Abstract
Members of the mammalian p160 family, such as GRIP1, are known as glucocorticoid receptor (GR) coactivators; at certain glucocorticoid response elements (GREs), however, GRIP1 acts as a GR corepressor. We characterized functional interactions of GR and GRIP1 in a repression complex where GR tethers to DNA-bound activator protein-1 (AP-1), as at the human collagenase-3 gene, and tested whether the identified interactions were similar or different at other response elements. At the AP-1 tethering GRE, we mapped the GRIP1 corepressor activity to a domain distinct from the two known GRIP1 activation domains; it exhibited intrinsic GR-independent repression potential when recruited to DNA via Gal4 DNA-binding domain. Interestingly, neither the domain nor the activity was detected in the other two p160 family members, SRC1 and RAC3. The same GRIP1 corepression domain was required for GR-mediated repression at the nuclear factor-κB (NF-κB) tethering GRE of the human IL-8 gene. In contrast, at the osteocalcin gene GRE, where GR represses transcription by binding to a DNA site overlapping the TATA box, both GRIP1 and SRC1 corepressed, and the GRIP1-specific repression domain was dispensable. Thus, in a single cell type, GR and GRIP1 conferred one mode of activation and two modes of repression by selectively engaging distinct surfaces of GRIP1 in a response element-specific manner.
Eukaryotic transcriptional regulation is accomplished by multiprotein complexes that assemble at response elements embedded in DNA sequences close to target promoters. The precise arrangements of sequences within response elements and the expression levels and activities of regulatory factors within a cell are key determinants of the composition and function of a given regulatory complex (1–3). Precisely how response elements affect the actions of a regulatory factor is not understood. What is clear, however, is that the effects can be profound: Within different regulatory complexes, a given factor may activate transcription, repress, or display no regulatory activity.
With respect to DNA binding, there are three contexts in which a regulatory factor can operate (4, 5): at “simple” response elements, the factor is the sole DNA-binding component of the regulatory complex; at “composite” response elements, the factor interacts functionally with at least one additional DNA-bound factor to nucleate regulatory complex assembly and action; and at “tethering” response elements, the factor does not itself bind specifically to DNA, but is recruited through interaction with another DNA-binding factor. Although simple response elements were the first to be described in experimental settings, composite and tethering elements likely predominate in natural genomes.
Steroid hormone receptors operate at all three types of response elements (5). In response to elevated hormone levels, the glucocorticoid receptor (GR), for example, associates with glucocorticoid response elements (GREs), leading to the assembly of other factors into functional regulatory complexes. At simple GREs that confer transcriptional activation, the p160 family members (SRC1, TIF2/GRIP1, and ACTR/RAC3/pCIP/AIB1) (6–11) interact with an activation function-2 that forms within the ligand-binding domain of GR (and other steroid receptors) in an agonist-dependent, antagonist-sensitive manner (12–14). The p160 factors carry two activation domains (AD1 and AD2) that recruit histone acetylases CREB-binding protein and p300, and an arginine methylase, CARM1, respectively (15–18). The p160 proteins include a nuclear receptor interaction domain (NID) containing three LxxLL motifs (NR boxes), which are differentially recognized by receptors; GR interacts preferentially with NR box3 (14, 19, 20). Genetic disruption of individual p160s in mice results in distinct phenotypes (21–25), suggesting that the different members may have distinct activities or preferences for particular receptors, but the nature and underlying mechanisms of these selectivities are not understood.
At the osteocalcin gene, GR represses transcription from a simple GRE that overlaps the TATA box in the promoter, presumably by occlusion of general transcription factor binding (26, 27); possible cofactor involvement at the osteocalcin GRE has not been investigated. In contrast, GR represses the collagenase-3 gene through a tethering GRE in which GR makes protein–protein contact with a DNA-bound activator protein-1 (AP-1); the AP-1 site is sufficient to confer both phorbol ester induction, through direct binding of activated AP-1, and glucocorticoid repression (28). In that context, TIF2/GRIP1 assembles in vivo into the regulatory complex in a GR- and glucocorticoid agonist-dependent, antagonist-sensitive manner (28). Importantly, however, GRIP1 potentiates GR-mediated repression of collagenase-3, rather than activation.
Together, these findings with a single regulator, GR, and a single cofactor, GRIP1, underscore the remarkable context dependence of transcriptional regulation, providing at three different response elements, one mode of activation and two modes of repression. What gives rise to these functional differences? In principle, the GR–GRIP1 interactions themselves may differ at different response elements; alternatively, context differences may be determined distal to the GR–GRIP1 interaction. Defining the points at which contexts diverge has the effect of “isolating” specific steps in regulatory complex assembly or conformation that produce selective functions. To begin to define the molecular determinants that distinguish different contexts, we chose to characterize some features of GRIP1 function and the GR–GRIP1 interaction in one context, the AP-1 tethering GRE. We then tested whether those features were similar or different when replacing GRIP1 with other p160 family members, or substituting the AP-1 element with different GREs.
Materials and Methods
Plasmids.
Previously described mammalian firefly luciferase reporters were: XG46TL, containing two copies of a simple GRE sequence from the mouse mammary tumor virus LTR; AP-1-Luc, containing a single AP-1 site (29); IL-8-Luc, containing a −1,481/+40 fragment of the IL-8 gene (30); and pOS-344-Luc, containing the −344/+33 fragment of the osteocalcin gene (27). The IL-8(κB)-Luc reporter was constructed by subcloning a 5′-AGTACGTGGAATTTCCTCT-3′ oligonucleotide (the IL-8-derived NF-κB site underlined) into the HindIII–SalI sites of pΔODLO plasmid (31). The 2xGal4/2xAP-1-Luc reporter was generated by subcloning a 5′-GTGAGTCAGAGACGTCTCTGAGTCACTGCA-3′ oligonucleotide (AP-1 sites underlined) into the PstI site of the 2xGal4-DLO plasmid (32). A β-actin-LacZ plasmid expressed β-galactosidase under control of human β-actin promoter.
