Abstract
We investigated the effect of actin filament barbed end uncapping on Arp2/3 complex function both in vivo and in vitro. Arp2/3 complex redistributes rapidly and uniformly to the lamellar edge of activated wild-type platelets and fibroblasts but clusters in marginal actin filament clumps in gelsolin-null cells. Treatment of gelsolin-null platelets with the negative dominant N-WASp C-terminal CA domain has no effect on their residual actin nucleation activity, placing gelsolin actin filament severing, capping, and uncapping function upstream of Arp2/3 complex nucleation. Actin filaments capped by gelsolin or the gelsolin homolog CapG fail to enhance Arp2/3 complex nucleation in vitro, but uncapping of the barbed ends of these actin filaments restores their ability to potentiate Arp2/3 complex nucleation. We conclude that Arp2/3 complex contribution to actin filament nucleation in platelets and fibroblasts importantly requires free barbed ends generated by severing and uncapping.
The elongation of polar actin filaments in the fast-growing (barbed-end) direction leads to membrane protrusion and crawling movements. The seminal studies of Oosawa and Kasai established that actin filament oligomers originate slowly from actin monomers after the addition of appropriate salts but that preexisting actin filaments with free barbed ends are powerful nucleation sites for filament elongation (1). Because approximately half of the actin in living cells is filamentous, theoretically many barbed ends are available as nucleation sites. A resting human blood platelet, for example, has ≈40% of its ≈550 μM actin assembled into 2,000–5,000 filaments (2). Blockade of these numerous barbed ends of actin filaments by abundant capping proteins explains the low level of barbed-end elongation documented in such resting cells.
Investigators now have proposed three mechanisms for initiating actin assembly after cell stimulation (3). In one, signaling intermediates lead to removal of barbed-end severing and capping proteins. In another, severing of actin filaments by proteins of the ADF/cofilin family exposes free barbed ends. The third proposal depends only on the activation of Arp2/3 complex (3). According to this model, activated Arp2/3 complex binds to and nucleates from the sides of actin filaments, where it also promotes de novo branching nucleation in the barbed-end direction, eliminating the need for free barbed ends (4). Some, however, have argued that Arp2/3 complex nucleates best off preexisting barbed ends, formed by uncapping or by spontaneous nucleation, a concept incompatible with “de novo” nucleation (5, 6). The ideas concerning Arp2/3 complex function derive from in vitro systems that use purified proteins, and investigators have not yet begun to characterize how Arp2/3 complex interacts with other actin regulatory proteins found in abundance at the leading edge.
We previously reported that the actin filament severing and capping protein gelsolin is a necessary downstream effector of Rac for extension of lamellae and cell motility (7–9). A large fraction of the actin nucleation activity of the blood platelet derives from the severing of the actin filaments by Ca2+-activated gelsolin and the subsequent uncapping of their barbed ends by membrane polyphosphoinositides downstream of Rac (2, 8). As a consequence, gelsolin-null platelets polymerize actin and spread poorly after stimulation of their thrombin and collagen receptors (7, 10, 11), as well as after chilling (12), a phenotype that results in vivo in a prolonged bleeding time (7). Similarly, gelsolin deficiency is associated with a markedly diminished, Rac-mediated ruffling response, locomotion, and actin assembly in fibroblasts (9, 13). Furthermore, gelsolin is required for optimal actin turnover in several different cell types, such as fibroblasts, Sertoli cells, and dendritic spines (13–15).
We provide here both in vivo and in vitro evidence for a critical role of gelsolin and free barbed ends for the regulation of Arp2/3 complex nucleation. Hence, Arp2/3 complex amplifies actin nucleation when gelsolin or CapG dissociates from the barbed ends of actin filaments. Severing and uncapping provides a mechanism that allows Arp2/3 complex to rapidly build off filament networks that already are formed at the leading edge.
Methods
Reagents.
The thrombin receptor PAR-1-activating peptide (TRAP; amino acids SFLLRNPNDKYEPF) was purchased from Bachem. The collagen-related peptide (CRP) specific for the collagen receptor GPVI was a gift from Michael J. Barnes, Richard W. Farndale, and C. Graham Knight (Department of Biochemistry, University of Cambridge, Cambridge, U.K.) (16). Polyclonal antibodies directed against the Arp3 and p34-Arc subunits of Arp2/3 complex were a gift from Matthew D. Welch (Department of Molecular and Cell Biology, University of California, Berkeley). Vectors encoding the GST fusion proteins containing the C-terminal VCA (GST-VCA; amino acids 392–505) and CA (GST-CA; amino acids 450–505) domains of N-WASp were a gift from Rajat Rohatgi and Marc W. Kirschner (Department of Cell Biology, Harvard Medical School, Boston). Arp2/3 complex was a gift from Fumihiko Nakamura (Division of Hematology, Brigham and Women's Hospital, Boston) (17). CapG was purified as described (18). Recombinant human plasma gelsolin was a gift from Biogen. Rabbit skeletal muscle actin was isolated and labeled with pyrene as described (2). All other reagents were also of the highest purity available (Sigma and Bio-Rad).
