Abstract
Peptidyl–tRNA hydrolase (encoded by pth) is an essential enzyme in all bacteria, where it releases tRNA from the premature translation termination product peptidyl–tRNA. Archaeal genomes lack a recognizable peptidyl–tRNA hydrolase (Pth) ortholog, although it is present in most eukaryotes. However, we detected Pth-like activity in extracts of the archaeon Methanocaldococcus jannaschii. The uncharacterized MJ0051 ORF was shown to correspond to a protein with Pth activity. Heterologously expressed MJ0051 enzyme catalyzed in vitro the cleavage of the Pth substrates diacetyl-[14C]lysyl–tRNA and acetyl-[14C]phenylalanyl–tRNA. On transformation of an Escherichia coli pthts mutant, the MJ0051 gene (named pth2) rescued the temperature-sensitive phenotype of the strain. Analysis of known genomes revealed the presence of highly conserved orthologs of the archaeal pth2 gene in all archaea and eukaryotes but not in bacteria. The phylogeny of pth2 homologs suggests that the gene has been vertically inherited throughout the archaeal and eukaryal domains. Deletions in Saccharomyces cerevisiae of the pth2 (YBL057c) or pth (YHR189w) orthologs were viable, as was the double deletion strain, implying that the canonical Pth and Pth2 enzymes are not essential for yeast viability.
Peptidyl–tRNA hydrolase (Pth) is an essential enzyme in bacteria (1–3). It cleaves peptidyl–tRNAs, premature protein synthesis products that dissociate from the ribosome, and allows the discharged tRNAs to reengage in protein synthesis. In mutants with limited Pth activity, peptidyl–tRNAs accumulate in the cell and reduce the availability of essential acylatable tRNAs below the limit compatible with protein synthesis; thus cell growth is impaired. However, the possibility that peptidyl–tRNA per se is toxic has not been ruled out. A significant proportion of the ribosomes that initiate mRNA translation interrupt protein synthesis before reaching the stop codon (4). These events of abortive translation may result in peptidyl–tRNA dissociation from the ribosome (5), forming the natural substrates for Pth. There are at least two documented instances of peptidyl–tRNA accumulation and growth inhibition in Escherichia coli mutants partly defective in Pth activity: minigene expression and overproduction of a nonfunctional protein. In each case, accumulation of peptidyl–tRNA occurs corresponding to the last sense codon, implying defects in translation termination (6–8).
Pth activity is ubiquitous. Orthologs of the E. coli pth gene have been identified in all known bacterial and some eukaryotic genomes (e.g., yeast, maize, mice, and human), and Pth activities were demonstrated in yeast long ago (9–11). Furthermore, the bacterial pth ortholog YHR189w of Saccharomyces cerevisiae complements a temperature-sensitive E. coli strain harboring a pthts mutant, although YHR189w is not essential for yeast viability (3). Biochemical studies with S. cerevisiae extracts revealed the presence of at least two types of Pth activities, distinguished by the different efficiency of cleaving various substrates (9). A Pth activity in yeast that acts only on acetyl–Phe–tRNA has also been reported (11). Thus, there may be several Pth-like activities in yeast.
Curiously, the known archaeal genomes do not contain any pth orthologs, raising the question whether Pth activity is dispensable for metabolism in these organisms or whether the activity is provided by a nonorthologous enzyme. Therefore we searched for a Pth activity in Methanocaldococcus jannaschii.
Materials and Methods
General.
E. coli lysyl–tRNA synthetase was prepared as described (12). Unfractionated E. coli RNA was purchased from Roche Applied Science. [14C]Lys (318 mCi/mmol) and [14C]Phe (469 mCi/mmol) were from Amersham Pharmacia Biosciences. RNase inhibitor, Complete protease inhibitor mixture tablets, and Expand High Fidelity PCR System were purchased from Roche Diagnostics. E. coli BL21-CodonPlus(DE3)-RIL was from Stratagene. Ni-NTA agarose was from Qiagen (Chatsworth, CA) and the TOPO TA cloning kit (pCR II-Topo Vector) from Invitrogen.
Purification of Pth Activity from M. jannaschii Cells.
Due to the low concentration of a very active protein in M. jannaschii or its high sensitivity to proteolytic cleavage, the purification of the Pth activity included pooling of fractions from different purification attempts. A general purification scheme is indicated below. Frozen M. jannaschii cells (700 g) were resuspended in 500 ml of buffer A (50 mM Tris⋅HCl, pH 8.0/10 mM MgCl2/10 mM 2-mercaptoethanol/2 mM benzamidine-HCl/two Complete protease inhibitor mixture tablets/10% glycerol), sonicated and centrifuged at 100,000 × g for 1 h at 4°C. This cell-free extract (S100) was dialyzed in buffer A and then applied to a DE52 (Whatman) column (35 cm × 20 cm2), preequilibrated with buffer A. The activity eluted in the flowthrough of the column. The active fractions were precipitated by ammonium sulfate (80% wt/vol), resuspended, and dialyzed into buffer B [20 mM potassium phosphate, pH 6.5/10 mM 2-mercaptoethanol/2 mM benzamidine⋅HCl/one Complete protease inhibitor mixture tablet (per 50 ml)/10% glycerol] and applied to a P11 (Whatman) column (20 cm × 20 cm2) preequilibrated with the same buffer. The fractions containing the activity were pooled, concentrated with polyethylene glycol 15,000–20,000 Mr, and dialyzed against buffer B. The dialyzed fractions were applied to a hydroxyl apatite column and eluted with a linear 20–500 mM potassium phosphate (pH 6.5) gradient. The active fractions were pooled, concentrated, dialyzed into fresh buffer B, and reapplied to the column. After elution with a second linear gradient of 20–375 mM phosphate, the active fractions were pooled, concentrated, and dialyzed against buffer C (50 mM sodium phosphate, pH 7.0/0.3 M NaCl/10% glycerol). The sample was then applied to a 320 ml Superdex 75PG column (Amersham Pharmacia Biosciences). The active protein(s) eluted after the 25-kDa standard (chymotrypsinogen A) and just before the 13.7-kDa standard (ribonuclease A).
