Abstract
Understanding the pattern of speciation in a group of plants is critical for understanding its morphological evolution. Lepidium is the genus with the largest variation in floral structure in Brassicaceae, a family in which the floral ground plan is remarkably stable. However, flowers in more than half of Lepidium species have reduced stamen numbers, and most of these also have reduced petals. The species with reduced flowers are geographically biased, distributed mostly in the Americas and Australia/ New Zealand. Previous phylogenetic studies using noncoding regions of chloroplast DNA and rDNA internal transcribed spacer were incongruent in most New World species relationships. These data, combined with the presence of many polyploid Lepidium species, implied a reticulate history of the genus but did not provide enough information to infer the evolutionary pattern of flower structures. To address this question more thoroughly, sequences of the first intron of a single copy nuclear gene, PISTILLATA, were determined from 43 species. Phylogenetic analysis of the PI intron suggests that many species in the New World have originated from allopolyploidization, and that this is correlated with floral reduction. Interspecific hybrids were generated to understand why allopolyploidization is associated with reduced flowers. The phenotypes of F1 flowers indicate allelic dominance of the absence of lateral stamens, suggesting that propagation of dominant alleles through interspecific hybridization could account for the abundance of the allopolyploid species without lateral stamens.
Through the study of model plant species, it is relatively well understood how organ identity is regulated in flowers (1–3). However, the underlying genetic mechanisms that lead to the remarkable variation in flower structures in angiosperms are not known. One way to address this question is to analyze floral variation in closely related species. Knowledge of the genetic bases of phenotypic variation between related species for which we know the phylogeny should lead to an understanding of the number of genetic changes underlying phenotypic evolution and other events that lead to the dispersal of certain phenotype(s).
The basic floral ground plan of Brassicaceae is well conserved, with four sepals, four petals, six stamens, and two carpels (Fig. 1A). However, within Lepidium L. (≈175 species worldwide), more than half of the species have only two or four stamens (Fig. 1 B and E–G), and in most of these species, petals are rudimentary (Fig. 1 B, and E, and F) (4–6). Species with reduced flower structures in Lepidium are more widespread in the Americas and Australia/New Zealand than in Eurasia and Africa. In addition, ploidy varies among species of Lepidium (7). However, hybridization, one possible origin of polyploidization, is reported rarely (8–10), and polyploid origins have not been identified experimentally. Questions regarding how species with reduced flowers became predominant in the Americas and Australia, and whether reduction in floral architecture occurred multiple times in parallel or intercontinental migration occurred repeatedly, have remained unanswered.
Fig 1.
Diverse floral forms of Lepidium and hybrid F1 flowers. (A) L. montanum has four medial and two lateral stamens and full-size petals, the ancestral condition. (B) L. hyssopifolium has two medial stamens and reduced petals. (C) L. oleraceum has two medial and two lateral stamens with full-size petals. (D) F1 flower of L. hyssopifolium × L. oleraceum. (E) L. oxytrichum has four medial stamens and very reduced petals. (F) L. pseudohyssopifolium. (G) L. virginicum. (H) F1 flower of L. pseudohyssopifolium × L. virginicum.
Studies of Lepidium phylogeny using the rDNA internal transcribed spacer (ITS) and two intergenic spacers and an intron of trnT-trnF in chloroplast DNA (cpDNA) clarified some relationships within the genus (11, 12). In ITS trees, species from Eurasia and Australia, which have the typical Brassicaceae floral structure, form sister lineages basal to the rest of the species (11), indicating that reduced floral structures are derived. ITS trees suggest that floral structures in Lepidium are rather fluid, and that many intercontinental migrations have occurred. Although cpDNA trees show a similar pattern of early lineages and agree with the ITS trees on the fluidity of the flower structures in Lepidium (12), species from similar geographic regions usually remain more closely related to one another than to species from other regions, implying radiations following a small number of intercontinental migrations, in contradiction to inferences from ITS trees (11, 12).
The incongruence of species relationships based on two separate molecular markers, one of which is maternally inherited (cpDNA) (13) and the other biparentally inherited, but with a potential for concerted evolution (ITS) (14, 15), and the large number of polyploid species suggest that the evolutionary history of Lepidium is reticulate. To better resolve the evolutionary pattern of floral structures in Lepidium, a phylogenetic study using a single-copy nuclear gene was required, because such markers better reflect biparental lineages of homoploid and allopolyploid hybrids and thus are powerful tools in reconstructing reticulate histories of plant groups (16–18).
