Abstract
Approximately 10% of the U.S. population ingests <50% of the current recommended daily allowance for zinc. We investigate the effect of zinc deficiency on DNA damage, expression of DNA-repair enzymes, and downstream signaling events in a cell-culture model. Low zinc inhibited cell growth of rat glioma C6 cells and increased oxidative stress. Low intracellular zinc increased DNA single-strand breaks (comet assay). Zinc-deficient C6 cells also exhibited an increase in the expression of the zinc-containing DNA-repair proteins p53 and apurinic endonuclease (APE). Repletion with zinc restored cell growth and reversed DNA damage. APE is a multifunctional protein that not only repairs DNA but also controls DNA-binding activity of many transcription factors that may be involved in cancer progression. The ability of the transcription factors p53, nuclear factor κB, and activator protein 1 (AP1) to bind to consensus DNA sequences was decreased markedly with zinc deficiency, as assayed by electrophoretic mobility-shift assays. Thus, low intracellular zinc status causes oxidative DNA damage and induces DNA-repair protein expression, but binding of p53 and important downstream signals leading to proper DNA repair are lost without zinc.
Low intake of vitamins and minerals could be major risk factors for several types of cancer, as suggested by both epidemiological and laboratory studies (1, 2). There is evidence that dietary deficiencies in vitamins and minerals result in single and double DNA-strand breaks and oxidative lesions that are similar to that of radiation-induced DNA damage (3–5). We hypothesize that inadequate nutrition is a significant environmental risk and bears a significant impact on the susceptibility to cancer. A significant portion of the North American population does not get adequate zinc (6). Ten percent of the U.S. population consumes less than half the recommended daily allowance for zinc, especially those who consume little meat and/or consume high phytate-containing food sources (7). Phytate (inositol hexaphosphate), which is found at high levels in cereal grains and legumes, forms a tight complex with zinc (or iron) and decreases its absorption. Marginal zinc intake could increase an individual's susceptibility to developing DNA damage and cancer.
Zinc is a component of >1,000 proteins including DNA-binding proteins with zinc fingers, copper/zinc superoxide dismutase (CuZnSOD), and several proteins involved in DNA-damage repair such as p53, which is mutated in half of human tumors (8). It can be hypothesized that insufficient zinc intake can impair antioxidant defenses and compromise DNA-repair mechanisms, making the cell highly susceptible to oxidative DNA damage. Thus, deficits in zinc intake could have a significant impact on the development of cancer. Epidemiological studies have shown decreased zinc status in cancer patients compared with healthy controls (9–12). It has also been suggested as a contributor to esophageal cancer in humans (9, 13). Zinc deficiency has been shown to cause esophageal tumors in rats (14) and also in conjunction with a single low dose of a nitrosamine (15, 16). Replenishment of zinc in zinc-deficient (ZnDF) rats has been shown to induce apoptosis in esophageal epithelial cells and thereby reduces the development of esophageal cancer (17). Zinc deficiency can also lead to increased oxidative damage to testicular cell DNA (18, 19). Together, these data strongly suggest that zinc deficiency itself may compromise the integrity of DNA.
The brain is highly sensitive to oxidative stress, and zinc deficiency has been linked to abnormalities in brain function. Zinc deficiency impairs cognitive function in experimental animals and humans (20–23). Zinc deficiency during early brain development causes malformations, and deficiency later in development causes microscopic abnormalities and also impairs function (24). In rats, zinc deficiency also can cause oxidant stress and physical breakdown of the blood–brain barrier (25). Thus, it is possible that the brain may be uniquely sensitive to oxidative DNA damage induced by zinc deficiency.
The overall objective of this study was to determine the effects of zinc deficiency on DNA integrity in a neural cell type by using a rat glioma cell line (C6 cells). To achieve this goal, we have examined the effects of low intracellular zinc on oxidative stress and DNA damage in rat glioma C6 cells. In addition, the expression of the DNA-repair protein apurinic endonuclease (APE, which is also known as Ref-1) and downstream signaling events such as activation/binding of p53, activator protein 1 (AP1), and nuclear factor κB (NFκB) were examined.
Materials and Methods
Cell Culture.
C6 rat glioma cells were obtained from the American Tissue Culture Collection (ATTC CCL-107). Cells were grown in DMEM (GIBCO Life Technology, Carlsbad, CA) and 10% CO2 at 37°C. ZnDF medium was prepared by using a chelation strategy. FBS was mixed at 4°C with 10% Chelex-100 overnight. Mineral levels were monitored by inductively coupled plasma spectroscopy. C6 rat glioma cells were seeded in 100-mm plates and grown in control medium (DMEM/10% FBS), zinc-adequate (ZnAD) medium (DMEM/10% Chelex FBS/4 μM ZnCl2) or ZnDF medium (DMEM/10% Chelex FBS). Mediua were replaced every 2–3 days. Cell counts were performed with the use of a Coulter counter.