Peptide Sequencing.
The peak sample was concentrated in polyethylene glycol to 30 μl, mixed with 4× SDS loading buffer, heated to 90°C for 5 min, and applied to 12% SDS/PAGE gel (20 × 20 cm). The gel was stained with Coomassie brilliant blue. A gel piece containing a 27-kDa protein was analyzed by matrix-assisted laser desorption ionization–time of flight analysis by the Procter & Gamble Pharmaceuticals Structural Biology Research Group. Eight peptides from a tryptic digest could be identified covering 62% of M. jannaschii MJ0051 protein (MVVVIR, AVDEWLR, MKMVVVIR, ELIDIYNK, IDKITGHLK, SEGLPCSIIR, VNSEKELIDIYNK, DAGHTQLEPGTLTAVAIGPEK). Four other hypothetical M. jannaschii proteins were also detected in the sample.
Cloning of the M. jannaschi MJ0051 Gene.
Genomic DNA was prepared from M. jannaschii cells by standard methods. The oligonucleotide primers were MJ0051NDE (5′-GAGGTTTAcatatgAAGATGGT-TGTAGTAA-3′) and MJ0051XHO (5′-AATTTActcgagTAAAAGTTTTAAATGTCC-3′). The lowercase letters indicate the NdeI and XhoI restriction sites; the translation initiation site is shown in bold and italics. ORF MJ0051 was amplified by PCR by using the primers above and the Expand High Fidelity PCR System. The PCR product of the expected size (≈345 bp) was gel purified, then cloned by using the TOPO TA cloning kit. Several clones were selected and analyzed by colony PCR by using TaqDNA polymerase and the forward and reverse primers provided in the Invitrogen kit. The MJ0051 gene was subcloned into pET20b (Invitrogen), which adds a C-terminal His-6-tag. The resulting plasmid pGR51C, which encodes the M. jannaschii Pth-His-6 enzyme, was then transformed into E. coli BL21-CodonPlus(DE3)-RIL for expression.
Purification of M. jannaschii Pth-His-6.
The E. coli strain BL21-CodonPlus(DE3)-RIL containing plasmid pGR51C was grown at 32°C in 1 liter of LB containing 0.1 mg/ml ampicillin with moderate shaking. Protein expression was induced by the addition of a final concentration of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the culture reached A600 = 0.6. After 5 h of induction, the culture was chilled on ice, and the cells were harvested by centrifugation. The cell pellet was resuspended in a lysis buffer (20 mM Tris⋅HCl, pH 8.0/500 mM NaCl/10 mM 2-mercaptoethanol/10% glycerol), sonicated, and flocculated at 65°C for 30 min to precipitate the majority of the E. coli proteins. The suspension was clarified by centrifugation at 100,000 × g for 1 h at 4°C, applied to a Ni-NTA agarose column, and washed with 50 ml of lysis buffer containing 50 mM imidazole. MJPth-His-6 was eluted from the column with lysis buffer supplemented with 750 mM imidazole. Fractions containing MJPth-His-6 (judged by SDS/PAGE analysis) were pooled and exchanged by dialysis into storage buffer containing 10 mM Tris⋅HCl, pH 7.2/10 mM 2-mercaptoethanol/50% glycerol, aliquoted, and stored at −72°C. Purity was judged to be >95% by SDS/PAGE analysis.
Preparation of Pth Substrate.
Aminoacylation of 8 A260 units E. coli total tRNA [containing tRNALys (40 pmol/A260) and tRNAPhe (30 pmol/A260)] was performed for 25 min at 37°C in 120 μl of buffer containing 100 mM Hepes⋅KOH, pH 7.0, 15 mM ATP, 10 mM MgCl2, 8 mM DTT, 35 mM KCl, 25 μM of a unlabeled 19-aa mix with either 15 μM [14C]Lys and 10 μM E. coli LysRS or with 10 μM [14C]Phe and a catalytic amount of E. coli S-100 extract. Lys–tRNA or Phe–tRNA was recovered by phenol/chloroform (equilibrated in 25 mM sodium acetate, pH 4.0) extraction and ethanol precipitation. The RNA was recovered by centrifugation and dissolved in 0.6 ml of diethyl pyrocarbonate (DEPC)-treated water. Acetylation of the aminoacyl–tRNA was achieved on ice for 1 h by the addition of 0.8 ml of 4 M potassium acetate, pH 5.0, and five aliquots of 20 μl of acetic anhydride at intervals of 12 min (13). After ethanol precipitation and centrifugation, the pellets were dissolved in 0.2 ml of DEPC-treated water, and the degree of acetylation was determined by the cupric reaction (14). The amount of substrate available for the Pth reaction was determined by spotting a sample onto a 3-mm filter disk (Whatman). The filters were then washed three times for 10–15 min in cold 10% (wt/vol) trichloroacetic acid supplemented with 3% (wt/vol) casamino acids, rinsed in cold 95% ethanol, and dried. The radioactivity on the filters was measured in a scintillation counter.