PISTILLATA (PI) acts to specify petal and stamen identity in Arabidopsis (2, 3, 19–21) and likely in all angiosperms (22–24). It is a single-copy gene, and its first intron is 997 bp long in Arabidopsis thaliana (20). Previous studies on the phylogenetic utility of PI first intron sequences in Sphaerocardamum and other Brassicaceae showed that it provides more phylogenetically informative characters than either ITS and chloroplast trnL intron sequences while supporting species relationships consistent with the other markers (25).
In this paper, the evolutionary pattern of Lepidium was inferred from the phylogeny based on PI first intron sequences. Tree results, along with analyses of flower structures in interspecific hybrids, provide new evidence that one mechanism for apparent parallel evolution, flower structure in the case of Lepidium, may be the propagation of dominant alleles through interspecific hybridization.
Materials and Methods
Taxon Sampling.
Forty-three taxa (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org) were chosen to represent all major geographic distributions: Asia/Europe, Africa, North America, South America, and Australia/New Zealand. Most taxa overlap with those used in cpDNA and ITS analyses (11, 12). Lepidium phlebopetalum and Lepidium leptopetalum were used as outgroups as these species form a clade (Monoploca s.str.) sister to two main lineages of Lepidium, i.e., Lepia s.l. and Lepidium s.str., in ITS trees (11). In cpDNA trees, all three main lineages are in an unresolved trichotomy.
DNA Isolation, PCR, and Sequencing.
DNA was isolated from fresh or dry leaves from individual plants (26). PI first intron sequences were amplified by PCR by using PI-ITF (5′-GAAATTATCTGGCAAGAAACTTTGGG-3′) and PI-ITR (5′-TCCTATCAATCTCATTGCTGAGGTTC-3′) as primers. Primers are from exon sequences flanking the first intron. The sequences are unique to PI such that nonspecific cloning of introns from paralogous MADS-box genes was not found in this study (27, 28). PCR used the Advantage cDNA PCR kit (CLONTECH) under the following conditions: initial denaturation of 2 min at 94°C; 35 cycles of 30 sec at 94°C, 30 sec at 60°C, and 1 min at 68°C; and final extension of 5 min at 68°C. PCR products were sequenced directly, and those that showed multiplicity or lacked clear base readings were cloned by using the TOPO TA cloning kit (Invitrogen). Of 43 species analyzed, multiple sequences were detected in 19 species, whereas 24 species had a single sequence. Six separate clones were sequenced from TA-cloned PCR products. If more than two distinct sequences were detected, more sequences were searched by finding clones having distinct restriction enzyme digestion patterns.
Phylogenetic Analysis.
Seventy-two sequences were aligned by using clustalx (29) and manually adjusted to minimize substitutions and to maximize gaps. Regions with several possible alternative alignments were eliminated, resulting in a final data matrix of 1,397 characters. The alignment is available on request. Gaps were treated as missing data. Uncorrected (“p”) pairwise divergence and GC content were calculated in paup* 4.0b 4b (30).
The data were analyzed in paup* 4.0b 4b by using maximum parsimony. Most parsimonious trees were obtained by using a heuristic search with 100 random additions of sequences with Tree Bisection and Reconnection (TBR) branch swapping. Bootstrap support values (31) were obtained from 200 replicates of full heuristic search with random addition of sequences with TBR swapping, and Max Trees set at 1,000 per replication.
Analysis of Interspecific Hybrids.
On the basis of ITS and cpDNA trees (11, 12), species that are closely related but have different stamen number or, alternatively, are distantly related but with the same stamen number, were chosen for artificial hybridization, and two sets of F1 hybrids were generated. One was a hybrid between Lepidium hyssopifolium and Lepidium oleraceum, the other a hybrid between Lepidium pseudohyssopifolium and Lepidium virginicum. F1 progeny of L. hyssopifolium and L. oleraceum were fertile, and stamen numbers of 397 F2 individuals were counted. To test the heritability of stamen distributions, F3 progeny of F2 individuals with differing numbers of stamens were counted: 22 F3 progeny from F2 plants that had two lateral stamens in all flowers; 237 F3 progeny from F2 plants that had two lateral stamens in most flowers; 44 F3 progeny F2 plants that had no lateral stamens in most flowers; and 40 F3 progeny from F2 plants that had no lateral stamens in any flowers. For each individual plant, stamen numbers were scored in 20–40 flowers.
Results
PI Intron Sequences and the Gene Tree.