Mineral Concentration.
Mineral concentrations of calcium, magnesium, iron, copper, and zinc were determined by using inductively coupled plasma/absorption emission spectrometry (Jarell-Ash Thermospec, Franklin, MA) with slight modification of a reported method (26). Briefly, either 1 ml of medium or cell pellets (1 × 107 cells) were incubated with 1 ml of 40% ultrapure nitric acid (VWR Scientific, West Chester, VA) overnight. After incubation, samples were diluted with deionized water to an 8% acid solution and analyzed by inductively coupled plasma/absorption emission spectrometry.
Assessment of DNA Damage.
Single-strand breaks in cells were determined by alkali single-cell gel electrophoresis (comet assay) as described by Singh et al. (27). The assay is based on alkaline lysis of labile DNA at sites of damage (i.e., from oxidation). Cells were suspended in 0.5% agarose and applied to microscope slides. Cells were subsequently lysed in comet assay lysis buffer (Trevigen, Gaithersburg, MD) for 1 h. After lysis, DNA was allowed to unwind in alkali buffer for 20 min before electrophoresis. Nuclear material then was stained with Sybr green (Molecular Probes). Fifty cells from four independent samples were scored blindly for tail migration intensity.
Assessment of Oxidant Production.
(i) Oxidant production was monitored by using the fluorescent probe 2′7′-dichlorodihydrofluorescein diacetate (DCFH, Molecular Probes). DCFH (10 μM) was added to cells after 5 days in control, ZnAD, or ZnDF medium. Cells were incubated for 15 min at 37°C. After this incubation period, cells were washed, trypsinized, and immediately analyzed by using a fluorescence-activated cell sorter (FACSort, Becton Dickinson). (ii) Nitrite formation was used as an indirect measure of nitric oxide (NO) production. Nitrite concentration in media from cells grown for 5 days in control, ZnAD, or ZnDF medium was assessed by using the Griess reagent as described by Hevel and Marletta (28). Samples were normalized against a blank containing DMEM/10% FBS to account for nitrates in the media.
Western Blot Analysis.
One million cells were harvested by trypsinization after 5 days in experimental medium and lysed in a 1.5% SDS lysis buffer. Samples were mixed with Laemmli buffer and boiled for 5 min. SDS electrophoresis was carried out under standard conditions. Protein was transferred from the SDS gel to poly(vinylidene difluoride) membranes (Millipore) at 350 mA for 2 h. Blots were blocked in 5% nonfat dry milk for 1 h at room temperature. Blots then were incubated with APE (Trevigen, 1:500 dilution), p50/p65, or the inhibitory subunit of NFκB (IκBα, Santa Cruz Biotechnology, 1:2,000 dilution). The blots were washed five times with PBS/0.2% Tween 20 and then incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG at a dilution of 1:5,000 for 1 h. Protein was detected using enhanced chemiluminescence reagents (NEN Life Science).
Subcellular Distribution.
Cytosolic and nuclear extracts were prepared from cells by using a Pierce NE-PER nuclear/cytoplasmic extraction kit. Protein concentrations were assessed by using the Bradford method.
AP1-, NFκB-, and p53-Binding Assay.
Binding of AP1, NFκB, and p53 was assessed in nuclear extracts by electrophoretic mobility-shift assay (EMSA). Double-stranded oligonucleotides for AP1, NFκB (Promega), or p53 (Santa Cruz Biotechnology) were labeled on the 3′ end with biotin by using the 3′-Biotin end-label kit from Pierce. EMSA was carried out by using the Lightshift kit from Pierce. Briefly, binding reactions containing 10 μg of nuclear protein, 10 mM Tris, 50 mM KCl, 1 mM DTT, 2.5% glycerol, 5 mM MgCl2, 0.05% Nonidet P-40, and 2 pmol of oligonucleotide probe were incubated for 20 min at room temperature. Specific binding was confirmed by using a 100- to 400-fold excess of unlabeled probe as specific competitor. Protein DNA complexes were separated by using a 6% nondenaturing acrylamide gel electrophoresis. Complexes were transferred to positively charged nylon membranes and UV-crosslinked in a Stratagene crosslinker. Gel shifts were visualized with a streptavidin-horseradish peroxidase followed by chemiluminescent detection.
SOD Activity.
CuZnSOD and manganese SOD activity was determined as described by Fridovich (29). Briefly, cells were lysed in 0.1% Triton X and under two subsequent freeze/rethaw cycles and stored on ice. SOD activity was determined from the percentage inhibition of the cytochrome c, xanthine–xanthine oxidase assay (29). The reduction of cytochrome c by superoxide generated from xanthine and xanthine oxidase is monitored by absorption at 418 nm.