Pth Assay and Verification of tRNA Integrity After the Pth Reaction.
Reactions were initiated by the addition of 150 μl of a prewarmed (37°C for 5 min) substrate mixture (230 mM Tris⋅HCl, pH 7.2/185 mM NH4Cl/0.70 mM sodium azide/either 227 μM-667 μM diacetyl-[14C]Lys–tRNA or acetyl-[14C]Phe–tRNA) to 300 μl of a reaction mixture [10 mM Tris⋅HCl, pH 7.2/20 mM NH4Cl/10 mM magnesium acetate/60 units of RNase inhibitor/either M. jannaschii S-100 extract (100 μg), M. jannaschii Pth-His-6 (13, 26, and 187 nM) or E. coli Pth (13 and 187 nM)]. The reaction was performed at 37°C. Aliquots (75 μl) were taken at different time points and spotted onto a 3-mm filter disk. The filters were washed, dried, and counted as described above. Recharging of the tRNA (9) was achieved by addition of a reaminoacylation mix containing 15 mM ATP and either 15 μM [14C]Lys and 10 μM E. coli LysRS or 10 μM [14C]Phe and a catalytic amount of E. coli S-100 extract to a Pth reaction aliquot taken after a 10-min incubation before chilling on ice. The tRNA was reaminoacylated at 37°C. The amount of aminoacyl–tRNA formed was quantified as described above.
Plasmid Constructions for Complementation Assay.
Genomic DNA was prepared from M. jannaschii cells by standard methods. The oligonucleotide primers were MJ0051ECO (5′-TTTGAGGTgaattcATGA-AGATGGTTGTAGTAAA-3′) and MJ0051HIN (5′-GATAAATAaagcttTTAAATTTATAAAAGTTTT-AAATC-3′). The lowercase letters indicate the EcoRI and HindIII restriction sites; the translation initiation site is shown in bold and italics. ORF MJ0051 was amplified by PCR by using the primers above and cloned into the TOPO TA cloning vector. The MJ0051 gene was subcloned into pKQV4 and named pGR518 (15). Genomic DNA from S. cerevisiae MHY500 cells (16) was obtained by standard methods. The oligonucleotide primers used were YBL057CEC (5′-TGACAGCGGgaattcATGATAACG-TCCTTTTTAA-3′), YBL057C87 (5′-TTGAGTCCgaattcATGAATGATATACCTGGAGAAG-3′), YBL057CHI (5′-GATGTATATaagcttTGTACCTCAATACAATTTCAAA-3′), YHR189WEC (5′-GGC-AGATAgaattcATGTCCGGTAAATGGAGACC-3′) and YHR189WHI (5′-GGTATTGTTaagcttGATG-GGCTATGAAATGTACTGG-3′). The lowercase letters indicate the EcoRI and HindIII restriction sites; the translation initiation site in bold and italics. ORF YBL057c was amplified by PCR with primers YBL057CEC/YBL057CHI. Deletion of 258 base pairs at the 5′ end and substitution of the TTG codon by ATG of ORF YBL057c were achieved by using primers YBL057C87/YBL57CHI. Primers YHR189WEC and YHR189WHI were used to PCR amplify the ORF YHR189w. The products of the expected sizes (≈645, ≈387, and ≈573 bp, respectively) were gel purified, then cloned by using the TOPO TA cloning kit. Several clones were selected and analyzed by colony PCR. The YBL057c, YBL057c deleted of 258 bp at the 5′ terminus, and YHR189w genes were subcloned into pKQV4 (15), and the constructions were called pGRAS, pGR87L, and pGRES, respectively.
Complementation of E. coli Strain C600 pthts.
Competent E. coli C600 pthts cells (2) were prepared and transformed with the pKQV4 vector containing either the wild-type E. coli pth gene (pGI01; ref. 17) or the M. jannaschii pth2 gene (pGR518), the yeast pth2 ortholog YBL057c (pGRAS); the yeast pth2 ortholog YBL057c with a 5′-terminal 258-bp deletion (pGR87L), or the bacterial pth ortholog YHR189w (pGRES), as well as with pKQV4 as empty vector control. Expression in each construct was controlled by the tac promoter, which is inducible with IPTG. The transformants were plated on LB agar containing 0.1 mg/ml ampicillin and 1 μM IPTG, and duplicate plates were incubated at 32 and 42°C.
Deletion of YHR189w and YBL057c in S. cerevisiae.
Yeast strains Y22883 (BY4743; Mat a/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15D0; ura3Δ0/ura3Δ0; YHR189w:kanMX4/YHR189w) and Y23083 (BY4743; Mat a/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0; YBL057c:kanMX4/YBL057c) were purchased from Research Genetics (Invitrogen) and used to make haploid deletion strains for pth (YHR189w) and pth2 (YBL057c). These haploid strains Δpth (Mat α; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YHR189w:kanMX4) and Δpth2 (Mat a; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YBL057c:kanMX4) were mated to produce the haploid double deletion strain Δpth/Δpth2 (Mat α; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YHR189w:kanMX4; YBL057c:kanMX4) and a haploid wild-type strain pth/pth2 (Mat α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YHR189w; YBL057c). Yeast strain manipulation as well as PCR verification of ORF knockouts was performed essentially as described (18). ORF specific primer pairs for pth and pth2 were designed according to published sequences in the Yeast Deletion Project (www-sequence.stanford.edu/group/yeast_deletion_project). Primers homologous to the KanMX4 cassette [forward (5′-TGATGTTGGACGAGTCGGAATG-3′) and reverse (5′-GTTTCAGAAAC-AACTCTGGC-3′)] were synthesized for verification of knockout alleles.