The length of the PI first intron ranged from 801 to 1,053 bp. Of 43 species analyzed, 24 had a single sequence, and 19 species had more than one sequence type. The average GC content was 28.0%, similar to that of other Brassicaceae species (25). Among 1,397 aligned characters, 517 characters were parsimony informative (37.0%), three times more informative characters than cpDNA sequences (12) and four times more than ITS sequences (11).
The strict consensus tree of 35,922 most parsimonious trees based on the first intron of PI is shown in Fig. 2. With the given outgroup, the ingroup clade was well supported (100% bootstrap value). The average pairwise divergence between ingroup and outgroup species was 19.9%, whereas among the ingroup species the value was 11.6%. Species in section Lepia s.l. (6) (Lepia heterophyllum, Lepia hirtum, Lepia campestre, and Lepia perfoliatum), which was sister to Lepidium s.str. (the remaining ingroup species) in ITS and cpDNA trees, formed a monophyletic group with 85% bootstrap support, but bootstrap support for its basal position is weak.
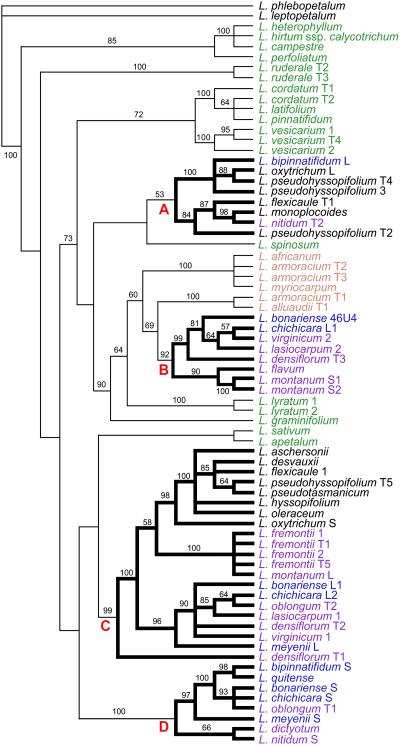
Fig 2.
Strict consensus of 35,922 most parsimonious PI intron gene trees (length = 1,664, confidence interval = 0.64, retention index = 0.86). Bootstrap values >50 are shown. Taxa on the tree are color-coded with their geographic distribution: green, Asia/Europe; orange, Africa; blue, South America; purple, North America; and black, Australia/New Zealand. The four clades that include phylogenetically distinct multiple PI intron sequences from individual species are denoted as clades A–D.
Phylogenetic analysis of the PI intron showed that multiple sequences found in many species were placed in different clades of the tree. For example, South American species Lepidium meyenii and North American species Lepidium lasiocarpum and L. virginicum have two sequence types that reside in two separate clades, and Lepidium bonariense has three sequence types in three distinct clades. Clades containing such sequences were designated as A, B, C, and D, and the geographic distributions of the source species were color-coded on each sequence (Fig. 2). These four clades include all of the taxa from North and South America and Australia/New Zealand (excluding the basal Australian species). Clades B, C, and D are strongly supported with high bootstrap values, whereas clade A is not.
The occurrence of multiple PI intron sequences from a single species in different clades could be from gene duplication/loss, lineage sorting, or hybridization (32, 33). On the basis of the pattern of PI intron trees, ploidy of species, and species relationships in cpDNA and ITS trees, we favor (allopolyploid) hybridization as the cause of many Lepidium species having multiple sequences residing in separate clades. For example, L. meyenii is octoploid and has two types of sequences, in clades C and D. In cpDNA trees, L. meyenii is in a clade with other South American species without any close relationship to North American species, but in ITS trees, it is closely related to North American species L. virginicum and L. lasiocarpum. Clade C in PI intron trees includes both L. meyenii and L. virginicum and L. lasiocarpum, as in ITS trees, and clade D contains all of the South American species that formed a clade in cpDNA trees (Fig. 2). L. virginicum and L. lasiocarpum provide a similar example. Thus, the PI intron trees reflect the patterns exhibited in both the ITS and cpDNA trees. Chromosome numbers are known for 11 of 13 species with multiple phylogenetically distinct PI intron sequences in clades A–D (Fig. 3). All 11 species are polyploids, and the number of their distinct sequences is the same as or less than their ploidal level. Thus, one interpretation is that species with multiple phylogenetically distinct PI intron sequences are allopolyploids. In this scenario, the cpDNA tree reflects maternal lineages, the ITS tree reflecting uniparental inheritance of rDNA, with the PI intron tree being a composite, reflecting nuclear inheritance of a single copy gene. Distances between clades and within a clade show that phylogenetically distinct intron sequences within a species are more divergent than sequences from different species within the same clade, suggesting that multiple sequences originated before speciation, consistent if they have become resident in the same species via an allopolyploidization event.