Results
The cell-culture medium FBS was rendered zinc deficient (ZnDF) by chelation with Chelex-100. Chelated medium was replenished to the original level of calcium, which was also removed from medium with Chelex. This ZnDF medium was compared with its control, ZnAD medium, which was also replenished with 4 μM zinc chloride; 4 μM zinc is equivalent to the zinc content found in control medium (DMEM/10% FBS, data not shown). Fig. 1 shows the cellular zinc levels in C6 cells after 5 days in culture in control, ZnAD, or ZnDF medium. A significant drop in cellular zinc is seen in cells fed ZnDF media, with no effect on other divalent metals such as copper or iron.
Fig 1.
Cellular copper, iron, and zinc concentrations in ZnDF C6 cells. Mineral levels were determined by inductively coupled plasma/absorption emission spectrometry as described in Materials and Methods. For each group, cells were fed control-DMEM (Control), ZnAD-DMEM (ZnAD), or ZnDF-DMEM (ZnDF) for a 5-day period. All samples are mean ± SE (n = 6). Significant differences between means were determined by one-way ANOVA. **, P < 0.01.
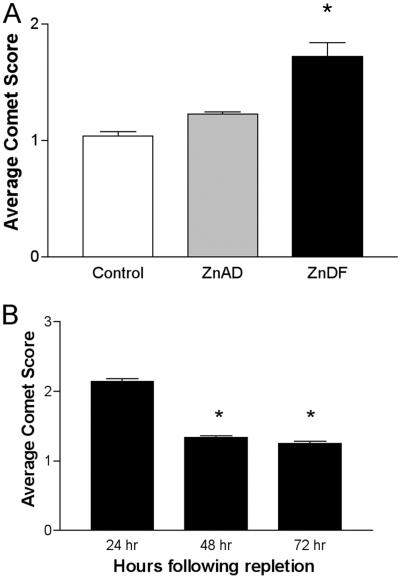
Zinc deficiency had a marked effect on the growth of C6 cells. The rate of growth of C6 cells was inhibited with zinc deficiency (data not shown). DNA damage was assessed by comet assay to determine single-strand breaks in control, ZnAD, or ZnDF C6 cells. As shown in Fig. 2A, zinc deficiency induced an increase in average comet score, indicating an increase in single-strand break damage in ZnDF C6 cells. However, it seems this DNA damage is reversible. If ZnDF cells were replenished with adequate zinc, average comet scores returned back to that of controls within 48 h after repletion (Fig. 2B). Growth and proliferation of cells were also restored with repletion of zinc (data not shown).
Fig 2.
Zinc deficiency induces single-strand breaks in C6 cells. (A) C6 cells were grown in control, ZnAD, or ZnDF medium for 5 days. Each sample is representative of an average mean comet score ± SE of four individual slides per treatment. (B) ZnDF cells were repleted with ZnAD medium. On each slide, 50 comets were scored blindly for tail moment. *, P < 0.05.
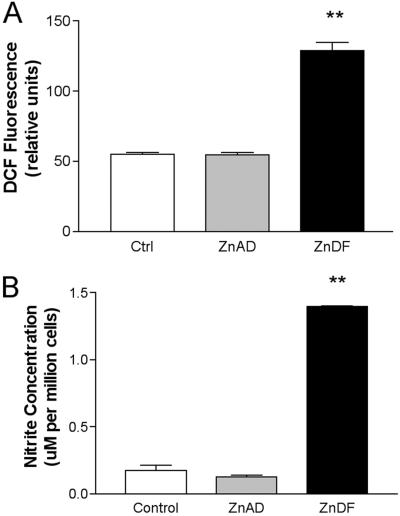
To determine whether zinc deficiency induces an oxidative stress, the nonfluorescent probe DCFH was used. DCFH becomes oxidized to 2′7′-dichlorofluorescein hydrochloride (DCF), a fluorescent product, and indicates increased oxidant production and oxidant stress. Fig. 3A shows a marked increase in DCF fluorescence in ZnDF C6 cells. These data confirm that zinc deficiency increases cellular oxidative stress. Zinc deficiency also seems to cause an increase in reactive nitrogen species such as NO. Nitrite production, a stable end product of NO, was used as an indirect measure of NO production. With zinc deficiency, we see a significant increase in nitrite production, suggesting an increase in NO (Fig. 3B).
Fig 3.
Zinc deficiency increases oxidant production and NO formation in ZnDF C6 cells. (A) DCFH probe was used to monitor oxidant production in C6 cells. DCFH was added to C6 cells grown in control, ZnAD, or ZnDF medium for 5 days. (B) Nitrite production was used as an indirect measure of NO production. Nitrite levels were assessed in media after 5 days in culture in control, ZnAD, or ZnDF medium. **, P < 0.01.