Phylogenetic Analysis of Pth2 Proteins.
The translated sequence of the M. jannaschii gene MJ0051 (GenBank accession no. NP_247015.1) was used to search the nonredundant protein database at the National Center for Biotechnology Information with the blastp program (Ver. 2.2.4) (19). Homologs (and their GenBank accession or reference nos.) were identified in the archaea Aeropyrum pernix (NP_148083.1), Archaeoglobus fulgidus (NP_070920.1), Ferroplasma acidarmanus (ZP_00001050.1), Halobacterium sp. (NP_444185), Methanobacterium thermoautotrophicum (NP_276808.1), Methanopyrus kandleri (NP_614880.1), Methanosarcina mazei (NP_632136.1), Pyrobaculum aerophilum (NP_558787.1), Pyrococcus abyssi (NP_126312.1), Pyrococcus furiosus (NP_579285.1), Sulfolobus solfataricus (NP_341731.1), Sulfolobus tokodaii (NP_376058.1), and Thermoplasma acidophilum (NP_393586.1). Eukaryal homologs were identified in Anopheles gambiae (EAA03889.1), Arabidopsis thaliana (AAM64624.1), Caenorhabditis elegans (NP_504461.1), Dictyostelium discoideum (AAL92211.1), Drosophila melanogaster (AAF53304.1), Encephalitozoon cuniculi (NP_584755.1), Homo sapiens (NP_057161.1), Neurospora crassa (11359529), Plasmodium falciparum (NP_702728.1) and S. cerevisiae (NP_009496.1). Additional homologs were identified in incomplete genomic sequences from Cryptosporidium parvum (University of Minnesota; www.cbc.umn.edu/ResearchProjects/AGAC/Cp/index.htm), Methanococcoides burtonii (Department of Energy Joint Genome Institute; www.jgi.doe.gov), Giardia lamblia (Marine Biological Laboratory; http://jbpc.mbl.edu/Giardia-HTML/), Haloferax volcanii (Integrated Genomics; www.integratedgenomics.com/), and Methanococcus maripaludis (University of Washington; www.genome.washington.edu/UWGC/Methanococcus).
Amino acid sequences were aligned automatically by using the clustalw program (Ver. 1.82) (20). From the alignment of 29 protein sequences, 116 positions in the conserved catalytic domain were deemed to be confidently aligned. These were analyzed by protein distance methods by using the protdist and neighbor programs (Ver. 3.6a2.1) (21) with the Jones, Taylor, and Thornton model of amino acid changes and a γ distribution of positional rates of change. Bootstrap proportions were calculated by using seqboot, protdist, neighbor, and consense programs (21) to create and evaluate 1,000 resampled alignments. Alternative trees were inferred by the protein maximum likelihood criteria by using the proml program (Ver. 3.6a2.1) (21) and by protein maximum parsimony (paup*, Ver. 4.0b, Sinauer, Sunderland, MA) to analyze the same alignment.
Results
M. jannaschii Extracts Contain Pth Activity.
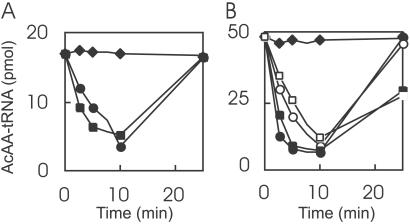
When a cell-free M. jannaschii S-100 extract was incubated with diacetyl-[14C]Lys–tRNA, an analog of the natural peptidyl–tRNA and a standard Pth substrate, a hydrolytic activity was detected (Fig. 1A). Thin-layer chromatographic analysis revealed one of the hydrolysis products to be diacetyl-[14C]lysine (data not shown). The other product, tRNA, could be reaminoacylated to a level similar to that of the initial substrate concentration (Fig. 1A). These results showed that the decrease in acid precipitable radioactivity during the course of the reaction was authentic hydrolysis and not degradation of the tRNA molecule. Thus, M. jannaschii harbors an enzymatic activity similar to that of E. coli Pth.
Fig 1.
Pth activity in M. jannaschii S-100 extract (A) and of the recombinant M. jannaschii Pth-His-6 enzyme (B). (A) The assay was performed at 37°C with M. jannaschii S-100 protein (100 μg, •) or purified E. coli Pth (13 nM, ▪), or without added enzyme (⧫). Each reaction contained 230 nM diacetyl-[14C]Lys–tRNA. An aliquot was taken after 10 min and reaminoacylated, as described in Materials and Methods. (B) The assay was performed at 37°C with 187 nM M. jannaschii MJPth-His-6 (open symbols) or purified E. coli Pth (filled symbols), or without added enzyme (⧫). Each reaction contained diacetyl-[14C]Lys–tRNA (○ and •) or acetyl-[14C]Phe–tRNA (□ and ▪). Aliquots were taken after 10 min and reaminoacylated, as described in Materials and Methods.
Purification of the M. jannaschii Protein Responsible for Pth Activity.