Platelet Preparation.
Blood was collected in 0.1 vol of Aster–Jandl anticoagulant from healthy volunteers by venous puncture and from wild-type and gelsolin-null (7) mice by retroorbital plexus bleeding. Platelets were isolated from platelet-rich plasma by using a metrizamide gradient (11).
Fibroblast Culture.
Lung fibroblasts were provided by Eric Osborn (Division of Hematology, Brigham and Women's Hospital). They were isolated from the lungs of 1- to 2-month-old mice and were maintained in DMEM (GIBCO) supplemented with 10% FCS. Cultured cells were used for all experiments by the second culture passage.
Immunogold Labeling.
Platelets were attached to coverslips coated previously with poly-L-lysine or 30 μg/ml anti-GPIbα IgGs in Tris-buffered saline. Platelet cytoskeletons were extracted with 0.75% Triton X-100 in PHEM buffer as described previously (19). Cytoskeletons of resting platelets were attached to glass coverslips by centrifugation in 0.75% Triton X-100 in PHEM buffer containing 0.1% glutaraldehyde. Cytoskeletons were fixed with 1% glutaraldehyde in PHEM buffer for 10 min, and unreacted aldehydes were blocked with 1 mg/ml sodium borohydride in PHEM for 1 min, treated with 0.1% SDS in PHEM for 1 min, and washed in 1% BSA in PBS, pH 8.2. Cytoskeletons were labeled with a mixture of rabbit anti-Arp3 and anti-p34-Arc antibodies, each at 4 μg/ml, washed with PBS/1% BSA, pH 8.2, and incubated with 10 nm of colloidal gold coated with goat anti-rabbit IgGs. Controls omitted the primary antibodies. Coverslips were washed, fixed with 1% glutaraldehyde, and prepared for electron microscopy as described (2). Wild-type and gelsolin-null mouse fibroblasts were grown on coverslips coated previously with 0.1% gelatin. After 24 h, cells were starved overnight in DMEM. Cells were stimulated or not with 20 ng/ml epidermal growth factor (EGF) for 30 min in DMEM. Cytoskeletons were fixed, prepared, and labeled as described above.
Immunofluorescence Analysis.
Platelets were centrifuged at 280 × g for 5 min onto poly-L-lysine-coated coverslips, fixed in 4% formaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 0.5% BSA in PBS. Arp2/3 complex sites were exposed by treating the fixed and blocked cytoskeletons with 0.1% SDS for 1 min. Specimens were washed in PBS and labeled with a mixture of rabbit anti-Arp3 and anti-p34-Arc antibodies, each at 4 μg/ml. Images were obtained by using a Zeiss Axiovert S100 microscope with a ×100 oil-immersion objective. The peptides used to raise the antibodies were used mixed with the anti-Arp2/3 complex antibodies as controls, each at 50 μg/ml, and blocked all detectable antibody binding (20).
SDS/PAGE and Immunoblot Analysis.
Platelets and fibroblasts were lysed with 0.1% Triton X-100 in PHEM buffer. Actin cytoskeletons were isolated by centrifugation at 100,000 × g for 30 min at 4°C in a Beckman Optima TL ultracentrifuge (Beckman Coulter). Triton X-100-insoluble and -soluble fractions were separated on 10% polyacrylamide gels, transferred onto an Immobilon-P membrane (Millipore), and probed with rabbit antibodies directed against the Arp3 and p34-Arc subunits of Arp2/3 complex as described (20).
Measurement of Actin Filament Ends in Permeabilized Cells.
Platelets were permeabilized with 0.25% octyl glucoside (OG) in PHEM buffer, and GST-CA was added just before the addition of agonist. The polymerization rate assay started by the addition of 185 μl of 100 mM KCl/2 mM MgCl2/0.5 mM ATP/0.1 mM EGTA/0.5 mM DTT/10 mM Tris, pH 7.4 (nucleation buffer), to 100 μl of platelet extract and the subsequent addition of 15 μl of 20 μM monomeric pyrene-labeled rabbit skeletal muscle actin. Pyrene-actin fluorescence was recorded by using a LS50B spectrofluorimeter (Perkin–Elmer). Excitation and emission wavelengths were 366 and 386 nm, respectively. The number of actin filament ends was determined as described (8). Wild-type and gelsolin-null mouse fibroblasts were grown on borosilicate glass culture tubes coated previously with 0.1% gelatin. After 24 h, cells were starved overnight in DMEM. Cells were stimulated with 20 ng/ml EGF for 30 min. Cells were extracted with 0.1% Triton X-100 in PHEM buffer, and the polymerization rate assay was started by the addition of 1 μM pyrene-labeled actin in nucleation buffer.
Actin Polymerization Assay with Purified Proteins.