To explore how zinc deficiency affects DNA-repair mechanisms, the expression and function of p53 and APE were determined. Western blot analysis revealed that zinc deficiency induced expression of p53 and APE in C6 cells, confirming that C6 cells may be experiencing an increase in DNA damage (Fig. 4).
Fig 4.
Zinc deficiency increases expression of p53 and APE. p53 and APE expression were determined by Western blot analysis in cells grown in control, ZnAD, or ZnDF medium for 5 days. SDS/PAGE was performed as outlined in Materials and Methods. Blots were stripped and blotted for actin to confirm each protein loading (data not shown). Each bar is representative of mean ± SE (n = 3). *, P < 0.05.
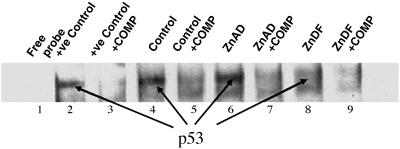
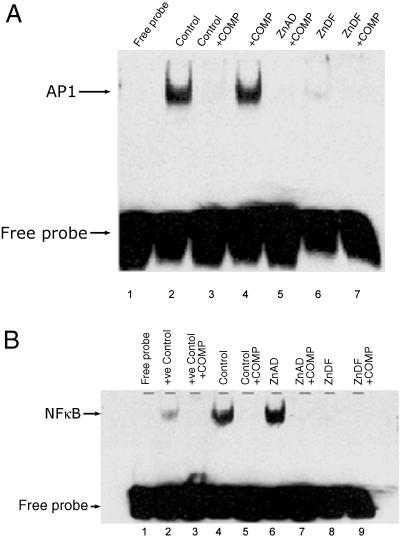
EMSAs were performed to examine the binding of control, ZnAD, and ZnDF nuclear extracts to p53-, AP1-, and NFκB-binding regions. Fig. 5 shows representative films of DNA–p53 complex interactions. Control and ZnAD (lanes 4 and 6) cells demonstrate a clear ability to bind to p53 probes. However, ZnDF cells have an apparent decrease in p53 binding (lane 8). The specificity of p53 binding was confirmed by using an excess of unlabeled oligonucleotide as a specific competitor (lanes 3, 5, 7, and 9). Zinc deficiency has a similar effect on AP1 and NFκB binding (Fig. 6).
Fig 5.
p53 is compromised in ZnDF C6 cells. EMSAs were performed with p53, AP1, or NFκB oligonucleotide probes to examine binding in nuclear extracts from cells grown in control, ZnAD, and ZnDF medium for 5 days. Lane 1 contains a free oligonucleotide probe, and lane 2 contains a positive control using HeLa cells (Promega) that contains active p53 binding. Lanes 3, 5, 7, and 9 represent nuclear extracts incubated with specific competitor (COMP, unlabeled p53 oligonucleotide) to confirm specificity of p53 binding. +ve, positive.
Fig 6.
AP1 and NFκB binding is compromised in ZnDF C6 cells. (A) AP1 binding: lane 1 contains a free oligonucleotide probe. (B) NFκB binding: lane 1 contains a free oligonucleotide probe, and lane 2 contains a positive control using HeLa cells (Promega) that contains active NFκB. Lanes 3, 5, 7, and 9 represent nuclear extracts incubated with unlabeled oligonucleotide (COMP) to confirm specificity of binding. +ve, positive.
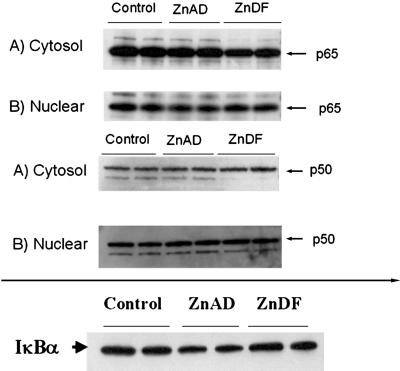
The data from the EMSA experiments suggest that binding of several transcription factors may be impaired with zinc deficiency. However, to confirm that zinc deficiency is truly affecting binding, we examined changes in nuclear translocation and inhibitory-unit degradation. To address these issues, subcellular fractionation and Western blots probing for NFκB subunits were performed. Fig. 7 demonstrates that there is no change in nuclear translocation of the p50 or p65 subunits of NFκB with zinc deficiency compared with ZnAD controls. In addition, Fig. 7 shows that there is no change in expression or degradation of IκBα. These results confirm that zinc deficiency is affecting the binding of these transcription factors with no effect on nuclear translocation or inhibitory-unit degradation.
Fig 7.