To identify the M. jannaschii gene responsible for the Pth-like activity, the enzyme activity was purified from the S-100 extract by a series of chromatographic steps. Several protein bands were recovered from the SDS gel electrophoretic separation of the most highly purified fraction; they were then analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry. A search of the M. jannaschii genome showed that one protein band (see Materials and Methods) corresponded to MJ0051, a hypothetical protein of unknown function, for which orthologous genes do not exist in bacteria. On the basis of the deduced sequence of 115 amino acids, the protein molecular mass was calculated to be 12.7 kDa. To confirm whether MJ0051 encodes an enzyme with Pth activity, the gene was cloned into pET20b with a C-terminal His-6-tag and transformed into the E. coli BL21- CodonPlus(DE3)-RIL strain for expression. The recombinant MJ0051 protein was purified to near homogeneity on a Ni-NTA column as judged by SDS/PAGE analysis (data not shown). A protein band of 13.5 kDa was identified, which corresponds to the calculated molecular mass of 12.7 kDa of MJ0051 plus the His-6-tag.
The Recombinant MJ0051 Protein Displays Pth Activity in Vitro.
Bacterial Pth enzymes cleave the ester bond between the tRNA and the peptide moiety of peptidyl–tRNA (9, 22, 23). To ascertain that the MJ0051 protein has a similar activity, two Pth substrates, diacetyl-[14C]Lys–tRNA and acetyl-[14C]Phe–tRNA, were tested. The enzyme was able to hydrolyze both substrates in a concentration- and time-dependent manner (Fig. 1B and data not shown). Most importantly, the deacylated tRNALys and tRNAPhe produced by the hydrolysis reaction could be reaminoacylated almost quantitatively, ruling out a nuclease-like activity in the recombinant protein. Therefore, the MJ0051 Pth-catalyzed hydrolysis is the result of a canonical Pth activity. Moreover, the specific activities measured for the M. jannaschii Pth-His-6 and the E. coli Pth proteins were comparable (9, 23–25). We named the protein with this enzymatic activity Pth2.
M. jannaschii pth2 Complements the Temperature-Sensitive pth Mutation of E. coli C600 pthts.
The availability of the E. coli C600 pthts mutant strain (2, 17, 26) provided the opportunity to test whether the archaeal pth2 gene would be active in E. coli and complement the temperature-sensitive phenotype. The E. coli C600 pthts strain was transformed with plasmids carrying either the M. jannaschii pth2 (pGR518) or the E. coli pth gene (pGI01). Each construct expresses pth under the Ptac IPTG inducible promoter and LacIQ repressor. The transformants were grown on LB/Amp/IPTG plates and incubated at 32 or 42°C. Both pth genes, but not the empty vector, were able to rescue growth at 42°C (Table 1). To eliminate the possibility that complementation was due to revertants, the clones were cured; the ampicillin-sensitive cured clones showed the temperature-sensitive phenotype. Therefore, complementation was brought about by the M. jannaschii pth2 gene. This was further confirmed by the ability of the M. jannaschii pth2 gene (in pGR518) to complement an E. coli pth deletion (27) strain (data not shown). Thus, the archaeal pth2 gene is functionally equivalent to the E. coli pth gene.
Table 1.
Complementation of the E. coli pthts mutant with pth and pth2 genes
| Plasmids
|
Gene
|
Incubation temperature, °C | Cured clones incubated at 42°C
|
|
|---|---|---|---|---|
| 32 | 42 | |||
| pKQV4 | None | + | − | ND |
| pGI01 | E. coli pth | + | + | – |
| pGR518 | MJ0051 | + | + | – |
| pGR87L | YBL057c | + | + | ND |
| pGRES | YHR189w | + | + | ND |
E. coli C600 pthts was transformed with plasmids containing the indicated genes, plated on LB plates, and incubated at the specified temperature. +, growth; −, no growth; ND, not determined.
The genes were cloned into pKQV4. The empty vector was a negative control.
The N-terminal 86 amino acids were deleted from the complete ORF.
The pth2 Gene Family Is Conserved in Archaea and Eukarya.
A search of the DNA sequence databases identified pth2 orthologs in all complete archaeal and eukaryal genome sequences. An alignment of Pth2 protein sequences revealed two domains. Most eukaryal homologs possess a highly diverged N-terminal domain that is not found in the archaeal homologs (Fig. 2). Computational analysis suggested that this domain might correspond to a potential transit signal (28), possibly for mitochondrial localization. The second domain, found in all Pth2 sequences, contains the UPF0099 domain of previously unknown function (http://pfam.wustl.edu). This catalytic domain has no significant sequence similarity to the larger (≈190 amino acids) canonical bacterial Pth proteins and lacks the characteristic motifs previously described for the E. coli Pth (27, 29). The very detailed crystal structure of the E. coli Pth (27) does not give any further insight into the archaeal protein.
Fig 2.
Alignment of archaeal-type Pth2 proteins. Sequences from the archaea M. jannaschii, P. furiosus, S. solfataricus, and A. pernix lack the highly diverged N-terminal domain found in eukaryal proteins from S. cerevisiae (YBL057c) and humans. Identical or highly conserved residues are shown in white against a black background, and similar residues are shown in black against a gray background. The beginning and the end of the UPF0099 domain are marked with arrows.