To determine the effect of barbed-end uncapping on Arp2/3 complex activity, gelsolin- and CapG-actin filament seeds were prepared by mixing 500 nM of each capping protein to 5 μM monomeric actin in nucleation buffer in the presence of 10 μM CaCl2, instead of EGTA. Polymerization was initiated by the addition of 1 μM pyrene-labeled actin to 10 nM Arp2/3 complex/50 nM GST-VCA in nucleation buffer in the presence or absence of 2 nM CapG- or 2 nM gelsolin-capped filaments. Experiments were performed in 10 μM CaCl2 or 100 μM EGTA to release CapG from the filament ends. To determine whether actin filaments nucleated by gelsolin regulate Arp2/3 complex activity, polymerization was initiated by the addition of 1 μM pyrene-labeled actin to 50 nM Arp2/3 complex/200 nM GST-VCA/10 μM CaCl2 with or without 5 nM gelsolin in nucleation buffer in the presence of 0.5 mM of the nonhydrolyzable ATP analog, AMP-PNP, instead of ATP. Pyrene-actin fluorescence was recorded as described above.
Results
Arp2/3 Complex Moves to the Edge of Spread Platelets.
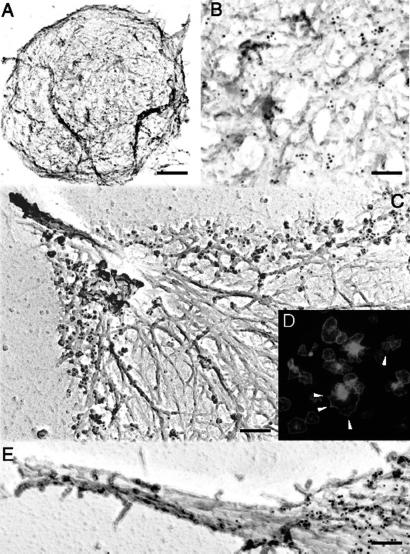
We evaluated the location of Arp2/3 complex in platelet spreading by immunogold electron microscopy and immunofluorescence by using well characterized antibodies directed against the Arp3 and p34-Arc subunits of Arp2/3 complex (21, 22). Immunoreactive Arp2/3 complex proteins distribute throughout the resting platelet, bound predominantly along the sides of actin filaments with a slight concentration near the cell rim (Fig. 1 A and B). As the active platelets spread, Arp2/3 complex redistributes into the most peripheral region of the cytoskeleton (Fig. 1 C and D) but is absent from the tips or shafts of filopods (Fig. 1E), consistent with previous observations in cultured cells (22–25). Some Arp2/3 complex also clusters in the center of activated platelets (Fig. 1D).
Fig 1.
Anti-Arp2/3 complex immunogold electron microscopy and immunofluorescence. Resting (A and B) or glass-stimulated (C–E) human platelet cytoskeletons were labeled with a mixture of polyclonal rabbit anti-Arp3 and anti-p34-Arc antibodies, followed by incubation with 10 nm of colloidal gold or FITC (D) coated with goat anti-rabbit IgGs. Cytoskeletons were fixed, frozen rapidly, freeze-dried, and metal-coated. [Bars = 0.5 (A) and 2 μm (B–E).]
Arp2/3 Complex Associates with the Platelet Cytoskeleton.
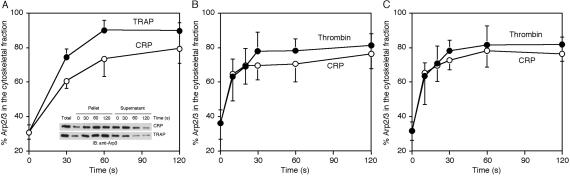
During platelet activation, many proteins move from a detergent-soluble to an insoluble fraction (26). Of the total platelet Arp2/3 complex, ≈30% associates with the Triton X-100-insoluble actin cytoskeleton of the resting cell (Fig. 2A). Stimulation of the platelet receptors PAR-1 with TRAP or GPVI with CRP increases the amount of Arp2/3 complex associated with the Triton X-100-insoluble actin cytoskeleton to 75–90% within 2 min.
Fig 2.
Arp2/3 complex redistribution into the platelet actin cytoskeleton. Human platelets (A) were activated with 25 μM TRAP (•) or 3 μg/ml CRP (○) for the indicated times. Wild-type (B) and gelsolin-null (C) mouse platelets were activated with 1 unit/ml thrombin (•) or 3 μg/ml CRP (○) for the indicated times. The amount of Arp2/3 complex in the cytoskeletal fraction was quantified by densitometric analysis of the immunoblots. Values are mean ± SD of three to five experiments.
Arp2/3 Complex Concentrates in Periodic Aggregates in Gelsolin-Null Platelets.