The pCDNA3-GRIP1 construct was described (28). pCDNA3-GRIP1 N1007 and N765 were generated by subcloning GRIP1 EcoRI–BspHI/blunt and EcoRI–XhoI fragments into the EcoRI–XhoI/blunt and EcoRI–XhoI sites of pCDNA3 (Invitrogen), respectively. GRIP1715–1007 and GRIP1648–1007 were constructed by subcloning BlpI/blunt-XbaI and DrdI/blunt-XbaI fragments of GRIP1 N1007 into the pCDNA6His (Invitrogen) EcoRI/blunt-XbaI sites. The GRIP1 NID fragment was excised with BamHI–XhoI from the pGex2TK-GRIP1563–765 (14) and subcloned into BamHI–XhoI sites of pCDNA6His. GRIP1 ΔRD was generated by subcloning the C-terminal BspHI/blunt-EcoRI/blunt fragment of GRIP1 into XhoI/blunt sites of pCDNA3-GRIP1, thereby releasing the internal XhoI–BspHI fragment of GRIP1. pSG5-GRIP1 ΔAD1/ΔAD2 (GRIP1 N1121Δ1056–1110; ref. 33) and Gal:GRIP1 expressing full-length GRIP1 fused to the Gal4 DNA-binding domain (DBD) were kindly provided by M. Stallcup (University of Southern California, Los Angeles). Gal: GRIP1631–1007 was generated by subcloning the DraIII/blunt-XbaI fragment of GRIP1 N1007 into the EcoRI/blunt-XbaI sites of pSG424 (34). The Gal:MAD construct was described (32). “Empty” pSG424 was constructed by releasing the HindIII/blunt-EcoRI/blunt Gal4DBD fragment and recircularizing the vector. The pCDNA3-SRC1 plasmid was constructed by subcloning the BspHI/blunt-XbaI fragment from pCR3.1-SRC1 (35) into EcoRV-XbaI sites of pCDNA3. The pCMX-RAC3 construct has been described (11). Reading frames of all fusion proteins were verified by sequencing.
Cells, Treatments, and Transient Transfections.
Parental U2OS human osteosarcoma and U2OS.G cells expressing rat GR (36) were cultured in DMEM (Invitrogen) supplemented with 10% FBS (HyClone) and 350 μg/ml G418 (Invitrogen). Dexamethasone (100 nM) and phorbol 12-myristate 13-acetate (PMA, 25 ng/ml) dilutions were made in 100% ethanol; “untreated” cells received an equivalent amount of 100% ethanol.
For reporter activity assays, cells were seeded into 24-well plates in DMEM/10% FBS at 20,000 cells per well and transfected the following day in FBS-free DMEM by using 0.8 μl of Lipofectamine and 1.6 μl of PLUS reagent (Invitrogen) per well according to the manufacturer's instructions. After transfection (3 h), cells were refed with DMEM/10% FBS, allowed to recover for 3 h, and refed with DMEM/10% FBS containing appropriate hormone dilutions. Twelve hours later, cells were lysed in 100 μl per well of 1× lysis buffer (PharMingen) and assayed for luciferase and β-galactosidase activity as described (31).
The expression of transfected GRIP1 derivatives was assessed by immunoblotting with Abs to TIF2 (Affinity Bioreagents, Neshanic Station, NJ), His-tag (Novagen), or Gal4DBD (Santa Cruz Biotechnology) by using standard protocols.
RNA Isolation, Reverse Transcription, and Real-Time PCR.
U2OS.G cells were treated in 10-cm dishes and washed with PBS, and total RNA was isolated by using QIAshredder and RNeasy-Mini kits (Qiagen, Valencia, CA). Random hexamer-primed cDNA was reverse-transcribed from 0.5 μg of total RNA by using the CLONTECH RT-for-PCR kit. Real-time PCR analysis was performed on a Prism 7700 Sequence Detection System (Applied Biosystems). IL-8 cDNA fragment was amplified with the 5′-ACCGGAAGGAACCATCTCACT and 5′-ATCAGGAAGGCTGCCAAGAG-3′ primer pair and the 5′-dT-FAM-TGTAAACATGACTTCCAAGCTGGCCGT-TAMRA-3′ probe (Synthegen). Samples were analyzed in duplicate and normalized by using a ribosomal RPL19 primer/probe set (5′-ATGTATCACAGCCTGTACCTG-3′, 5′-TTCTTGGTCTCTTCCTCCTTG-3′, and 5′-dT-FAM-AGGTCTAAGACCAAGGAAGCACGCAA-TAMRA-3′) run in parallel. Data were transformed by using the ∂∂Ct method (Applied Biosystems user bulletin no. 2, 1997).
Results
GRIP1 corepression at an AP-1 tethering GRE. TIF2/GRIP1 serves as a GR corepressor at collagenase-3, as well as in the simplified context of an AP-1-Luc reporter (28) introduced into U2OS.G cells (36); in this setting, the activity of transfected GRIP1 can be observed over the baseline established by the endogenous protein. The only transcriptional regulatory domains previously described in GRIP1 were two coactivation domains, AD1 and AD2 (33, 37, 38), which act in the context of an inducible GRE. We therefore set out to define a part of GRIP1 that mediated GR corepression from an AP-1-Luc reporter.