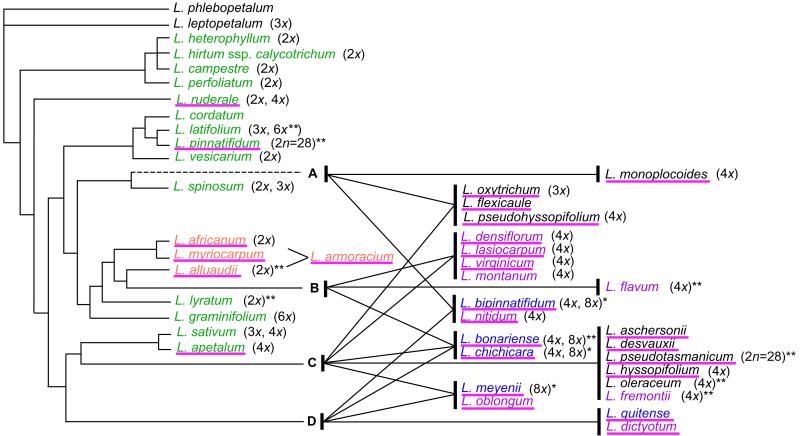
Fig 3.
Pattern of speciation in Lepidium based on the PI intron gene tree and the evolution of lateral stamen arrangements. For taxa in clades A–D that originated from allopolyploid hybridization, genome compositions of each species are shown by lines between species and genome source. Clade A is marked with a dashed line due to its weak bootstrap support. Species lacking lateral stamens are underlined, and their geographic distribution is color-coded as in Fig. 2. Ploidy of species is listed; 2x represents diploid (x = 8 for Lepidium). Most ploidy values are from the Index to Plant Chromosome Numbers in Monographs in Systematic Botany (Missouri Botanical Garden). Ploidy marked with * is from ref. 42; **, from an unpublished compilation of chromosome data (I. A. Al-Shehbaz and K.M.).
Although many New World species have only one type of sequence in each clade, some species such as Lepidium fremontii, Lepidium montanum, L. pseudohyssopifolium, and Lepidium densiflorum have more than one type of sequence within a clade (Fig. 2). Because L. fremontii and L. montanum are self-incompatible, the similar sequences probably have their origin in allelic variation. In contrast, L. pseudohyssopifolium and L. densiflorum are autogamous, and gene duplication may be involved for their multiple sequences in the same clade.
We also found several species from the Old World (Lepidium ruderale, Lepidium cordatum, Lepidium vesicarium, Lepidium armoracium, and Lepidium lyratum) that have multiple sequences. However, all of their multiple sequences are closely related to each other, and we did not find any species having multiple sequences residing in phylogenetically distinct lineages. Multiple sequences possibly caused by hybridization were detected in only one species, L. armoracium. Sequences of L. armoracium are imbedded in the Lepidium africanum/Lepidium myriocarpum and Lepidium alluaudii lineages, suggesting that L. armoracium might be a hybrid between species from the same geographic region. Otherwise, because most of these Old World species are diploid, multiple sequences in a single species appear to be derived from allelic diversity or gene duplication (Fig. 2).
Species Tree and Floral Structure.
On the basis of the allopolyploid hybridization hypothesis derived from PI intron trees, the putative species tree of Lepidium was constructed (Fig. 3). The topology of backbone was constructed on the basis of the relationships among species having single or only closely related multiple PI intron sequences, all of which are from the Old World. Onto the backbone, species residing in clades A, B, C, and D were placed, and the origins of their multiple sequences were demarcated. On the basis of this analysis, there appear to be AC, BC, AD, CD, and BCD allopolyploids, implying multiple independent hybridization events. There is at least one species with only a single sequence in each of the clades, A–D. However, these species are nested inside the clades, and those with known ploidal levels are all polyploids, implying they may have originated after the hybridization events but have lost one homeologue via gene conversion or loss (34).
Species lacking lateral stamens (underlined in Fig. 3) are more common among the allopolyploids, and most allopolyploids lack lateral stamens. Among 13 allopolyploid species from the New World, 12 species do not have lateral stamens (92%). Among 17 putative nonhybrid species from Asia/Europe and Africa, six species have no lateral stamens (35%). Therefore, species lacking lateral stamens are significantly more common among allopolyploid species in the New World. Species from the New World with a single PI sequence or multiple sequences in the same clade were not counted in this comparison (Fig. 3), because the PI intron data does not support an allopolyploid origin, although other data suggest that most may be of hybrid origin (see Discussion). Among several aspects of floral structures that vary in Lepidium, we focused on the status of lateral stamens in this analysis, because the reduction in medial stamens varies in penetrance and is genetically separable from the absence of lateral stamens (5, 35).