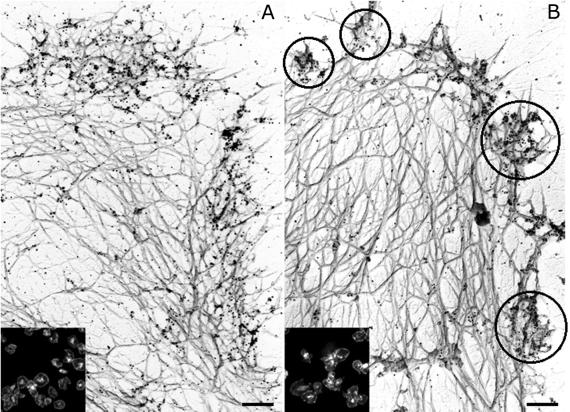
Arp2/3 complex localizes uniformly around the edge of the spread wild-type mouse platelets (Fig. 3A). Although the proportion of immunochemically detectable Arp2/3 complex redistributing into the Triton X-100-insoluble cytoskeleton of stimulated gelsolin-null platelets (Fig. 2C) is indistinguishable from the transfer observed in wild-type mouse platelets (Fig. 2B), the relocation of Arp2/3 complex within the cortex is not uniform as in activated wild-type platelets. Arp2/3 complex, instead, appears in actin filament aggregates at the cell rim in the cytoskeleton of activated gelsolin-null platelets (Fig. 3B).
Fig 3.
Actin cytoskeleton of mouse platelets activated on glass. Wild-type (A) and gelsolin-null (B) mouse platelets activated on anti-GPIbα-IgG-coated glass coverslips were labeled with a mixture of polyclonal rabbit anti-Arp3 and anti-p34-Arc antibodies, followed by incubation with 10 nm of colloidal gold or FITC (Insets) coated with goat anti-rabbit IgGs. Cytoskeletons were fixed, frozen rapidly, freeze-dried, and metal-coated. (Bars = 0.2 μm.) Clusters are indicated in circles.
Arp2/3 Complex Contributes to Platelet Actin Filament Nucleation Activity.
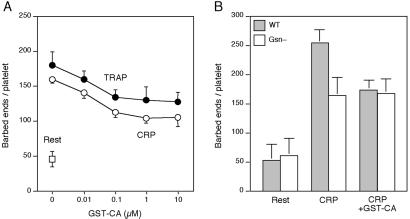
Net actin assembly after ligation of the PAR-1 and GPVI receptors temporally follows the appearance of ≈500 and ≈350 actin filament barbed-end nucleation sites per platelet, respectively (8, 11). We have addressed the role of Arp2/3 complex in the production of barbed-end nucleation sites in OG-permeabilized platelets by using the N-WASp-derived peptide GST-CA that sequesters and inhibits Arp2/3 complex (27). GST-CA inhibits by approximately half the barbed-end actin nucleation activity induced by the chemoattractant peptide formyl-Met-Leu-Phe in OG-permeabilized neutrophils (28) or by chilling in OG-permeabilized platelets (12). Fig. 4A shows that ligation of PAR-1 or of GPVI, respectively, induced the exposure of 178 ± 20 (mean ± SD, n = 3) and 158 ± 5 (n = 4) barbed-end actin filament nuclei in OG-permeabilized platelets 1 min after adding the agonist, compared with 46 ± 11 (n = 7) measured in OG-permeabilized resting platelets. This demonstrates that OG-permeabilized platelets can respond to stimulation by the collagen receptor GPVI. GST-CA inhibited barbed-end actin nucleus production induced by either PAR-1 or GPVI in a dose-dependent manner to a maximum of ≈40% at concentrations ≥1 μM (Fig. 4A). GST alone had no effect on the actin assembly response of OG-permeabilized platelets (8), and GST-CA (3 μM) had no effect on the number of barbed-end nuclei measured in resting cells (data not shown).
Fig 4.
Role of Arp2/3 complex in platelet actin assembly. (A) GST-CA is added at the indicated concentrations to OG-permeabilized platelets before activation (□) with 25 μM TRAP (•) or 3 μg/ml CRP (○) for 1 min. (B) GST-CA (3 μM) is added to OG-permeabilized wild-type (filled bars) and gelsolin-null (open bars) mouse platelets before activation with 3 μg/ml CRP for 1 min.
Arp2/3 Complex Nucleation Activity Requires Gelsolin Expression in Platelets.
We investigated the relative roles of gelsolin and Arp2/3 complex in the actin nucleation activity of OG-permeabilized wild-type and gelsolin-null mouse platelets (Fig. 4B). In wild-type platelets, CRP induced the exposure of 254 ± 23 (mean ± SD, n = 7) barbed-end nuclei/platelet after 1 min of activation, compared with 53 ± 28 (n = 3) barbed-end nuclei measured in OG-permeabilized platelets at rest. GST-CA (3 μM) inhibited nucleus production by ≈40% in CRP-challenged mouse platelets (174 ± 16, n = 8), resembling its effect in OG-permeabilized human platelets. In gelsolin-null platelets, CRP induced ≈45% fewer barbed-end nuclei/platelets (165 ± 31, n = 8). However, GST-CA, at concentrations maximally effective in inhibiting actin nucleation in wild-type permeabilized platelets, was without effect (168 ± 26, n = 8).
Arp2/3 Complex Distribution Requires Gelsolin Expression in Fibroblasts.