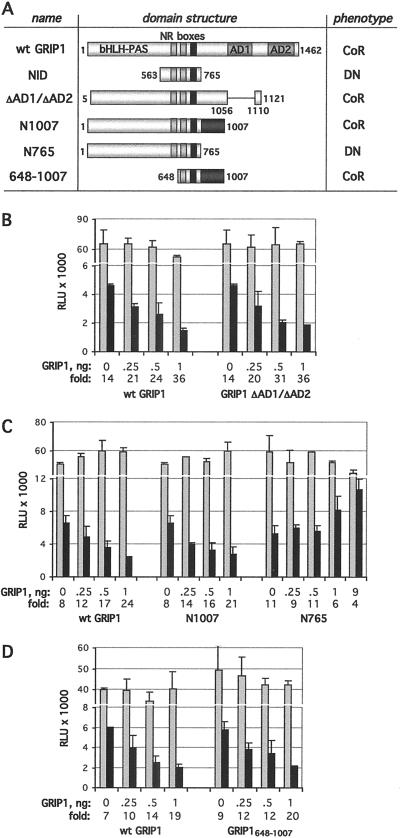
Fig. 1 A–C show that GRIP1 coactivation domains AD1 and AD2 are dispensable in this context, because two GRIP1 derivatives lacking both AD1 and AD2 (GRIP1 ΔAD1/ΔAD2 and N1007) were as potent in supporting GR-mediated repression as was full-length GRIP1. In contrast, GRIP1 N765, a C-terminal truncation at the end of the NID (residues 563–765), as well as the NID alone (28), failed to corepress and appeared to act in a dominant-negative manner on further overexpression (Fig. 1 A and B). The dominant-negative effect of NID and N765 is consistent with these derivatives interacting with GR and displacing endogenous TIF2/GRIP1 from the GR repression complex. Thus, residues 765–1,007 immediately downstream of the NID are essential for corepression.
Fig 1.
Mapping a GRIP1 corepression domain. (A) The domain structure of WT and mutant GRIP1 derivatives. The basic helix–loop–helix (bHLH)/Per-ARNT-Sim (PAS) domain, NR boxes (NR box3 is shown in black), AD1, and AD2 are diagrammed. GRIP1 derivatives act as corepressors (CoR) or dominant-negatives (DN), as indicated, with respect to GR-mediated repression of AP-1. Residues 765–1,007 (shaded) are required for corepression. U2OS.G cells were transfected with indicated amounts of WT (wt) GRIP1 or ΔAD1/ΔAD2 (B), N1007 and N765 (C), or GRIP1648–1007 (D) along with 40 ng per well each of the AP-1-Luc reporter and β-actin-LacZ plasmid. Total amounts of transfected DNA were equalized with pCDNA3. Cells were treated overnight with 25 ng/ml PMA in the absence (gray) or presence (black) of 100 nM dexamethasone, and reporter activity was measured, normalized to β-galactosidase activity, and expressed as relative luminescence units (RLU). The y axis is broken to better visualize reporter activity in the absence and presence of Dex. Fold repression in each case is shown.
We next deleted the N-terminal half of GRIP1 including a part of the NID, NR box1. The resultant GRIP1648–1007 construct, expressed as an N-terminally His-tagged derivative, corepressed the AP-1-Luc reporter (Fig. 1 A and D). Further truncation eliminating NR box2 (GRIP1715–1007) abolished corepressor activity although the protein was expressed (not shown). We conclude that GRIP1648–1007 is sufficient to serve as a corepressor for GR at the AP-1 tethering GRE because it includes both a GR interaction surface and a corepression domain.
Repression and Corepression by GRIP1.
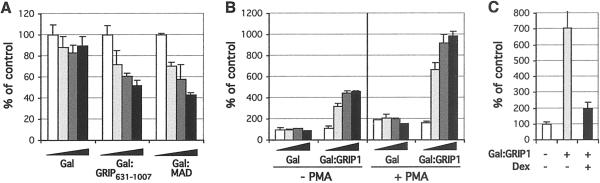
A transcriptional coregulator could in principle function in three ways: it might (i) display factor recruitment or enzymatic activities in the absence of the cognate regulator; (ii) bind to a regulator and alter its activity; or (iii) be induced to display an activity by a regulator to which it binds. Importantly, these functions are not mutually exclusive. However, only the first scheme predicts that the coregulator would affect transcription without interacting with a cognate regulatory factor. Consequently, we fused GRIP1631–1007, similar to the GR corepression-competent fragment defined above (Fig. 1A), to the Gal4DBD and tested its function on a luciferase reporter regulated by a composite element containing two Gal4 and two AP-1-binding sites (2xGal4/2xAP-1-Luc). In parental GR-deficient U2OS cells, Gal:GRIP1631–1007 repressed AP-1 activity ≈2-fold in the absence (not shown) or presence (Fig. 2A) of PMA; expression of the Gal4DBD alone had no effect. Thus, GRIP1631–1007 includes an intrinsic repression domain that can function independent of GR. In a parallel experiment, a similar 2-fold repression was conferred by a Gal4DBD fusion to the max dimerizer (MAD) corepression domain (Gal:MAD; Fig. 2A), shown previously to carry intrinsic repression activity (39, 40). Of note, Gal:GRIP1631–1007 did not affect the constitutive activity of a reporter controlled by two Gal4-binding sites (2xGal4-Luc), suggesting that it represses activated rather than basal transcription (not shown).