Floral Structures of Interspecific Hybrids.
To investigate the genetic basis of reduced flower structures, two sets of interspecific hybrids were generated. One is a hybrid between L. hyssopifolium (female) and L. oleraceum (male). L. hyssopifolium has two medial stamens with inconspicuous petals (Fig. 1B), and L. oleraceum has two medial and two lateral stamens with full-size petals (Fig. 1C). These species are closely related based on PI intron data (0.2% sequence divergence; Fig. 2) as well as other data (11, 12). Examination of flowers of F1 plants indicated that the alleles conferring lack of lateral stamens are semidominant. Approximately 80% of F1 flowers had two medial stamens and no lateral stamens (Fig. 1D), whereas 20% had one lateral stamen and two medial stamens. The second interspecific hybrid was derived from crossing L. pseudohyssopifolium and L. virginicum. These species are allopolyploids with separate evolutionary histories (Figs. 2 and 3). Both species have flowers with two medial stamens and no lateral stamens (Fig. 1 F and G), and in the F1 plants, 100% of flowers had two medial stamens and no lateral stamens (Fig. 1H).
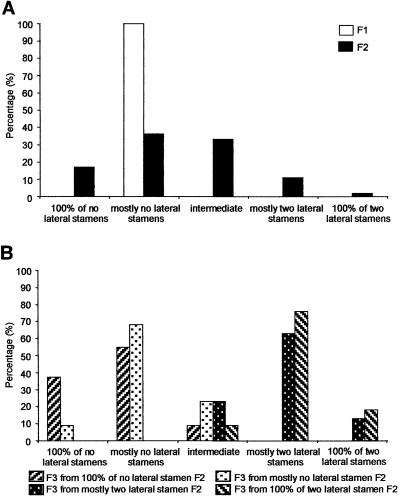
Because the F1 hybrid of L. hyssopifolium and L. oleraceum was self-fertile, we examined the segregation of floral phenotypes in subsequent generations. The distribution of stamen numbers in the F2 was quantitative, implicating multiple loci in the reduction of lateral stamens (Fig. 4A) (36, 37). Stamen numbers fell largely into three classes: two medial stamens and no lateral stamens, two medial stamens and one lateral stamen, and two medial stamens and two lateral stamens. About 17% of F2 plants had flowers with only two medial stamens, and only 2% of plants had flowers that always had two medial and two lateral stamens. The rest were intermediate, although about 55% were individuals with primarily two medial stamens. The heredity of the stamen characters observed in the F2 was tested by counting stamens in F3 plants derived from F2 plants with four different stamen distributions (Fig. 4B). Although still quantitative, stamen distributions of F2 plants were highly reflected in F3 plants. F3 plants derived from F2 plants with primarily two lateral stamens had mostly two lateral stamens, and F3 plants derived from F2 plants without lateral stamens mostly lacked lateral stamens.
Fig 4.
Stamen distribution in progeny of L. hyssopifolium and L. oleraceum hybrids. Stamen distribution of F1 and F2 progeny (A) and F3 progeny (B). Based on stamen numbers observed in F2 and F3 plants, stamen characters were divided into five classes: 100% no lateral stamens, mostly no lateral stamens, intermediate, mostly two lateral stamens, and 100% two lateral stamens. When an individual has a mixture of flowers with various number of lateral stamens (0, 1, or 2), it is classified as intermediate.
Discussion
PI Intron Tree and Speciation Patterns in Lepidium.
Hybridization and polyploidization are common in flowering plants and are known to play a significant role in their evolution (38–40). In Brassicaceae, polyploidy is common throughout the family (37% of the species), and some genera (e.g., Crambe, Moricandia, Vella) appear to be exclusively polyploid (41). In Lepidium, Lepidium oreganum was suspected as the hybrid derivative of Lepidium nitidum and the Lepidium latipes–Lepidium dictyotum–Lepidium oxycarpum complex based on morphological intermediacy and shared habitats of putative parental species (8–10), but its possible hybrid origin was not investigated further.