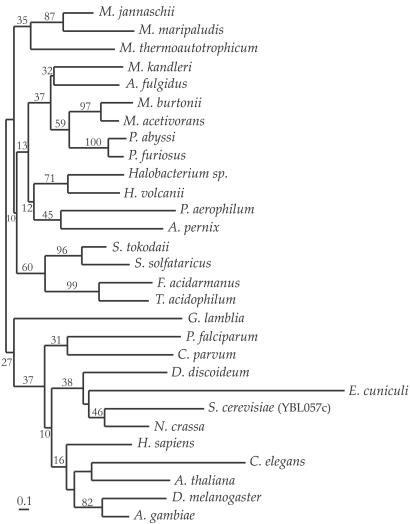
Phylogeny of pth2 Genes in Archaea and Eukarya.
The phylogeny of pth2 genes was inferred from an alignment of the catalytic domain of 29 archaeal and eukaryal protein sequences (Fig. 3). All complete archaeal and eukaryal genomes encode pth2 homologs and the aligned sequences range from 31% to 88% similarity. Despite low bootstrap support in parts of the phylogeny due to the short lengths of Pth2 sequences, the tree resolves separate archaeal and eukaryal lineages. Therefore, the phylogeny suggests that pth2 has been vertically inherited in both domains, and the Pth2 enzyme appears to be closely tied to the archaeal and eukaryal translational apparatus. Phylogenies inferred by alternative methods (protein maximum likelihood or protein parsimony) are substantially consistent with that shown in Fig. 3.
Fig 3.
Phylogeny of archaeal-type Pth2 proteins inferred by using the neighbor-joining method. The tree is rooted by using the eukaryal homologs as an outgroup. Bootstrap percentages are indicated for branches supported by a plurality of bootstrap replicates. (Bar = 10 amino acid replacements per 100 positions.)
At Least Two Genes Encode Pth Activity in S. cerevisiae.
Sequence analysis suggested the S. cerevisiae ORF YBL057c to be the ortholog of M. jannaschii MJ0051 (Fig. 2). To verify that the yeast protein also possesses Pth activity, we checked for the ability of the YBL057c gene to rescue growth of the E. coli C600 pthts strain at 42°C. The intact yeast gene was unable to accomplish this. However, removal of the putative organellar targeting sequence (see above) of the YBL057c gene (the 5′-terminal 258 bp corresponding to the N-terminal 86 amino acids of the Pth2 protein) resulted in a truncated gene that rescued the temperature sensitivity of the E. coli pthts strain (Table 1). This result suggests that the archaeal-type Pth enzyme may be localized in the mitochondrion.
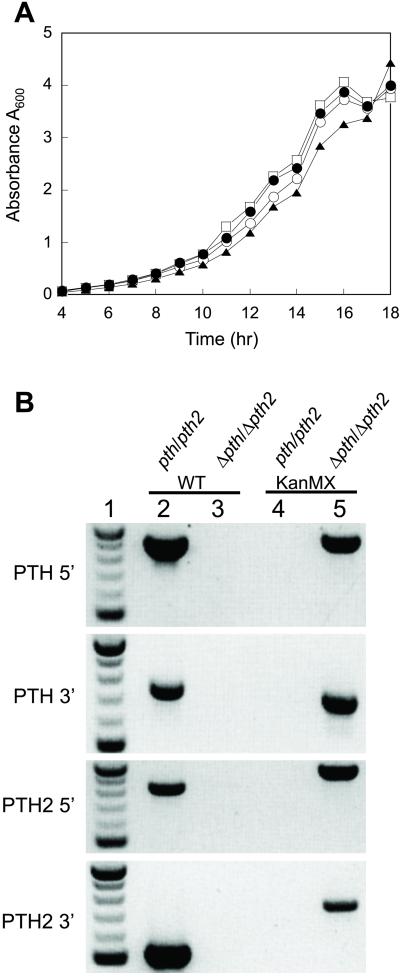
As was shown recently (3), the S. cerevisiae gene YHR189w is orthologous to the E. coli pth gene. We also demonstrated that it was able to support growth at the nonpermissive temperature when transformed into E. coli C600 pthts (Table 1). Therefore, yeast contains at least two potentially functionally active Pths, with the archaeal-type enzyme possibly located in the mitochondrion. We then wanted to ascertain the essentiality of these two enzymes for S. cerevesiae by examining the growth of strains carrying deletions in either pth (YHR189w) or pth2 (YBL057c) or in a double deletion strain. In line with previous reports, the deletion of neither YHR189w (3) nor YBL057c (30) affected the ability of a haploid yeast strain to grow on medium containing glucose as the carbon source. Also, pth and pth2, separately, were shown to be nonessential ORFs in yeast (3, 31, 32). Thus, neither the archaeal- nor the bacterial-type Pth activities are individually required for yeast growth. Guided by these results, we attempted to delete both pth and pth2 simultaneously. To our surprise, the pth/pth2 deletion strain, obtained by crossing two haploid deletion strains, was viable (Fig. 4A). Furthermore, the double deletion strain also did not exhibit slow growth on glycerol (data not shown), a hallmark of mitochondrial dysfunction. PCR analysis (Fig. 4B) confirmed that both the YHR189w and YBL057c genes were deleted in their entirety. Thus, both of these Pth enzymes are dispensable in S. cerevisiae. Furthermore, yeast mitochondria may have an additional mechanism of peptidyl–tRNA hydrolysis, or no need for hydrolysis under these growth conditions. It remains to be seen whether there are additional Pth-like activities in yeast, and whether they are essential for growth.
Fig 4.