Fig 2.
GRIP1 corepressor activity in heterologous context. (A) GRIP1 repression domain is active in the absence of GR. Parental U2OS cells were transfected with increasing amounts of pSG424 plasmid expressing Gal4DBD (Gal), Gal4DBD fused to GRIP1631–1007 (Gal:GRIP1631–1007) or to MAD repression domain (Gal:MAD), along with the 2xGal4/2xAP-1-Luc reporter and β-actin-LacZ. Total amount of DNA was equalized with empty pSG424. Reporter activity was measured in the presence of PMA, normalized to β-galactosidase activity, and expressed as the percentage of activity observed for empty pSG424. (B) Full-length GRIP1 fused to Gal4DBD is a transcriptional activator. U2OS cells were transfected with increasing amounts of Gal4DBD (Gal) or Gal:GRIP1, and the activity of 2xGal4/2xAP-1-Luc reporter was assayed with/without PMA and normalized as described in A. (C) Liganded GR blocks GRIP1-mediated transcriptional activation. U2OS.G cells were transfected with Gal:GRIP1 or empty pSG424, 2xGal4-Luc reporter, and β-actin-LacZ. Reporter activity in the presence of PMA−/+Dex, as indicated, was assayed as described in A.
Interestingly, replacement of the GRIP631–1007 fragment by full-length GRIP1 in the Gal4DBD fusion construct yielded a potent activator (Gal:GRIP1, Fig. 2B). This result is consistent with previous studies showing that AD1 and AD2 carry intrinsic activation activity (37) and suggests either that activation activity exceeds repression activity in this context or that intrinsic repression activity is inhibited in full-length Gal:GRIP1 in the absence of a cognate regulator.
Because the regulatory outcome of the GR–GRIP1 interaction depends on the response element to which GR is bound, we tested the intrinsic effect of GR, not bound to a response element, on the activity of the Gal:GRIP1 fusion protein. For these experiments, we expressed Gal:GRIP1 in U2OS.G cells together with a 2xGal4-Luc luciferase reporter. Induction by the Gal:GRIP1 fusion protein was strongly repressed by dexamethasone (Fig. 2C). Thus, when tethered to DNA by protein–protein contacts with AP-1 or Gal:GRIP1, GR interaction with GRIP1 appeared to evoke repression activity; in contrast, GR interaction with GRIP1 at idealized palindromic GREs resulted in transcriptional activation. It is intriguing that GR is a homodimer when bound to the palindromic GRE, but a monomer in absence of DNA binding (5), perhaps suggesting that the GR oligomeric state may affect its signaling to GRIP1.
Actions of Other p160 Proteins at the AP-1 Tethering GRE.
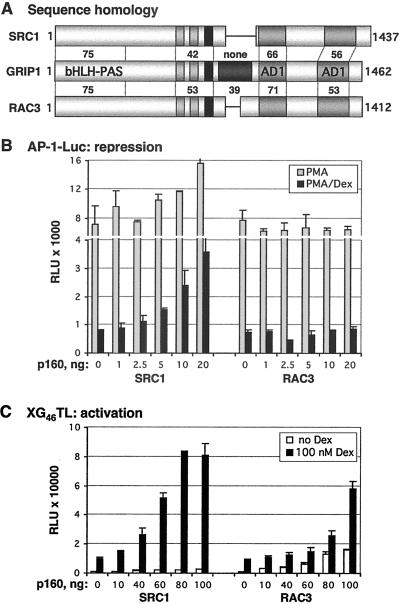
Given the evidence that the GRIP1 corepression domain can fold and function intrinsically in certain contexts, we wished to determine whether the other members of the p160 family, SRC1 and RAC3, are similarly active as GR corepressors at the AP-1 tethering GRE. Sequence comparisons revealed that the three factors share strong amino acid similarity throughout their basic helix–loop–helix (bHLH)/Per-ARNT-Sim (PAS), NID, and coactivation domains, AD1 and AD2 (Fig. 3A), and functional studies demonstrate that all are GR coactivators at simple palindromic GREs (6, 7, 10). Strikingly, however, the GRIP1 corepression region outside of the NID (residues 765–1,007, Fig. 1A) bears little resemblance to other p160s: amino acid similarity between GRIP1 and RAC3 within this region is only ≈39%, whereas SRC1 lacks most of the domain (Fig. 3A). Consistent with this observation, SRC1 and RAC3 failed to corepress with GR at the AP-1 tethering GRE in U2OS.G cells (Fig. 3B). Furthermore, overexpressed SRC1 antagonized glucocorticoid repression, indicating that it may function as a dominant-negative by displacing endogenous TIF2/GRIP1 from the repression complex; RAC3 lacked detectable activity in parallel experiments. Of importance, both SRC1 and RAC3 served as GR coactivators at an mouse mammary tumor virus simple GRE under the same conditions, demonstrating that both proteins were functional and capable of interacting with GR (Fig. 3C). Thus, in the context of an AP-1 tethering GRE in U2OS.G cells, GRIP1 is the only p160 family member that functions as a GR corepressor.
Fig 3.