As inferred from ITS and cpDNA results, PI intron trees clearly suggest that many Lepidium species in the Americas and Australia are the result of allopolyploid hybridization. Five major groups of allopolyploid hybrids from 13 species were identified (Fig. 3), four of which are composed of two genomes, with one group, including L. bonariense and Lepidium chichicara, having three component genomes. These South American species are reported as either tetraploids or octoploids (Fig. 3). The L. chichicara accession used in this analysis is octoploid (42); however, without knowing the genome composition of the tetraploid populations, the relationships between the tetraploid and octoploid accessions are unclear.
Because 10 of 13 identified allopolyploid species are from America, most hybridization events apparently have occurred in the Americas, with some Australian species, such as L. pseudohyssopifolium, Lepidium oxytrichum, and Lepidium flexicaule, likely arising from a separate allopolyploid hybridization event. The origin of the remaining Australian/New Zealand species is enigmatic. In these species, we detected only a single PI intron sequence in each species, all of which (except for the Lepidium monoplocoides sequence) are in clade C, nested within American species. However, the positions of these species in the ITS and cpDNA trees are incongruent, suggesting that they are also likely to be hybrids. Consistent with this interpretation, L. hyssopifolium and L. oleraceum are tetraploids, and preliminary comparative mapping of F2 progeny by restriction fragment length polymorphism (RFLP) analysis suggests that they are allotetraploids (J.-Y.L. and J.L.B., unpublished work). RFLP analysis with PI also found two loci in both species, meaning one of the homeologues of PI was not detected by PCR due to either rapid mutation of coding sequences in the process of pseudogenization or homogenization of sequences of two homeologous loci via concerted evolution or recombination (34). Further study using another single-copy nuclear marker may help to clarify the origin of these Australian species.
Although PI intron trees were useful in identifying many allopolyploid species and their possible origins, several relationships among putative backbone species are significantly different from the cpDNA and ITS results. In both cpDNA and ITS trees, Lepidium spinosum and Lepidium sativum are sister to each other with more than 90% bootstrap support, but in PI intron trees, they are located in distantly related clades, and eight additional steps were required to force their sister relationship. Likewise, Lepidium pinnatifidum is sister to L. ruderale in cpDNA and L. cordatum in ITS trees, but the latter two species form a clade with L. vesicarium and Lepidium latifolium in the PI intron gene tree, not with L. ruderale (Fig. 2). These conflicting species relationships specially of L. spinosum and L. sativum might illustrate lineage sorting of PI in some of Lepidium species, although hybridization cannot be ruled out as another possible cause considering variable ploidal levels in those species (Fig. 3).
Hybridization and Floral Morphology.
Most traditional classifications of Lepidium relied on similarities in morphology such as floral structures and fruit and embryo shape along with geographical distribution to ascertain subgeneric relationships (5, 6). However, molecular data based on ITS, cpDNA, and PI intron sequences do not support such groupings. Although morphological similarities could have arisen via convergent evolution, another possibility is introgression of morphological traits via allopolyploid hybridization (43). This latter mechanism has not been documented to play a role in the morphological evolution in natural species, but given the extent of hybridization in angiosperms (44), it seems likely. The patterns of hybridization and floral morphology in Lepidium support the hypothesis that the preponderance of species with reduced floral forms could have been produced by introgression by way of allopolyploid speciation.
Along with the PI intron gene tree data, interspecific hybrid analyses support the hypothesis that hybridization events may be responsible for the large proportion of species with reduced floral forms in Lepidium. In the cross between L. oleraceum and L. hyssopifolium, alleles responsible for the absence of lateral stamens are dominant to those for their presence. Because the hybrid generated is a homoploid hybrid, the stability of the F1 floral phenotype in subsequent generations could not be demonstrated. However, F2 and F3 analyses demonstrate that the patterns of stamen distributions are genetically inherited, with the distribution of phenotypes in the F2 and F3 suggesting a multigenic origin (36, 37). It is likely that the presence of lateral stamens in L. oleraceum evolved from an ancestral condition in which they were absent and thus represents a reversion event rather than one of the original reduction events. Unfortunately, repeated attempts to hybridize, via embryo rescue techniques, a six-stamen species with a two-stamen species failed (J.-Y.L. and J.L.B., unpublished work). However, another hybrid derived from L. virginicum and L. pseudohyssopifolium also supports the idea of allelic dominance of absence of lateral stamens. Both parental species are allopolyploids and lack lateral stamens, but they have different evolutionary histories as L. virginicum contains the B and C genomes whereas L. pseudohyssopifolium contains the A and C genomes. That all of the F1s lack lateral stamens can be due either to a dominance of alleles controlling this trait or to the fact that the same genetic loci, possibly due to the common C genome, are involved in the reductions in these species.