Zinc deficiency has no effect on nuclear translocation of NFκB subunits p50 or p65 or IκBα degradation. Western blot analysis was used to determine protein levels of p50, p65, and IκBα in nuclear and cytoplasmic fractions as described in Materials and Methods. SDS/PAGE was performed in nuclear and cytoplasmic fractions from cells grown in control, ZnAD, or ZnDF medium. Blots were stripped and blotted for actin to confirm each protein loading (data not shown). Results are representative of two individual experiments.
Discussion
Zinc deficiency is shown to have a marked effect on the DNA integrity of a neural-derived cell line (Figs. 2–4). The mechanism by which zinc deficiency induces DNA damage in these cells is most likely a combined effect of increased oxidative stress and an impairment of DNA-repair signal pathways (Figs. 5–7). Together, these disruptions make the cell highly susceptible to oxidative DNA damage. C6 cells showed a unique sensitivity to zinc deficiency. In response to low zinc, C6 cells showed decreased proliferation, increased oxidative stress (Fig. 3), and increased single-strand breaks (Fig. 2). C6 cells showed the most marked response to zinc deficiency when compared with other cell lines including human lung fibroblasts (IMR90), small airway epithelial cells, prostate epithelial cells, and cancerous prostate epithelial cells (PC3). In all these other cell types, cell proliferation was not impaired severely. In addition, the degree of oxidative stress and DNA damage was not as severe as that of found in ZnDF C6 cells (data not shown). These results suggest that neural-derived cells may be more sensitive to the effects of oxidative stress.
The antioxidant function of zinc and ability of zinc to induce oxidative stress has been shown by others. However, the actual source of these oxidants still remains unknown. Zinc deficiency induces both reactive oxygen species (increased DCF fluorescence) and reactive nitrogen species (increased nitrite) in C6 cells (Fig. 3). The increase in oxidative stress cannot be explained by a loss in CuZnSOD activity. Instead, CuZnSOD activity is induced slightly rather than decreased with zinc deficiency (data not shown). Thus, the mechanisms by which zinc deficiency induces oxidative stress is unclear. One possibility is that zinc deficiency may impair mitochondrial function, causing excess free radical species to be leaked through the electron transport chain. In other studies looking at human lung fibroblasts, we have found using microarrays that several subunits involved in the electron transport chain are down-regulated with zinc deficiency (30). Another possibility is that with zinc deficiency, the CuZnSOD has a toxic “gain of function.” In amyotrophic lateral sclerosis (ALS), a small proportion of this disease is due to a mutation in CuZnSOD that causes the protein to lose zinc (31–33). In response to this loss, the CuZnSOD retains its WT dismutase activity but seems to gain a peroxidase activity and increases the production of the highly reactive peroxynitrite molecule. Excess production of peroxynitrite ultimately leads to the death of the motor neurons. It is possible that dietary zinc deficiency is affecting these C6 cells in the same manner.
An increase in DCF fluorescence (Fig. 3) suggests that there is an increase in oxidative stress with zinc deficiency. Oxidative stress may play a significant role in the pathology associated with zinc deficiency; oxidative base modifications are highly prevalent with zinc deficiency and could lead to the single-strand breaks seen in Fig. 2A. This increase in oxidative base modifications should increase the likelihood of mutations. Other studies have also shown an increase in DNA damage with lowered zinc status (34). In infant rhesus monkeys, maternal dietary zinc deficiency increases 8-hydroxy-2′-deoxyguanosine levels in infant liver (35). An increase in levels of 8-oxo-2′-deoxyguanosine with zinc deficiency has also been demonstrated in rat testes (36). In ZnDF rats, there is an increase in h-ras and p53 mutations in response to a normally noncarcinogenic dose of N-nitrosomethylbenzylamine (16).
One major contributing factor to the increase in DNA damage with zinc deficiency could be an impairment of DNA repair. Replenishing the ZnDF C6 cells with zinc rapidly reversed DNA damage in these cells (Fig. 3). Therefore, it can be hypothesized that repair mechanisms were “turned back on” with the addition of zinc. However, it is also possible that damaged cells were replaced with new, undamaged cells, because repletion also restored cell growth. To examine the effects of zinc deficiency on DNA repair, we examined several important candidate DNA-repair proteins. The tumor-suppressor protein p53 plays an important role in coordinating events leading to appropriate DNA repair. p53 plays a role in modulating cell-cycle progression, apoptosis, DNA repair, and cell proliferation/differentiation (37, 38). Over 50% of human malignancies contain a mutation in p53 (39). Interestingly, the majority of these mutations are found in the region of the gene that encodes for the DNA-binding region of p53 (40, 41). This binding region contains zinc, and to coordinate the events related to DNA repair, p53 must be able to bind to specific DNA-binding domains to transcriptionally activate downstream targets involved in DNA repair. To test the ability of p53 to function properly and bind to downstream targets with zinc deficiency, EMSAs were performed. We can detect an increase in p53 expression (Fig. 4), yet Fig. 5 shows a marked decrease in the ability of ZnDF nuclear extracts to bind to consensus p53 oligonucleotides. In other experiments, we also showed an increase in p53 protein expression with zinc deficiency (30). This up-regulation of p53 expression is most likely in response to DNA damage induced with zinc deficiency. Thus, although there is an increase in p53 expression with zinc deficiency, this p53 is dysfunctional, and hence DNA repair is compromised.