GRIP1 corepressor activity at the AP-1 tethering GRE is not conserved across the p160 family. (A) GRIP1 corepression domain is unique in the p160 family. Diagrammed are the p160 proteins: SRC1, GRIP1, and RAC3. Numbers indicate the amino acid similarity of SRC1 and RAC3 to GRIP1 throughout their basic helix–loop–helix (bHLH)/Per-ARNT-Sim (PAS), NID, RD, AD1, and AD2 domains, as assessed by using blast software (National Center for Biotechnology Information). (B and C) The effects of SRC1 and RAC3 on GR repression (B) and activation (C). Indicated amounts of SRC1 and RAC3 were transfected into U2OS.G cells and AP-1-Luc- (B) or XG46TL- (C) reporter activity was measured as described in Fig. 1.
Overall, our studies define two physical differences in the functionally distinct interactions between GR and GRIP1 at the AP-1 tethering GRE, where GR represses transcription, and the mouse mammary tumor virus simple GRE, where GR activates. At the AP-1 site, GRIP1 appears to function through a repression domain adjacent to or encompassing the NID, and other p160 family members are inactive; in contrast, at a simple GRE, GRIP1 acts via two activation domains, and all three p160 family members are functional.
GRIP1 Corepression at an NF-κB Tethering GRE.
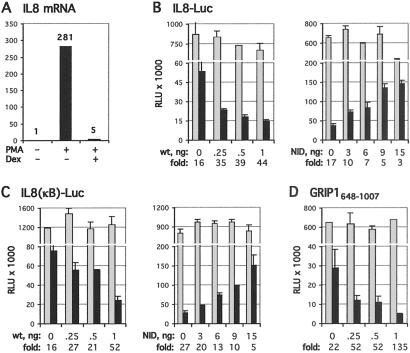
Having established two contexts for GR–GRIP1 interactions that lead to glucocorticoid repression or activation, we examined two additional response element contexts to assess whether yet further “molecular landmarks” distinguish the functional interactions of this regulator–coregulator pair. In search of a second element at which GR represses transcription via a tethering mechanism, we tested whether the IL-8 gene is active and regulated in U2OS.G cells. NF-κB elements enhance transcription of many inflammatory cytokine genes, such as IL-8, and glucocorticoid inhibition of NF-κB activity has been characterized in multiple cell types (32, 41–44). Fig. 4A shows that treatment of U2OS.G cells with PMA led to a >250-fold increase in IL-8 mRNA level and that dexamethasone fully repressed the response; indeed, regulated cytokine expression has been reported previously in osteoblast-like cells (45, 46). As expected, a luciferase-reporter gene linked either to the −1,481/+40 fragment upstream of the IL-8-coding sequence or to the isolated NF-κB site from that upstream region (Fig. 4 B and C, respectively) were similarly responsive to PMA induction (not shown) and glucocorticoid repression. As with the AP-1 tethering GRE, overexpression of full-length GRIP1 potentiated repression, whereas the GRIP1 NID functioned as a dominant-negative (Fig. 4 B and C). Further mapping demonstrated that GRIP1648–1007 (Fig. 4D) but not GRIP1715–1007 (not shown) was a fully functional GR corepressor at the IL-8-derived NF-κB tethering GRE. Overexpression of other p160 family members, SRC1 and RAC3, had no effect on GR-mediated repression in this context (not shown). Thus, at our current level of analysis, we have not detected differences in the GR–GRIP1 functional interactions at two distinct tethering GREs, the collagenase-3 AP-1 site and the IL-8 NF-κB site.
Fig 4.
GRIP1 potentiates GR-mediated repression of NF-κB. (A) IL-8 gene is active in U2OS.G cells. U2OS.G cells were treated for 2 h with PMA−/+Dex, as indicated, and the IL-8 mRNA level was assessed by real-time PCR, as described in Materials and Methods, and expressed relative to untreated control cells. (B–D) GRIP1 residues 648–1,007 mediate corepression at the NF-κB tethering GRE. Indicated amounts of wt GRIP1, GRIP1 NID (B and C) or GRIP1648–1007 (D) (equalized with pCDNA3) were transfected into U2OS.G cells, along with the IL-8-Luc (B) or IL-8(κB)-Luc (C and D) reporter and β-actin-LacZ. Cell treatments and reporter activities were measured as described in Fig. 1.
p160 Corepression Activities at the Osteocalcin GRE.
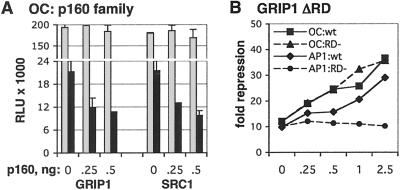
In a third context for GR-mediated repression, we examined the simple GRE in the osteocalcin gene; in this case, the GRE overlaps the TATA box such that GR binding to DNA occludes binding by TBP (26, 27). As shown in Fig. 5A, dexamethasone treatment repressed a luciferase reporter driven by the osteocalcin promoter in U2OS.G cells; of interest, both GRIP1 and SRC1, displayed GR corepressor activity under these conditions. Because SRC1 lacks the GR corepression domain functional in GRIP1 at AP-1 and NF-κB tethering GREs, this result suggests that distinct p160 corepression surfaces may operate at the osteocalcin GRE. To test this idea, we constructed GRIP1 ΔRD, which lacks residues (767–1,006) encompassing the GRIP1 corepression domain for tethering GREs. Fig. 5B demonstrates that GRIP1 ΔRD was fully active as a GR corepressor at the osteocalcin simple GRE, but, as expected, failed to corepress at the AP-1 tethering GRE. Thus, these results reveal the difference between the two types of response elements at which GR represses transcription: at the AP-1 and NF-κB tethering GREs, GRIP1 is the only p160 that confers repression, by using a distinct repression domain, whereas both GRIP1 and SRC1 can potentiate repression of osteocalcin and the GRIP1 repression domain is dispensable for this activity.