If allelic dominance is responsible for the propagation of the reduced stamen flowers to allopolyploid species, one of the parental lineages of each of the allopolyploids should be a species without lateral stamens. Among the four PI clades, only the B clade shows a well-supported relationship to extant descendants of a possible ancestor, the African lineages whose species lack lateral stamens. For the rest of the genomes, their sister lineages are unclear due to poor bootstrap support and conflicting species relationships with cpDNA and ITS trees. More exhaustive taxon sampling from the Old World and additional nuclear gene studies to reevaluate the origin of species on the backbone tree (Fig. 3) are needed to determine potential parental species of allopolyploid species, if they are extant.
On the basis of the phylogeny and genetic analysis of Lepidium, a pathway of floral evolution is suggested (Fig. 5). The PI species tree suggests there may be four independent lateral stamen reductions in the Old World. Whether the same genetic pathway is responsible for the reduction of the lateral stamens of the Old World species is not known, but construction of a hybrid between an AD species (e.g., L. nitidum) and a BC species (e.g., L. virginicum) would be informative. In the course of migration to the New World, several independent hybridization events occurred, and allelic dominance could have led to the dispersion of species whose floral structure lacks lateral stamens. Why then are allopolyploids dominant only in the New World? Among the Old World Lepidium, species with reduced flower structures are often self-fertilizing and are successful colonizing species. Because allopolyploidization is one mechanism to increase the gene pool to avoid the deleterious outcome of inbreeding, a speculative hypothesis is that this may have been a factor for promoting hybridization events during the rapid radiation of the genus in the Americas and Australia. In this scenario, outcrossing species such as L. fremontii and L. oleraceum would be derived from self-fertilizing ancestors, in contrast to the more common pattern of derived autogamy.
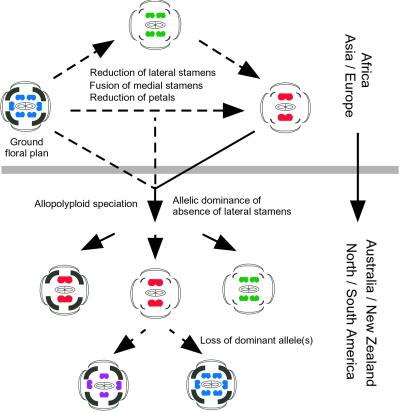
Fig 5.
Model for the evolution of floral forms in Lepidium. The change in ground floral plan from the ancestral condition (four medial and two lateral stamens and four petals) has apparently occurred in the Old World, resulting in flowers with two medial stamens, no lateral stamens, and reduced petals. Species with reduced floral forms migrated to the Americas and participated in allopolyploid hybridizations. Dominance of alleles conferring an absence of lateral stamens results in hybrid species having flowers without lateral stamens. Most of these species are autogamous, and thus natural selection may favor a reduction in petals in these species. A few allopolyploid species do develop lateral stamens, presumably due to the loss of the dominant allele(s) after hybridization. The paths experimentally shown in this paper are marked with solid lines and others are marked with dotted lines.
Given that allelic differences govern variation in floral structures, what genes might be involved in the process of floral reduction? Because the variations involve petals and stamens, it was suggested that genes specifically involved in organ formation, identity, and growth of those two whorls might have been altered (4, 35). The plausible candidates are the B class genes, APETALA3 and PI, and genes that regulate B class gene activity (2, 3, 19, 20, 45, 46). However, mapping studies using interspecific hybrids did not implicate these genes in stamen reduction (J.-Y.L. and J.L.B., unpublished results). To understand how the multigenic and genetically separable reductions of petals and stamens coevolved, mapping of such traits in interspecific crosses would be informative.
Supplementary Material
Acknowledgments
We greatly appreciate advice on data analyses and critical reviews from Sang-Hun Oh, Sandra Floyd, Daniel Potter, and Mike Sanderson and assistance in hybrid analyses by Dale Cox. We thank David Smyth, Charles Gasser, Yuval Eshed, Stuart Baum, Andreas Franzke, and the members of the Bowman laboratory for fruitful comments and insights. We also thank three anonymous reviewers for improving this manuscript. We thank people and institutions who kindly provided plant material, including Ellen Dean at the University of California Davis Herbarium. This work was supported by a Beckman Young Investigator Award (to J.L.B.).