The DNA-binding activity of p53 is largely mediated by a conformation-sensitive structure in the central portion of the protein (residues 102–292) (41). Mutations in this region cause an “unfolding” of this structure and a loss of binding activity. Other researchers have found also that the removal of zinc, either by chemical chelation or feeding ZnDF medium, alters the expression of p53 (42, 43). Direct chemical chelation also seems to reversibly alter p53 conformation, with the loss of DNA-binding activity (44). The finding that dietary zinc deficiency in cells increases p53 protein levels (Fig. 4) but also compromises p53 DNA-binding activity (Fig. 5) is noteworthy, because it shows that p53 can be disabled by poor nutrition as well as through a mutation.
To examine other DNA-repair pathways, we also investigated the expression of APE, an important endonuclease in base-excision repair (45). DNA base-excision repair is a major pathway responsible for the repair of both cellular alkylation and oxidative DNA damage. A critical step in this pathway involves the cleavage of damaged sites in DNA by APE. APE is a multifunctional protein that not only repairs AP sites but also controls DNA-binding activity, via redox mechanisms, of numerous transcription factors that are involved in cancer promotion and progression (such as AP-1, NFκB, and p53) (46). In addition, APE levels appear elevated in a number of cancers (47–49). Zinc deficiency increases the expression of APE in C6 cells, most likely in response to DNA damage induced by zinc deficiency (Fig. 4). Increases in APE expression are expected to be followed by increases in the binding activity of several redox-sensitive transcription factors such as NFκB and AP1. However, this does not hold true in ZnDF C6 cells. Instead, we see a marked reduction in both NFκB and AP1 binding with zinc deficiency (Fig. 6). These transcription factors play important roles in controlling oxidative stress response and cell proliferation (50, 51), and their inactivation of binding impairs the ability of the cell to respond to oxidative stress and damage. Mackenzie et al. (52) have also demonstrated recently alterations in NFκB activation with low intracellular zinc in human neuroblastoma IMR32 cells. However, the precise mechanisms by which zinc affects transcription factor binding remain unclear. The direct influence of zinc deficiency on zinc-finger binding and/or oxidation of critical cysteine groups needs to be explored.
It is well known that exposure to environmental stresses such as ionizing radiation and carcinogenic chemicals can result in significant damage to DNA. There is now increasing evidence that vitamin and mineral deficiencies, such as zinc deficiency, can also damage DNA by the same mechanisms (1). Approximately 10% of the U.S. population ingests <50% of the recommended daily allowance for zinc and are at risk at for marginal zinc deficiency because of low consumption of zinc and/or high phytate intake (1). This study confirms that low intracellular zinc induces oxidative stresses but at the same time compromises the ability of the cell to deal with this stress. Zinc deficiency renders the cell highly susceptible to oxidative DNA damage by both inducing oxidative stresses and impairing DNA-repair mechanisms. Thus, zinc deficiency has a highly detrimental effect on DNA integrity and emphasizes the importance of good nutrition in the prevention of cancer.
Acknowledgments
This work was supported by National Institutes of Health Training Grant 5 T32 ES07075 to E.H. and National Foundation for Cancer Research Grant 00-CHORI, Wheeler Fund for the Biological Sciences at the University of California, Berkeley, the Ellison Medical Foundation Grant SS-042-99, and National Institute of Environmental Health Sciences Center Grant P30 ES01896 to B.N.A.