Fig 5.
p160 corepression at the octeocalcin GRE. (A) GRIP1 and SRC1 potentiate GR-mediated repression of osteocalcin. U2OS.G cells were transfected with a pOS-344-Luc reporter, β-actin-LacZ, and indicated amounts of GRIP1 or SRC1 (equalized with pCDNA3). Reporter activity was assessed in the absence (gray) or presence (black) of Dex as described in Fig. 1. (B) GRIP1 repression domain is dispensable for corepression at the osteocalcin GRE. U2OS.G cells were transfected with indicated amounts of WT (wt) GRIP1 or GRIP1 ΔRD (RD−), and the activities of the pOS-344-Luc (OC) and AP-1-Luc (AP1) reporters were assessed in the absence or presence of Dex and plotted as “fold repression” at each amount of transfected GRIP1 derivative.
Discussion
It is well established that response elements play a major role in defining distinct contexts for combinatorial regulation (5). Thus, a typical genomic response element contains multiple orderly arranged protein-binding sequences, which together with the levels and activities of participating cellular factors generate a particular multicomponent assemblage that results in a specific regulatory effect. However, precisely how response elements specify prescribed activities remains poorly understood. Our strategy for approaching this issue was to characterize the functional interaction of a particular “factor pair,” a regulator and a coregulator that operate together in one context, and then to test whether its functional interaction is the same or different at other response elements. Under constant cell and growth conditions, a difference detected in the factor pair interaction would imply that it was imposed by the response element. This approach allows one to “map” at the molecular level context effects that lead to functional differences, and having isolated such distinctions, to investigate their underlying mechanisms.
The factor pair we chose was GR and TIF2/GRIP1, members of two well characterized protein families. A wealth of biochemical, molecular, and structural data supports the role of p160 proteins as ligand-dependent coactivators for steroid receptors, capable of recruiting HAT and MT activities into functional regulatory complexes, which in turn can modify substrates in the vicinity of target promoters and contribute to enhanced transcription. Sequence similarities, receptor interaction studies and reporter gene activation assays suggested that the three p160 factors might be functionally redundant. However, RAC3 and GRIP1 knockout mice exhibit distinct reproductive phenotypes, both strikingly different from the relatively mild phenotype of the SRC1 null animal. In addition, RAC3 but not its sister factors, has been observed in the cytoplasm in some circumstances, implying a layer of regulation not observed with other p160s (47). Thus, the activation functions of the p160 proteins appear to share related but distinguishable properties.
Here, we characterized GR-GRIP1 factor pair activities at an AP-1 element in U2OS.G cells, a simplified response element context in which GR tethers to AP-1 in an agonist-dependent manner and collaborates with GRIP1 to repress transcription (28). Because the surfaces specifying GR–GRIP1 interaction were indistinguishable from those described at inducible GREs (19, 28), we tested for other differences that might distinguish the activation and repression contexts. We found that SRC1 and RAC3 lack corepressor activity at the AP-1 tethering GRE, whereas all three p160s are functional coactivators at a simple GRE (Fig. 3). Indeed, the GRIP1 corepressor activity mapped to a region of the protein not previously functionally assigned, which lacks evident similarity to other sequenced proteins, including p160 family members (Fig. 3A). Interestingly, this segment of GRIP1 displayed repressor activity in the absence of GR when expressed as a Gal4DBD fusion protein (Fig. 2A), implying that its folding and function require neither other GRIP1 domains, nor interaction with or proximity to GR.
In principle, the coactivation and corepression activities of GRIP1 could operate simultaneously, with the net effect dependent on which function is “stronger” in the context of a given regulatory complex. However, ΔAD1/ΔAD2 and full-length GRIP1 display similar corepressor activities at the AP-1 tethering GRE (Fig. 1B), implying that the coactivation domains are inactive when the corepression function is engaged. Conversely, GRIP1 ΔRD and full-length GRIP1 exhibit similar coactivation activities at an mouse mammary tumor virus GRE (not shown), suggesting that the corepression function is disabled in activation contexts. Thus, although the GRIP1 coactivation and corepression domains can fold and function independently when expressed as separate fragments, it appears that these activities “toggle” in the full-length protein, giving rise to either coactivation or corepression, but not both. It will be interesting to determine the mechanism of this molecular switch.
Within the p160 family, the GRIP1-selective corepressor activity at tethering elements implies that the relative levels or activities of individual p160s in a given cell or tissue might determine the magnitude of GR repression of AP-1- or NF-κB-dependent transcription. Thus, in cells expressing more SRC1 than GRIP1, glucocorticoid-mediated repression would be less pronounced than in cells expressing a preponderance of GRIP1, even if the levels of GR itself were similar. Although the relative importance of p160 levels in defining cell type-specific differences in GR action has not been examined, these results may have implications for the management of diseases in which glucocorticoid repression of proliferative or inflammatory signaling is a therapeutic goal.
As with GR repression at AP-1 and NF-κB tethering GREs, GRIP1 has also been implicated as a corepressor for the estrogen receptor at a TNFα response element, another tethering context (48). Similar to our results with tethering GREs, GRIP1 amino acid residues 912–1,003 are required for the corepressor activity, whereas SRC1 and RAC3 appear not to affect corepression at the TNF-RE (J. An and D.C.L., unpublished observations). Thus, the factor pair analysis in contexts in which steroid receptors repress transcription via tethering, identifies a particular segment of GRIP1 as a distinctive functional surface. In contrast, within the architecturally distinct GR repression complexes at the osteocalcin GRE, the functional p160 surfaces apparently differ, as SRC1 and GRIP1 display similar activities (Fig. 5A). Although cofactors that facilitate glucocorticoid repression of the osteocalcin gene in vivo have not been examined, our results imply that p160s may merely stabilize hormone or DNA binding by GR, or that a region distinct from the described corepression domain may actively confer repression in this context.