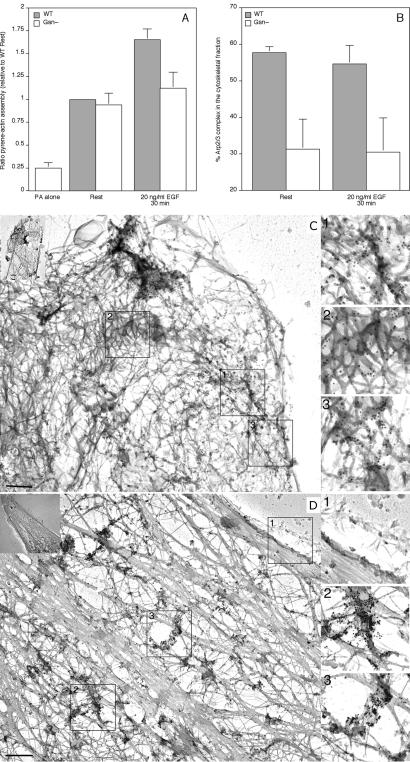
To investigate whether the role of actin filament uncapping on Arp2/3 complex activity was restricted only to platelets, we investigated Arp2/3 complex redistribution in EGF-stimulated wild-type and gelsolin-null mouse lung fibroblasts (Fig. 5). Fig. 5A shows gelsolin-null lung fibroblasts to have an actin assembly response decreased by ≈80% compared with wild-type cells, when stimulated with 20 ng/ml EGF, consistent with previous observations in dermal fibroblasts (9). In gelsolin-null fibroblasts, ≈30% of total Arp2/3 complex associated with the Triton X-100-insoluble actin cytoskeleton, compared with ≈55% in wild-type cells (Fig. 5B). The amount of cytoskeletal Arp2/3 complex did not change within 30 min of stimulation with 20 ng/ml EGF. Sixty to 70% of cellular actin polymerized in both cell types (data not shown). Similar to platelets, fibroblast Arp2/3 complex redistributes uniformly into the cortical region of the cytoskeleton of serum-starved (data not shown) and EGF-stimulated fibroblasts expressing gelsolin (Fig. 5C). In gelsolin-null cells (Fig. 5D), the relocation of Arp2/3 complex within the cortex is not uniform, as in stimulated wild-type cells. Instead, Arp2/3 complex localizes to distinct foci at the cortex, resembling the phenotype observed in platelets.
Fig 5.
Arp2/3 complex redistribution in mouse fibroblasts. Wild-type and gelsolin-null mouse lung fibroblasts were activated with 20 ng/ml EGF for 30 min. Cells were extracted with 0.1% Triton X-100 in PHEM buffer, and the polymerization rate assay was started by the addition of 1 μM pyrene-labeled actin in nucleation buffer (A). Results are expressed as the ratio of pyrene-actin monomers assembled into filaments relative to wild-type resting values and are mean ± SD of three experiments. The amount of Arp2/3 complex in the cytoskeletal fraction was quantified by densitometric analysis of the immunoblots (B). Values are mean ± SD of three experiments. Wild-type (C) and gelsolin-null (D) mouse lung fibroblasts were stimulated with 20 ng/ml EGF for 30 min. Cytoskeletons were labeled with a mixture of polyclonal rabbit anti-Arp3 and anti-p34-Arc antibodies, followed by incubation with 10 nm of colloidal gold coated with goat anti-rabbit IgGs. Cytoskeletons were fixed, frozen rapidly, freeze-dried, and metal-coated. (Bars = 2 μm.) Higher magnifications of regions 1, 2, and 3 are shown in Insets.
Free Barbed-End Availability Regulates Arp2/3 Complex Function in Vitro.
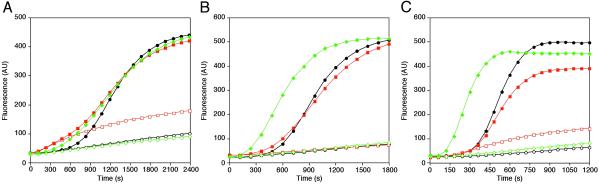
During platelet activation, gelsolin releases from the barbed ends of actin filaments after severing and capping them (29). To determine whether barbed-end uncapping can regulate Arp2/3 complex-mediated actin filament nucleation activity, Arp2/3 complex was activated in vitro by adding the N-WASp-derived peptide GST-VCA (27) to actin monomers in the presence of filaments capped at their barbed ends with gelsolin or CapG, a gelsolin-related protein. As described previously (30), gelsolin in Ca2+-containing buffers is a potent nucleator of actin filaments in the pointed direction (Fig. 6 A and C). Actin filaments nucleated by gelsolin or CapG failed to shorten the activation lag for GST-VCA-activated Arp2/3 complex in the presence of 10 μM CaCl2, consistent with previous observations, implicating a requirement for free barbed ends for Arp2/3 complex function (5, 6). Once activated by Ca2+ and bound to actin filament barbed ends, reduction of the Ca2+ concentration by chelation removes CapG (31) but not gelsolin (32) from the barbed ends. Actin filaments nucleated by CapG and subsequently uncapped by the presence of 100 μM EGTA shortened the lag of Arp2/3 complex-mediated nucleation; actin filaments nucleated by gelsolin and then chelated did not have this effect (Fig. 6B).