Abbreviations
SOD, superoxide dismutase
CuZnSOD, copper/zinc SOD
ZnDF, zinc-deficient
APE, apurinic endonuclease
AP1, activator protein 1
NFκB, nuclear factor κB
ZnAD, zinc-adequate
DCFH, 2′7′-dichlorodihydrofluorescein diacetate
DCF, 2′7′-dichlorofluorescein hydrochloride
IκBα, the inhibitory subunit of NFκB
EMSA, electrophoretic mobility-shift assay
References
- 1.Ames B. N. & Wakimoto, P. (2002) Nat. Rev. Cancer 2, 694-704. [DOI] [PubMed] [Google Scholar]
- 2.Ames B. N. (2001) Mutat. Res. 475, 7-20. [DOI] [PubMed] [Google Scholar]
- 3.Ames B. N. & Gold, L. S. (2000) Mutat. Res. 447, 3-13. [DOI] [PubMed] [Google Scholar]
- 4.Blount B. C., Mack, M. M., Wehr, C. M., MacGregor, J. T., Hiatt, R. A., Wang, G., Wickramasinghe, S. N., Everson, R. B. & Ames, B. N. (1997) Proc. Natl. Acad. Sci. USA 94, 3290-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott R. L., Elliott, M. C., Wang, F. & Head, J. F. (1993) Ann. N.Y. Acad. Sci. 698, 159-166. [DOI] [PubMed] [Google Scholar]
- 6.King J. C., Shames, D. M. & Woodhouse, L. R. (2000) J. Nutr. 130, 1360S-1366S. [DOI] [PubMed] [Google Scholar]
- 7.Wakimoto, P. & Block, G. (2001) J. Gerontol. A Biol. Sci. Med. Sci. 56 (Special no. 2), 65–80. [DOI] [PubMed]
- 8.Walsh C. T., Sandstead, H. H., Prasad, A. S., Newberne, P. M. & Fraker, P. J. (1994) Environ. Health Perspect. 102, Suppl. 2, 5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z. F., Kurtz, R. C., Yu, G. P., Sun, M., Gargon, N., Karpeh, M., Jr., Fein, J. S. & Harlap, S. (1997) Nutr. Cancer 27, 298-309. [DOI] [PubMed] [Google Scholar]
- 10.Prasad A. S., Beck, F. W., Doerr, T. D., Shamsa, F. H., Penny, H. S., Marks, S. C., Kaplan, J., Kucuk, O. & Mathog, R. H. (1998) J. Am. Coll. Nutr. 17, 409-418. [DOI] [PubMed] [Google Scholar]
- 11.Federico A., Iodice, P., Federico, P., Del Rio, A., Mellone, M. C. & Catalano, G. (2001) Eur. J. Clin. Nutr. 55, 293-297. [DOI] [PubMed] [Google Scholar]
- 12.Doerr T. D., Marks, S. C., Shamsa, F. H., Mathog, R. H. & Prasad, A. S. (1998) Nutrition 14, 489-495. [DOI] [PubMed] [Google Scholar]
- 13.Lipman T. O., Diamond, A., Mellow, M. H. & Patterson, K. Y. (1987) J. Am. Coll. Nutr. 6, 41-46. [DOI] [PubMed] [Google Scholar]
- 14.Newberne P. M., Broitman, S. & Schrager, T. F. (1997) Pathobiology 65, 253-263. [DOI] [PubMed] [Google Scholar]
- 15.Fong L. Y. & Magee, P. N. (1999) Cancer Lett. (Shannon, Irel.) 143, 63-69. [DOI] [PubMed] [Google Scholar]
- 16.Fong L. Y., Lau, K. M., Huebner, K. & Magee, P. N. (1997) Carcinogenesis 18, 1477-1484. [DOI] [PubMed] [Google Scholar]
- 17.Fong L. Y., Nguyen, V. T. & Farber, J. L. (2001) J. Natl. Cancer Inst. 93, 1525-1533. [DOI] [PubMed] [Google Scholar]
- 18.Oteiza P. L., Olin, K. L., Fraga, C. G. & Keen, C. L. (1996) Proc. Soc. Exp. Biol. Med. 213, 85-91. [DOI] [PubMed] [Google Scholar]
- 19.Oteiza P. I., Clegg, M. S., Zago, M. P. & Keen, C. L. (2000) Free Radical Biol. Med. 28, 1091-1099. [DOI] [PubMed] [Google Scholar]
- 20.Hansen C. R., Malecha, M., Mackenzie, T. B. & Kroll, J. (1983) Biol. Psychiatry 18, 395-401. [PubMed] [Google Scholar]
- 21.Black M. M. (1998) Am. J. Clin. Nutr. 68, 464S-469S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandstead H. H., Frederickson, C. J. & Penland, J. G. (2000) J. Nutr. 130, 496S-502S. [DOI] [PubMed] [Google Scholar]
- 23.Salgueiro M. J., Zubillaga, M. B., Lysionek, A. E., Caro, R. A., Weill, R. & Boccio, J. R. (2002) Nutrition 18, 510-519. [DOI] [PubMed] [Google Scholar]