In sum, our study demonstrates the value of the factor pair approach for analyzing multicomponent complexes. That is, by comparing the structural and functional interactions of two factors that act in regulatory complexes in different response element contexts, it is possible to identify molecular features that serve as context determinants without first identifying all of the interacting components. In this way, the factor pair serves as a biological probe of context, and the surfaces identified provide foci for mechanistic study.
Acknowledgments
We thank M. Stallcup and M. Garabedian for providing expression plasmids and U2OS.G cells, respectively, K. Zarember for help with real-time PCR, members of the Yamamoto laboratory for helpful discussions, and B. Black, R. Derynck, A. Frankel, K. Guy, and D. Pearce for critical comments on the manuscript. I.R. was supported by the Parker B. Francis Fellowship for Pulmonary Research and by a Special Fellowship from the Leukemia and Lymphoma Society. H.F.L. is a recipient of a Postdoctoral Fellowship from the National Institutes of Health. Research support was from the National Institutes of Health and the National Science Foundation.
Abbreviations
GR, glucocorticoid receptor
GRE, glucocorticoid response element
AP-1, activator protein-1
NF-κB, nuclear factor-κB
NID, nuclear receptor interaction domain
AD, activation domain
DBD, DNA-binding domain
MAD, max dimerizer
References
- 1.Hu X. & Lazar, M. A. (2000) Trends Endocrinol. Metab. 11, 6-10. [DOI] [PubMed] [Google Scholar]
- 2.Freedman L. P. (1999) Cell 97, 5-8. [DOI] [PubMed] [Google Scholar]
- 3.Malik S. & Roeder, R. G. (2000) Trends Biochem. Sci. 25, 277-283. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto K. R., Darimont, B. D., Wagner, R. L. & Iniguez-Lluhi, J. A. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 587-598. [DOI] [PubMed] [Google Scholar]
- 5.Lefstin J. A. & Yamamoto, K. R. (1998) Nature 392, 885-888. [DOI] [PubMed] [Google Scholar]
- 6.Hong H., Kohli, K., Trivedi, A., Johnson, D. L. & Stallcup, M. R. (1996) Proc. Natl. Acad. Sci. USA 93, 4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onate S. A., Tsai, S. Y., Tsai, M. J. & O'Malley, B. W. (1995) Science 270, 1354-1357. [DOI] [PubMed] [Google Scholar]
- 8.Voegel J. J., Heine, M. J., Zechel, C., Chambon, P. & Gronemeyer, H. (1996) EMBO J. 15, 3667-3675. [PMC free article] [PubMed] [Google Scholar]
- 9.Anzick S. L., Kononen, J., Walker, R. L., Azorsa, D. O., Tanner, M. M., Guan, X. Y., Sauter, G., Kallioniemi, O. P., Trent, J. M. & Meltzer, P. S. (1997) Science 277, 965-968. [DOI] [PubMed] [Google Scholar]
- 10.Chen H., Lin, R. J., Schiltz, R. L., Chakravarti, D., Nash, A., Nagy, L., Privalsky, M. L., Nakatani, Y. & Evans, R. M. (1997) Cell 90, 569-580. [DOI] [PubMed] [Google Scholar]
- 11.Li H., Gomes, P. J. & Chen, J. D. (1997) Proc. Natl. Acad. Sci. USA 94, 8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiau A. K., Barstad, D., Loria, P. M., Cheng, L., Kushner, P. J., Agard, D. A. & Greene, G. L. (1998) Cell 95, 927-937. [DOI] [PubMed] [Google Scholar]
- 13.Williams S. P. & Sigler, P. B. (1998) Nature 393, 392-396. [DOI] [PubMed] [Google Scholar]
- 14.Darimont B. D., Wagner, R. L., Apriletti, J. W., Stallcup, M. R., Kushner, P. J., Baxter, J. D., Fletterick, R. J. & Yamamoto, K. R. (1998) Genes Dev. 12, 3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torchia J., Rose, D. W., Inostroza, J., Kamei, Y., Westin, S., Glass, C. K. & Rosenfeld, M. G. (1997) Nature 387, 677-684. [DOI] [PubMed] [Google Scholar]
- 16.Yao T. P., Ku, G., Zhou, N., Scully, R. & Livingston, D. M. (1996) Proc. Natl. Acad. Sci. USA 93, 10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D., Ma, H., Hong, H., Koh, S. S., Huang, S. M., Schurter, B. T., Aswad, D. W. & Stallcup, M. R. (1999) Science 284, 2174-2177. [DOI] [PubMed] [Google Scholar]
- 18.Ma H., Baumann, C. T., Li, H., Strahl, B. D., Rice, R., Jelinek, M. A., Aswad, D. W., Allis, C. D., Hager, G. L. & Stallcup, M. R. (2001) Curr. Biol. 11, 1981-1985. [DOI] [PubMed] [Google Scholar]
- 19.Ding X. F., Anderson, C. M., Ma, H., Hong, H., Uht, R. M., Kushner, P. J. & Stallcup, M. R. (1998) Mol. Endocrinol. 12, 302-313. [DOI] [PubMed] [Google Scholar]
- 20.Bledsoe R. K., Montana, V. G., Stanley, T. B., Delves, C. J., Apolito, C. J., McKee, D. D., Consler, T. G., Parks, D. J., Stewart, E. L., Willson, T. M., et al. (2002) Cell 110, 93-105. [DOI] [PubMed] [Google Scholar]