Fig 6.
Free barbed-end availability regulates Arp2/3 complex activity in vitro. Polymerization was initiated by the addition of 1 μM pyrene-labeled actin to 2 nM CapG-capped (green diamonds) or 2 nM gelsolin-capped filaments (orange squares) or buffer (•) with (filled symbols) or without (open symbols) 10 nM GST-VCA-activated Arp2/3 complex in nucleation buffer in the presence of 10 μM CaCl2 (A) or 100 μM EGTA (B). Polymerization was initiated by the addition of 1 μM pyrene-labeled actin to 5 nM gelsolin (orange squares), 5 nM F-actin (green diamonds), or buffer (•) with (filled symbols) or without (open symbols) 50 nM GST-VCA-activated Arp2/3 complex in nucleation buffer in the presence of 10 μM CaCl2 and 0.5 mM AMP-PNP, instead of ATP (C).
The polarized elongation of actin filaments requires the addition of ATP monomers to their barbed ends. Once assembled, ATP monomers hydrolyze to ADP monomers, releasing Pi. It has been proposed that Arp2/3 complex binds specifically to the ATP cap at the fast-growing end of filaments because uncapped filaments rich in ATP or in ATP analogues, such as AMP-PNP, markedly potentiate Arp2/3 complex binding and filament branch formation (33). Therefore, one explanation for our findings is that capping proteins inhibit Arp2/3 complex binding and activation by preventing the exposure of ATP-actin monomers at the barbed end of filaments. To explore this possibility, we generated gelsolin-capped actin filaments containing AMP-PNP (Fig. 6C). As observed with filaments generated with hydrolyzable ATP, these capped filaments failed to shorten the lag period of GST-VCA-activated Arp2/3 complex, whereas uncapped AMP-PNP filaments stimulated Arp2/3 complex nucleation.
Discussion
Arp2/3 complex was detectable at the edge of spread platelets, consistent with its location in the actin networks of cultured cells (22–25). However, it concentrates in periodic aggregates at the cortex of gelsolin-null platelets and fibroblasts, and the Arp2/3 complex inhibitor GST-CA (27), at concentrations maximally effective in inhibiting actin nucleation in wild-type permeabilized platelets, had no inhibitory effect in permeabilized gelsolin-null platelets. Taken together, the data indicate that gelsolin contributes to the reorganization of Arp2/3 complex and in the activation of its actin nucleation activity. Actin assembly in gelsolin-null platelets and fibroblasts is markedly deficient in response to stimuli that signal through Rac and membrane polyphosphoinositides (7, 9, 11, 12). Activated Rac is the most potent small GTPase stimulator of actin nucleation in permeabilized platelets (8) and localizes at the edge of the lamellae of spread platelets (34). Lamellipodial Arp2/3 complex localization in the platelet, therefore, suggests regulation by Rac (35, 36). Interestingly, platelets lacking the Cdc42 effector WASp normally activate Arp2/3 complex, assemble actin, and change shape (20). As proposed by others based on in vitro studies (5, 6), Arp2/3 complex alone appears to be a weak nucleator of actin assembly in cells until gelsolin or other capping proteins dissociate from the cellular actin filaments to provide free barbed ends and potentiate Arp2/3 complex-driven events. Another abundant barbed-end capping protein of the platelet, CapZ, appears to lack this capacity, because gelsolin-null platelets have normal CapZ levels. This finding is in agreement with the conclusion that CapZ terminates platelet actin assembly, whereas gelsolin initiates it (10).
Actin filaments assembled in the presence of activated Arp2/3 complex in vitro branch (37, 38), but this fact does not mean that the branching originates off actin filament sides. Here, we show that sides are not sufficient to activate Arp2/3 complex and that free barbed ends are required for its nucleation activity as proposed by others (5, 6). Arp2/3 complex-mediated assembly of AMP-PNP actin was faster than polymerization of actin prepared conventionally with ATP (33). The interpretation of this result was that Arp2/3 complex promotes de novo nucleation only off the sides of actin monomers in their ATP-like conformation (33). However, other interpretations are possible, for example, nuclei of AMP-PNP actin may be more stable than ATP-actin nuclei, resulting in more barbed ends. Our finding that barbed-end capping completely blocks Arp2/3 complex function is incompatible with nucleation off actin filament sides. Our results, however, do not rule out preferential branching off barbed ends with ATP monomers. Severing and capping by gelsolin family members alter the actin filament structure (39) and might prevent its interaction with Arp2/3 complex. In support of this hypothesis, gelsolin binding displaces phalloidin from actin filament barbed ends (40). Uncapping would be expected to release this constraint, allowing for Arp2/3 complex binding and the activation of its side-based nucleation. This observation is not inconsistent with branching because once the original filament end and the Arp2/3 complex nuclei elongate, a branch would form.