- 24.Bhatnagar S. & Taneja, S. (2001) Br. J. Nutr. 85, Suppl. 2, S139-S145. [DOI] [PubMed] [Google Scholar]
- 25.Noseworthy M. D. & Bray, T. M. (2000) Proc. Soc. Exp. Biol. Med. 223, 175-182. [DOI] [PubMed] [Google Scholar]
- 26.Verbanac D., Milin, C., Domitrovic, R., Giacometti, J., Pantovic, R. & Ciganj, Z. (1997) Biol. Trace Elem. Res. 57, 91-96. [DOI] [PubMed] [Google Scholar]
- 27.Singh N. P., McCoy, M. T., Tice, R. R. & Schneider, E. L. (1988) Exp. Cell. Res. 175, 184-191. [DOI] [PubMed] [Google Scholar]
- 28.Hevel J. M. & Marletta, M. A. (1994) Methods Enzymol. 233, 250-258. [DOI] [PubMed] [Google Scholar]
- 29.Fridovich I. (1970) J. Biol. Chem. 245, 4053-4057. [PubMed] [Google Scholar]
- 30.Ho E., Courtemanche, C., Killilea, D. K. & Ames, B. N. (2001) J. Trace Elem. Exp. Med. 14, 282-283. [Google Scholar]
- 31.Beckman J. S., Carson, M., Smith, C. D. & Koppenol, W. H. (1993) Nature 364, 584. [DOI] [PubMed] [Google Scholar]
- 32.Crow J. P., Sampson, J. B., Zhuang, Y., Thompson, J. A. & Beckman, J. S. (1997) J. Neurochem. 69, 1936-1944. [DOI] [PubMed] [Google Scholar]
- 33.Beckman J. S., Estevez, A. G., Crow, J. P. & Barbeito, L. (2001) Trends Neurosci. 24, S15-S20. [DOI] [PubMed] [Google Scholar]
- 34.Powell S. R. (2000) J. Nutr. 130, 1447S-1454S. [DOI] [PubMed] [Google Scholar]
- 35.Olin K. L., Shigenaga, M. K., Ames, B. N., Golub, M. S., Gershwin, M. E., Hendrickx, A. G. & Keen, C. L. (1993) Proc. Soc. Exp. Biol. Med. 203, 461-466. [DOI] [PubMed] [Google Scholar]
- 36.Oteiza P. I., Olin, K. L., Fraga, C. G. & Keen, C. L. (1995) J. Nutr. 125, 823-829. [DOI] [PubMed] [Google Scholar]
- 37.Levine A. J. (1997) Cell 88, 323-331. [DOI] [PubMed] [Google Scholar]
- 38.Lane D. P. (1992) Nature 358, 15-16. [DOI] [PubMed] [Google Scholar]
- 39.Hollstein M., Sidransky, D., Vogelstein, B. & Harris, C. C. (1991) Science 253, 49-53. [DOI] [PubMed] [Google Scholar]
- 40.Cho Y., Gorina, S., Jeffrey, P. D. & Pavletich, N. P. (1994) Science 265, 346-355. [DOI] [PubMed] [Google Scholar]
- 41.Pavletich N. P., Chambers, K. A. & Pabo, C. O. (1993) Genes Dev. 7, 2556-2564. [DOI] [PubMed] [Google Scholar]
- 42.Fanzo J. C., Reaves, S. K., Cui, L., Zhu, L., Wu, J. Y., Wang, Y. R. & Lei, K. Y. (2001) Am. J. Physiol. 281, C751-C757. [DOI] [PubMed] [Google Scholar]
- 43.Meplan C., Richard, M. J. & Hainaut, P. (2000) Oncogene 19, 5227-5236. [DOI] [PubMed] [Google Scholar]
- 44.Hainaut P. & Mann, K. (2001) Antioxid. Redox Signal. 3, 611-623. [DOI] [PubMed] [Google Scholar]
- 45.Fritz G. (2000) Int. J. Biochem. Cell Biol. 32, 925-929. [DOI] [PubMed] [Google Scholar]
- 46.Evans A. R., Limp-Foster, M. & Kelley, M. R. (2000) Mutat. Res. 461, 83-108. [DOI] [PubMed] [Google Scholar]
- 47.Xu Y., Moore, D. H., Broshears, J., Liu, L., Wilson, T. M. & Kelley, M. R. (1997) Anticancer Res. 17, 3713-3719. [PubMed] [Google Scholar]
- 48.Thomson B., Tritt, R., Davis, M. & Kelley, M. R. (2001) J. Pediatr. Hematol. Oncol. 23, 234-239. [DOI] [PubMed] [Google Scholar]
- 49.Puglisi F., Barbone, F., Tell, G., Aprile, G., Pertoldi, B., Raiti, C., Kelley, M. R., Damante, G., Sobrero, A., Beltrami, C. A. & Di Loreto, C. (2002) Oncol. Rep. 9, 11-17. [PubMed] [Google Scholar]
- 50.Shaulian E. & Karin, M. (2001) Oncogene 20, 2390-2400. [DOI] [PubMed] [Google Scholar]
- 51.Siebenlist U., Franzoso, G. & Brown, K. (1994) Annu. Rev. Cell Biol. 10, 405-455. [DOI] [PubMed] [Google Scholar]
- 52.Mackenzie G. G., Zago, M. P., Keen, C. L. & Oteiza, P. I. (2002) J. Biol. Chem. 13, 34610-34617. [DOI] [PubMed] [Google Scholar]