- 21.Qi C., Zhu, Y., Pan, J., Yeldandi, A. V., Rao, M. S., Maeda, N., Subbarao, V., Pulikuri, S., Hashimoto, T. & Reddy, J. K. (1999) Proc. Natl. Acad. Sci. USA 96, 1585-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J., Liao, L., Ning, G., Yoshida-Komiya, H., Deng, C. & O'Malley, B. W. (2000) Proc. Natl. Acad. Sci. USA 97, 6379-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J., Qiu, Y., DeMayo, F. J., Tsai, S. Y., Tsai, M. J. & O'Malley, B. W. (1998) Science 279, 1922-1925. [DOI] [PubMed] [Google Scholar]
- 24.Weiss R. E., Xu, J., Ning, G., Pohlenz, J., O'Malley, B. W. & Refetoff, S. (1999) EMBO J. 18, 1900-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gehin M., Mark, M., Dennefeld, C., Dierich, A., Gronemeyer, H. & Chambon, P. (2002) Mol. Cell. Biol. 22, 5923-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stromstedt P. E., Poellinger, L., Gustafsson, J. A. & Carlstedt-Duke, J. (1991) Mol. Cell. Biol. 11, 3379-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer T., Gustafsson, J. A. & Carlstedt-Duke, J. (1997) DNA Cell Biol. 16, 919-927. [DOI] [PubMed] [Google Scholar]
- 28.Rogatsky I., Zarember, K. A. & Yamamoto, K. R. (2001) EMBO J. 20, 6071-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogatsky I., Waase, C. L. & Garabedian, M. J. (1998) J. Biol. Chem. 273, 14315-14321. [DOI] [PubMed] [Google Scholar]
- 30.Warny M., Keates, A. C., Keates, S., Castagliuolo, I., Zacks, J. K., Aboudola, S., Qamar, A., Pothoulakis, C., LaMont, J. T. & Kelly, C. P. (2000) J. Clin. Invest. 105, 1147-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iniguez-Lluhi J. A., Lou, D. Y. & Yamamoto, K. R. (1997) J. Biol. Chem. 272, 4149-4156. [DOI] [PubMed] [Google Scholar]
- 32.Nissen R. M. & Yamamoto, K. R. (2000) Genes Dev. 14, 2314-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen D., Huang, S. M. & Stallcup, M. R. (2000) J. Biol. Chem. 275, 40810-40816. [DOI] [PubMed] [Google Scholar]
- 34.Sadowski I. & Ptashne, M. (1989) Nucleic Acids Res. 17, 7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenster G., Spencer, T. E., Burcin, M. M., Tsai, S. Y., Tsai, M. J. & O'Malley, B. W. (1997) Proc. Natl. Acad. Sci. USA 94, 7879-7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogatsky I., Trowbridge, J. M. & Garabedian, M. J. (1997) Mol. Cell. Biol. 17, 3181-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voegel J. J., Heine, M. J., Tini, M., Vivat, V., Chambon, P. & Gronemeyer, H. (1998) EMBO J. 17, 507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma H., Hong, H., Huang, S. M., Irvine, R. A., Webb, P., Kushner, P. J., Coetzee, G. A. & Stallcup, M. R. (1999) Mol. Cell. Biol. 19, 6164-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayer D. E., Laherty, C. D., Lawrence, Q. A., Armstrong, A. P. & Eisenman, R. N. (1996) Mol. Cell. Biol. 16, 5772-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laherty C. D., Yang, W. M., Sun, J. M., Davie, J. R., Seto, E. & Eisenman, R. N. (1997) Cell 89, 349-356. [DOI] [PubMed] [Google Scholar]
- 41.De Bosscher K., Vanden Berghe, W., Vermeulen, L., Plaisance, S., Boone, E. & Haegeman, G. (2000) Proc. Natl. Acad. Sci. USA 97, 3919-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heck S., Bender, K., Kullmann, M., Gottlicher, M., Herrlich, P. & Cato, A. C. (1997) EMBO J. 16, 4698-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito K., Barnes, P. J. & Adcock, I. M. (2000) Mol. Cell. Biol. 20, 6891-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liden J., Delaunay, F., Rafter, I., Gustafsson, J. & Okret, S. (1997) J. Biol. Chem. 272, 21467-21472. [DOI] [PubMed] [Google Scholar]
- 45.Chaudhary L. R. & Avioli, L. V. (1994) Calcif. Tissue Int. 55, 16-20. [DOI] [PubMed] [Google Scholar]
- 46.Siddiqi A., Burrin, J. M., Wood, D. F. & Monson, J. P. (1998) J. Endocrinol. 157, 453-461. [DOI] [PubMed] [Google Scholar]
- 47.Wu R. C., Qin, J., Hashimoto, Y., Wong, J., Xu, J., Tsai, S. Y., Tsai, M. J. & O'Malley, B. W. (2002) Mol. Cell. Biol. 22, 3549-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.An J., Ribeiro, R. C., Webb, P., Gustafsson, J. A., Kushner, P. J., Baxter, J. D. & Leitman, D. C. (1999) Proc. Natl. Acad. Sci. USA 96, 15161-15166. [DOI] [PMC free article] [PubMed] [Google Scholar]