(A) Growth of S. cerevisiae strains with pth and pth2 genes deleted. Yeast strains with pth (○), pth2 (•), and pth, pth2 (▴) genes deleted were compared with the growth of the parental wild-type strain (□) at 30°C. (B) Confirmation of the genotype of the Δpth/Δpth2 double deletion strain by PCR. Negative image of 1.5% agarose gels stained with ethidium bromide. Lane 1 contains a DNA size marker in 100-bp increments from 500 bp to 1 kb. Lanes 2 and 4 show colony PCR products of the wild-type (pth/pth2) strain, and lanes 3 and 5 show PCR results for the double deletion strain (Δpth/Δpth2). Each intact or knockout allele is verified at both the 5′ and 3′ ends of the gene by using four unique primers (see Materials and Methods). ORF specific primers (PTH 5′, PTH 3′, PTH2 5′, and PTH2 3′) used in combination with two internal primers for each wild-type ORF (WT) or the KanMX deletion cassette produce a PCR product when the wild-type ORF is intact or deleted, respectively.
Discussion
Is Pth Activity Essential?
Our thinking has been guided by our knowledge of a single essential enzyme present in all bacteria. However, multiple Pth activities, possibly even with different specificities, have long been known in yeast. Given that the eukaryotic cell has more than one protein synthesizing system (e.g., cytosolic and mitochondrial), the presence of multiple enzymes in a eukaryote may not be surprising. That the two canonical Pth enzymes identified so far are not essential for yeast viablity may point to the existence of even greater diversity in Pth activities. It is likely that some eukaryotic specific pth lineage may also exist that is distinct from the other two discovered thus far.
Although Pth is essential for viability of bacteria, there is no a priori reason for the existence of this enzyme. Peptidyl transferase, the enzyme catalyzing peptide bond formation on the ribosome, could possibly carry out this function. Mechanistically, cleavage of peptidyl–tRNA is a reaction competing with the elongation function of peptidyl transferase (reflecting an attack of water vs. aminoacyl–tRNA); it is plausible that an appropriate conformation of the peptidyl transferase could “clean up” peptidyl–tRNAs stalled on the ribosome.
Evolution of pth and pth2 Genes.
Cells have evolved a number of hydrolytic enzymes that augment the capacity of aminoacyl–tRNA synthetases and the ribosome to proofread and salvage misacylated tRNAs (33, 34). One of the most common such enzymes is the canonical Pth protein, which removes toxic peptidyl–tRNA side products. This protein is remarkable both for its selectivity in recognizing peptidyl–tRNA complexes and its lack of specificity for the peptide moiety. The crystal structure of Pth showed it to have the same three-layer α/β/α fold as a zinc-dependent aminopeptidase and purine nucleoside phosphorylases (27). Although these hydrolases have significantly different reaction mechanisms, their shared structure and topology suggests that they evolved from a common ancestor.
A phylogeny inferred from bacterial and eukaryal Pth sequences shows that the gene has been vertically inherited in most lineages and is ubiquitous in bacteria (D.E.G. and D.S., unpublished results). Nevertheless, the genomes of several eukaryotes, including C. elegans, D. melanogaster, and P. falciparum, contain no recognizable pth homologs. Remarkably, homologs in plants and green algae do not share a recent common ancestor with the pth genes of fungi and animals. Although this phylogeny indicates that green algae and plants recruited a pth gene from bacteria, there is no specific relationship between these pth genes and those of cyanobacterial relatives of a chloroplast ancestor. These results, combined with the absence of pth homologs in archaea and some eukaryotes, suggest that several nonhomologous systems manifest Pth activity.
In this paper, we show that the Pth2 enzyme is one such alternative Pth. Parts of the pth2 phylogeny are poorly resolved due to the sequences' short lengths and significant differences in rates of change among amino acid positions. Still, this phylogeny is compatible with phylogenies of the ribosome and other translational apparatus components (35–37). Genetic complementation and deletion experiments show that Pth and Pth2 are functionally interchangeable in E. coli and even dispensable in yeast. Therefore, one would expect the two enzymes to be evolutionarily modular: important for growth, but loosely coupled to the cell's translational apparatus (38). Yet, instead of the horizontal gene transfer events described by phylogenies of many modular genes, both pth and pth2 phylogenies show remarkable evolutionary conservation. This discrepancy suggests that significant unrealized differences exist in the bacterial, archaeal, and eukaryal translation machineries.
Acknowledgments
We thank Alan Curno and Kenneth Greis for peptide sequencing; Constantinos Stathopoulos for help and encouragement; Dmitry Klimenko for advice and technical assistance on the gel filtration column; Alaron Lewis for S. cerevisiae genomic DNA; and Helen Toogood and Debra Tumbula-Hansen for comments on the manuscript. This work was supported by grants from the National Institute of General Medical Sciences (to D.S.), the Department of Energy (to D.S.), the National Aeronautics and Space Administration (to D.S.), the National Science Foundation (to D.E.G.), and Consejo Nacional de Ciencia y Tecnología (CONACyT) (Grants 28401N and 37759N) of México (to G.G.). G.R.-S. was a doctoral loan fellow of CONACyT, and L.R.C.-V. was a postdoctoral loan fellow of Consejo del Sistema Nacional de Educación Tecnológica (México).
