (2002) Nat. Rev. Drug Discov. 1, 503-514. [DOI] [PubMed] [Google Scholar]
- 2.Knauert M. P. & Glazer, P. M. (2001) Hum. Mol. Genet. 10, 2243-2251. [DOI] [PubMed] [Google Scholar]
- 3.Giovannangeli C. & Helene, C. (2000) Curr. Opin. Mol. Ther. 2, 288-296. [PubMed] [Google Scholar]
- 4.Faria M., Wood, C. D., Perrouault, L., Nelson, J. S., Winter, A., White, M. R., Helene, C. & Giovannangeli, C. (2000) Proc. Natl. Acad. Sci. USA 97, 3862-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takasugi M., Guendouz, A., Chassignol, M., Decout, J. L., Lhomme, J., Thuong, N. T. & Helene, C. (1991) Proc. Natl. Acad. Sci. USA 88, 5602-5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Havre P. A., Gunther, E. J., Gasparro, F. P. & Glazer, P. M. (1993) Proc. Natl. Acad. Sci. USA 90, 7879-7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang G., Levy, D. D., Seidman, M. M. & Glazer, P. M. (1995) Mol. Cell. Biol. 15, 1759-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasquez K. M., Wensel, T. G., Hogan, M. E. & Wilson, J. H. (1996) Biochemistry 35, 10712-10719. [DOI] [PubMed] [Google Scholar]
- 9.Wang G., Seidman, M. M. & Glazer, P. M. (1996) Science 271, 802-805. [DOI] [PubMed] [Google Scholar]
- 10.Vasquez K. M., Narayanan, L. & Glazer, P. M. (2000) Science 290, 530-533. [DOI] [PubMed] [Google Scholar]
- 11.Chan P. P., Lin, M., Faruqi, A. F., Powell, J., Seidman, M. M. & Glazer, P. M. (1999) J. Biol. Chem. 274, 11541-11548. [DOI] [PubMed] [Google Scholar]
- 12.Faruqi A. F., Seidman, M. M., Segal, D. J., Carroll, D. & Glazer, P. M. (1996) Mol. Cell. Biol. 16, 6820-6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruqi A. F., Datta, H. J., Carroll, D., Seidman, M. M. & Glazer, P. M. (2000) Mol. Cell. Biol. 20, 990-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Z., Macris, M. A., Faruqi, A. F. & Glazer, P. M. (2000) Proc. Natl. Acad. Sci. USA 97, 9003-9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen P. E., Egholm, M., Berg, R. H. & Buchardt, O. (1993) Anticancer Drug Des. 8, 53-63. [PubMed] [Google Scholar]
- 16.Egholm M., Buchardt, O., Christensen, L., Behrens, C., Freier, S. M., Driver, D. A., Berg, R. H., Kim, S. K., Norden, B. & Nielsen, P. E. (1993) Nature 365, 566-568. [DOI] [PubMed] [Google Scholar]
- 17.Bentin T. & Nielsen, P. E. (1996) Biochemistry 35, 8863-8869. [DOI] [PubMed] [Google Scholar]
- 18.Larsen H. J. & Nielsen, P. E. (1996) Nucleic Acids Res. 24, 458-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peffer N. J., Hanvey, J. C., Bisi, J. E., Thomson, S. A., Hassman, C. F., Noble, S. A. & Babiss, L. E. (1993) Proc. Natl. Acad. Sci. USA 90, 10648-10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egholm M., Christensen, L., Dueholm, K. L., Buchardt, O., Coull, J. & Nielsen, P. E. (1995) Nucleic Acids Res. 23, 217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen P. E., Egholm, M. & Buchardt, O. (1994) J. Mol. Recognit. 7, 165-170. [DOI] [PubMed] [Google Scholar]
- 22.Praseuth D., Grigoriev, M., Guieysse, A. L., Pritchard, L. L., Harel-Bellan, A., Nielsen, P. E. & Helene, C. (1996) Biochim. Biophys. Acta 1309, 226-238. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen P. E., Egholm, M., Berg, R. H. & Buchardt, O. (1993) Nucleic Acids Res. 21, 197-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faruqi A. F., Egholm, M. & Glazer, P. M. (1998) Proc. Natl. Acad. Sci. USA 95, 1398-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta H. J., Chan, P. P., Vasquez, K. M., Gupta, R. C. & Glazer, P. M. (2001) J. Biol. Chem. 276, 18018-18023. [DOI] [PubMed] [Google Scholar]
- 26.Glazer P. M., Sarkar, S. N., Chisholm, G. E. & Summers, W. C. (1987) Mol. Cell. Biol. 7, 218-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones C. J. & Wood, R. D. (1993) Biochemistry 32, 12096-12104. [DOI] [PubMed] [Google Scholar]
- 28.Vasquez K. M., Christensen, J., Li, L., Finch, R. A. & Glazer, P. M. (2002) Proc. Natl. Acad. Sci. USA 99, 5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan J., Narayanan, L., Rockwell, S. & Glazer, P. M. (2000) Cancer Res. 60, 4372-4376. [PubMed] [Google Scholar]
- 30.Goncz K. K., Kunzelmann, K., Xu, Z. & Gruenert, D. C. (1998) Hum. Mol. Genet. 7, 1913-1919. [DOI] [PubMed] [Google Scholar]
- 31.Campbell C. R., Keown, W., Lowe, L., Kirschling, D. & Kucherlapati, R. (1989) New Biol. 1, 223-227. [PubMed] [Google Scholar]
- 32.Wang G., Chen, Z., Zhang, S., Wilson, G. L. & Jing, K. (2001) Nucleic Acids Res. 29, 1801-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood R. D. (1996) Annu. Rev. Biochem. 65, 135-167. [DOI] [PubMed] [Google Scholar]
- 34.Vasquez K. M., Marburger, K., Intody, Z. & Wilson, J. H. (2001) Proc. Natl. Acad. Sci. USA 98, 8403-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petit C. & Sancar, A. (1999) Biochimie 81, 15-25. [DOI] [PubMed] [Google Scholar]
- 36.Batty D. P. & Wood, R. D. (2000) Gene 241, 193-204. [DOI] [PubMed] [Google Scholar]
- 37.Capecchi M. R. (1989) Science 244, 1288-1292. [DOI] [PubMed] [Google Scholar]