Arp2/3 complex incorporation into the active platelet cytoskeleton does not necessarily correlate with the cellular actin assembly response. The aggregation of Arp2/3 complex with actin filaments observed at the cell edge in the absence of gelsolin implies that Arp2/3 complex still moves to the plasma membrane in response to signaling pathways. Few barbed ends are exposed, however, and Arp2/3 complex clusters at these few barbed ends there. This peripheral targeting of Arp2/3 complex alone is insufficient to activate its nucleation activity, and clustering may occur as a consequence of there being a limited number of free filament barbed ends initially available for Arp2/3 complex binding. Hence, gelsolin does not appear to be required for Arp2/3 complex activation, but for its function, which is to amplify the barbed ends exposed at the leading edge.
Arp2/3 complex promotes barbed-end-directed actin filament assembly associated with intracellular propulsion of microorganisms such as Listeria and Shigella (21, 41, 42). However, both Listeria and Shigella generally infect nonmotile cells and may hijack Arp2/3 complex as the only source of barbed-end nucleation activity in such cells. Increasing gelsolin levels in vivo accelerates Listeria motility (43), indicating that even in the Listeria actin-based motility system, gelsolin is capable of enhancing Arp2/3 complex nucleation activity. In platelets and fibroblasts, Arp2/3 complex primarily amplifies barbed ends produced by gelsolin and other severing and capping proteins downstream of Rac. The advancing lamellipodia requires widespread expansion of the actin network. Gelsolin is able to sever actin filaments rapidly and, in the presence of polyphosphoinositides generated by cell activation, dissociates rapidly to produce large numbers of free barbed ends dispersed across the plasma membrane. Actin filament severing and uncapping by gelsolin allows Arp2/3 complex to build off filament networks that are formed already, thus allowing the cell to generate the broad-based expansion of the actin cytoskeleton network required to advance its leading edge.
Acknowledgments
We thank our colleagues and collaborators for sharing reagents, Natasha Isaac for excellent technical assistance, and Karen Vengerow for editorial help. This work was supported by National Institutes of Health Grants HL-56252, HL-56949, and AI-32634, Edwin W. Hiam, and the Edwin S. Webster Foundation.
Abbreviations
CRP, collagen-related peptide
GST-CA, GST fusion protein containing N-WASp C-terminal CA domain (amino acids 450–505)
GST-VCA, GST fusion protein containing N-WASp C-terminal VCA domain (amino acids 392–505)
OG, octyl glucoside
TRAP, thrombin receptor PAR-1-activating peptide
References
- 1.Oosawa F. & Kasai, M. (1962) J. Mol. Biol. 4, 10-21. [DOI] [PubMed] [Google Scholar]
- 2.Hartwig J. (1992) J. Cell Biol. 118, 1421-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condeelis J. (2001) Trends Cell Biol. 11, 288-293. [DOI] [PubMed] [Google Scholar]
- 4.Welch M. & Mullins, R. (2002) Annu. Rev. Cell Dev. Biol. 18, 247-288. [DOI] [PubMed] [Google Scholar]
- 5.Pantaloni D., Boujemaa, R., Didry, D., Gounon, P. & Carlier, M. (2000) Nat. Cell Biol. 2, 385-391. [DOI] [PubMed] [Google Scholar]
- 6.Boujemaa-Paterski R., Gouin, E., Hansen, G., Samarin, S., Le Clainche, C., Didry, D., Dehoux, P., Cossart, P., Kocks, C., Carlier, M. & Pantaloni, D. (2001) Biochemistry 40, 11390-11404. [DOI] [PubMed] [Google Scholar]
- 7.Witke W., Sharpe, A., Hartwig, J., Azuma, T., Stossel, T. & Kwiatkowski, D. (1995) Cell 81, 41-51. [DOI] [PubMed] [Google Scholar]
- 8.Hartwig J., Bokoch, G., Carpenter, C., Janmey, P., Taylor, L., Toker, A. & Stossel, T. (1995) Cell 82, 643-653. [DOI] [PubMed] [Google Scholar]
- 9.Azuma T., Witke, W., Stossel, T., Hartwig, J. & Kwiatowski, D. (1998) EMBO J. 17, 1362-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barkalow K., Witke, W., Kwiatkowski, D. & Hartwig, J. (1996) J. Cell Biol. 134, 389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falet H., Barkalow, K., Pivniouk, V., Barnes, M., Geha, R. & Hartwig, J. (2000) Blood 96, 3786-3792. [PubMed] [Google Scholar]