Abbreviations
IPTG, isopropyl-β-d-thiogalactopyranoside
Pth, peptidyl–tRNA hydrolase
References
- 1.Menninger J. R. (1979) J. Bacteriol. 137, 694-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.García-Villegas M. R., De La Vega, F. M., Galindo, J. M., Segura, M., Buckingham, R. H. & Guarneros, G. (1991) EMBO J. 10, 3549-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menez J., Buckingham, R. H., de Zamaroczy, M. & Campelli, C. K. (2002) Mol. Microbiol. 45, 123-129. [DOI] [PubMed] [Google Scholar]
- 4.Manley J. L. (1978) J. Mol. Biol. 125, 407-432. [DOI] [PubMed] [Google Scholar]
- 5.Jørgensen F. & Kurland, C. G. (1990) J. Mol. Biol. 215, 511-521. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez-Sanchez J., Valadez, J. G., Herrera, J. V., Ontiveros, C. & Guarneros, G. (1998) EMBO J. 17, 3758-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinçbas V., Heurgue-Hammard, V., Buckingham, R. H., Karimi, R. & Ehrenberg, M. (1999) J. Mol. Biol. 291, 745-759. [DOI] [PubMed] [Google Scholar]
- 8.Menez J., Heurgue-Hamard, V. & Buckingham, R. H. (2000) Nucleic Acids Res. 28, 4725-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kössel H. & RajBhandary, U. L. (1968) J. Mol. Biol. 35, 539-560. [DOI] [PubMed] [Google Scholar]
- 10.Jost J. P. & Bock, R. M. (1969) J. Biol. Chem. 244, 5866-5873. [PubMed] [Google Scholar]
- 11.Coloma A. & Heredia, C. F. (1978) Eur. J. Biochem. 92, 597-603. [DOI] [PubMed] [Google Scholar]
- 12.Brevet A., Chen, J., Leveque, F., Blanquet, S. & Plateau, P. (1995) J. Biol. Chem. 270, 14439-14444. [DOI] [PubMed] [Google Scholar]
- 13.Haenni A. L. & Chapeville, F. (1966) Biochim. Biophys. Acta 114, 135-148. [DOI] [PubMed] [Google Scholar]
- 14.Schofield P. & Zamecnik, P. C. (1968) Biochim. Biophys. Acta 155, 410-416. [DOI] [PubMed] [Google Scholar]
- 15.Strauch M. A., Spiegelman, G. B., Perego, M., Johnson, W. C., Burbulys, D. & Hoch, J. A. (1989) EMBO J. 8, 1615-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen P., Johnson, P., Sommer, T., Jentsch, S. & Hochstrasser, M. (1993) Cell 74, 357-369. [DOI] [PubMed] [Google Scholar]
- 17.Cruz-Vera L. R., Toledo, I., Hernández-Sánchez, J. & Guarneros, G. (2000) J. Bacteriol. 182, 1523-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giaever G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenstein J., (2001) PHYLIP (Phylogeny Inference Package), Ver. 3.6a2.1 (Department of Genetics, Univ. of Washington, Seattle).
- 22.Cuzin F., Kretchmer, N., Greenberg, R. E., Hurwitz, R. & Chapeville, F. (1967) Proc. Natl. Acad. Sci. USA 58, 2079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Groot N., Panet, A. & Lapidot, Y. (1968) Biochem. Biophys. Res. Commun. 31, 37-42. [DOI] [PubMed] [Google Scholar]
- 24.Kössel H. (1970) Biochim. Biophys. Acta 204, 191-202. [DOI] [PubMed] [Google Scholar]
- 25.Menninger J. R., Mulholland, M. C. & Stirewalt, W. S. (1970) Biochim. Biophys. Acta 217, 496-511. [DOI] [PubMed] [Google Scholar]
- 26.Atherly A. G. & Menninger, J. R. (1972) Nat. New Biol. 240, 245-246. [DOI] [PubMed] [Google Scholar]
- 27.Schmitt E., Mechulam, Y., Fromant, M., Plateau, P. & Blanquet, S. (1997) EMBO J. 16, 4760-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen H., Engelbrecht, J., Brunak, S. & von Heijne, G. (1997) Protein Eng. 10, 1-6. [DOI] [PubMed] [Google Scholar]
- 29.De La Vega F. M., Galindo, J. M., Old, I. G. & Guarneros, G. (1996) Gene 169, 97-100. [DOI] [PubMed] [Google Scholar]
- 30.Scherens B., el Bakkoury, M., Vierendeels, F., Dubois, E. & Messenguy, F. (1993) Yeast 9, 1355-1371. [DOI] [PubMed] [Google Scholar]
- 31.Winzeler E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 32.Lucau-Danila A., Wysocki, R., Roganti, T. & Foury, F. (2000) Yeast 16, 547-552. [DOI] [PubMed] [Google Scholar]
- 33.Blanquet S., Mechulam, Y. & Schmitt, E. (2000) Curr. Opin. Struct. Biol. 10, 95-101. [DOI] [PubMed] [Google Scholar]
- 34.Ibba M. & Söll, D. (1999) Science 286, 1893-1897. [DOI] [PubMed] [Google Scholar]
- 35.Woese C. R. (1987) Microbiol. Rev. 51, 221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woese C. R., Olsen, G. J., Ibba, M. & Söll, D. (2000) Microbiol. Mol. Biol. Rev. 64, 202-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansmann S. & Martin, W. (2000) Int. J. Syst. Bacteriol. 50, 1655-1663. [DOI] [PubMed] [Google Scholar]
- 38.Woese C. R. (2000) Proc. Natl. Acad. Sci. USA 97, 8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]