- 12.Hoffmeister K., Falet, H., Toker, A., Barkalow, K., Stossel, T. & Hartwig, J. (2001) J. Biol. Chem. 276, 24751-24759. [DOI] [PubMed] [Google Scholar]
- 13.McGrath J., Osborn, E., Tardy, Y., Dewey, C., Jr. & Hartwig, J. (2000) Proc. Natl. Acad. Sci. USA 97, 6532-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guttman J., Janmey, P. & Vogl, A. (2002) J. Cell Sci. 115, 499-505. [DOI] [PubMed] [Google Scholar]
- 15.Star E., Kwiatkowski, D. & Murthy, V. (2002) Nat. Neurosci. 5, 239-246. [DOI] [PubMed] [Google Scholar]
- 16.Morton L., Hargreaves, P., Farndale, R., Young, R. & Barnes, M. (1995) Biochem. J. 306, 337-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura N., Osborn, E., Janmey, P. & Stossel, T. (2002) J. Biol. Chem. 277, 9148-9154. [DOI] [PubMed] [Google Scholar]
- 18.Southwick F. & DiNubile, M. (1986) J. Biol. Chem. 261, 14191-14195. [PubMed] [Google Scholar]
- 19.Hartwig J. & DeSisto, M. (1991) J. Cell Biol. 112, 407-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falet H., Hoffmeister, K., Neujahr, R. & Hartwig, J. (2002) Blood 100, 2113-2122. [PubMed] [Google Scholar]
- 21.Welch M., Iwamatsu, A. & Mitchison, T. (1997) Nature 385, 265-269. [DOI] [PubMed] [Google Scholar]
- 22.Welch M., DePace, A., Verma, S., Iwamatsu, A. & Mitchison, T. (1997) J. Cell Biol. 138, 375-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailly M., Macaluso, F., Cammer, M., Chan, A., Segall, J. & Condeelis, J. (1999) J. Cell Biol. 145, 331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svitkina T. & Borisy, G. (1999) J. Cell Biol. 145, 1009-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flanagan L., Chou, J., Falet, H., Neujahr, R., Hartwig, J. & Stossel, T. (2001) J. Cell Biol. 155, 511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox J., Reynold, C. & Boyles, J. (1992) Methods Enzymol. 215, 42-77. [DOI] [PubMed] [Google Scholar]
- 27.Rohatgi R., Ma, L., Miki, H., Lopez, M., Kirchhausen, T., Takenawa, T. & Kirschner, M. (1999) Cell 97, 221-231. [DOI] [PubMed] [Google Scholar]
- 28.Glogauer M., Hartwig, J. & Stossel, T. (2000) J. Cell Biol. 150, 785-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lind S., Janmey, P., Chaponnier, C., Herbert, T. & Stossel, T. (1987) J. Cell Biol. 105, 833-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin H., Hartwig, J., Maruyama, K. & Stossel, T. (1981) J. Biol. Chem. 256, 9693-9697. [PubMed] [Google Scholar]
- 31.Young C., Southwick, F. & Weber, A. (1990) Biochemistry 29, 2232-2240. [DOI] [PubMed] [Google Scholar]
- 32.Janmey P., Chaponnier, C., Lind, S., Zaner, K., Stossel, T. & Yin, H. (1985) Biochemistry 24, 3714-3723. [DOI] [PubMed] [Google Scholar]
- 33.Ichetovkin I., Grant, W. & Condeelis, J. (2002) Curr. Biol. 12, 79-84. [DOI] [PubMed] [Google Scholar]
- 34.Azim A., Barkalow, K., Chou, J. & Hartwig, J. (2000) Blood 95, 959-964. [PubMed] [Google Scholar]
- 35.Nobes C. & Hall, A. (1995) Cell 81, 53-62. [DOI] [PubMed] [Google Scholar]
- 36.Hall A. (1998) Science 279, 509-513. [DOI] [PubMed] [Google Scholar]
- 37.Volkmann N., Amann, K., Stoilova-McPhie, S., Egile, C., Winter, D., Hazelwood, L., Heuser, J., Li, R., Pollard, T. & Hanein, D. (2001) Science 293, 2456-2459. [DOI] [PubMed] [Google Scholar]
- 38.Robinson R., Turbedsky, K., Kaiser, D., Marchand, J., Higgs, H., Choe, S. & Pollard, T. (2001) Science 294, 1679-1684. [DOI] [PubMed] [Google Scholar]
- 39.Dawson, J. F., Sablin, E. P., Spudich, J. A. & Fletterick, R. J. (2002) J. Biol. Chem., 10.1074/jbc.M209160200.
- 40.Allen P. & Janmey, P. (1994) J. Biol. Chem. 269, 32916-32923. [PubMed] [Google Scholar]
- 41.Welch M., Rosenblatt, J., Skoble, J., Portnoy, D. & Mitchison, T. (1998) Science 281, 105-108. [DOI] [PubMed] [Google Scholar]
- 42.Loisel T., Boujemaa, R., Pantaloni, D. & Carlier, M. (1999) Nature 401, 613-616. [DOI] [PubMed] [Google Scholar]
- 43.Laine R., Phaneuf, K., Cunningham, C., Kwiatkowski, D., Azuma, T. & Southwick, F. (1998) Infect. Immunol. 66, 3775-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]