Abbreviations
PI, PISTILLATA
ITS, internal transcribed spacer
cpDNA, chloroplast DNA
References
- 1.Coen E. S. & Meyerowitz, E. M. (1991) Nature 353, 31-37. [DOI] [PubMed] [Google Scholar]
- 2.Bowman J. L., Smyth, D. R. & Meyerowitz, E. M. (1989) Plant Cell 1, 37-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman J. L., Smyth, D. R. & Meyerowitz, E. M. (1991) Development (Cambridge, U.K.) 112, 1-20. [DOI] [PubMed] [Google Scholar]
- 4.Endress P. K. (1992) Int. J. Plant Sci. 153, S106-S122. [Google Scholar]
- 5.Thellung A. (1906) Neue Denkschr. Allg. Schweiz. Naturforsch. Ges. 41, 1-304. [Google Scholar]
- 6.Hewson H. (1981) Brunonia 4, 217-308. [Google Scholar]
- 7.Al-Shehbaz I. A. (1986) J. Arnold Arbord. Harv. Univ. 67, 265-311. [Google Scholar]
- 8.Howell J. T. (1934) Leafl. West. Bot. 1, 92-94. [Google Scholar]
- 9.Hitchcock C. L. (1936) Madrono 3, 265-320. [Google Scholar]
- 10.Hitchcock C. L. (1945) Lilloa 11, 75-143. [Google Scholar]
- 11.Bowman J. L., Brüggemann, H., Lee, J.-Y. & Mummenhoff, K. (1999) Int. J. Plant Sci. 160, 917-929. [DOI] [PubMed] [Google Scholar]
- 12.Mummenhoff K., Brüggemann, H. & Bowman, J. L. (2001) Am. J. Bot. 88, 2051-2063. [PubMed] [Google Scholar]
- 13.Song K., Tang, T. C. & Osborn, T. C. (1993) Theor. Appl. Genet. 86, 811-821. [DOI] [PubMed] [Google Scholar]
- 14.Wendel J. F., Schnabel, A. & Seelanan, T. (1995) Proc. Natl. Acad. Sci. USA 92, 280-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sang T., Crawford, D. J. & Stuessy, T. F. (1995) Proc. Natl. Acad. Sci. USA 92, 6813-6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Small R. L., Ryburn, J. A., Cronn, R. C., Seelanan, T. & Wendel, J. F. (1998) Am. J. Bot. 85, 1301-1315. [PubMed] [Google Scholar]
- 17.Song G., Sang, T., Lu, B.-R. & Hong, D.-Y. (1999) Proc. Natl. Acad. Sci. USA 96, 14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson D. & Sang, T. (2001) Proc. Natl. Acad. Sci. USA 98, 3915-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jack T., Brockmann, L. L. & Meyerowitz, E. M. (1992) Cell 68, 683-697. [DOI] [PubMed] [Google Scholar]
- 20.Goto K. & Meyerowitz, E. M. (1994) Genes Dev. 8, 1548-1560. [DOI] [PubMed] [Google Scholar]
- 21.Riechmann J. L., Krizek, B. A. & Meyerowitz, E. M. (1996) Proc. Natl. Acad. Sci. USA 93, 4793-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambrose B. A., Lerner, D. R., Ciceri, P., Padilla, C. M., Yanofsky, M. F. & Schmidt, R. J. (2000) Mol. Cell 5, 569-579. [DOI] [PubMed] [Google Scholar]
- 23.Kyozuka J., Kobayashi, T., Morita, M. & Shimamoto, K. (2000) Plant Cell Physiol. 41, 710-718. [DOI] [PubMed] [Google Scholar]
- 24.Kramer E. M., Dorit, R. L. & Irish, V. F. (1998) Genetics 149, 765-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey C. D. & Doyle, J. J. (1999) Mol. Phylogenet. Evol. 13, 20-30. [DOI] [PubMed] [Google Scholar]
- 26.Doyle J. J. & Doyle, J. L. (1987) Phytochem. Bull. 19, 11-15. [Google Scholar]
- 27.Theissen G., Kim, J. T. & Saedler, H. (1996) J. Mol. Evol. 43, 484-516. [DOI] [PubMed] [Google Scholar]
- 28.Alvarez-Buylla E. R., Pelaz, S., Liljegren, S. J., Gold, S. E., Burgeff, C., Ditta, G. S., Ribas de Pouplana, L., Martinez-Castilla, L. & Yanofsky, M. F. (2000) Proc. Natl. Acad. Sci. USA 97, 5328-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swofford D. L., (2000) paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.0b 4b.
- 31.Felsenstein J. (1985) Evolution (Lawrence, KS) 39, 783-791. [DOI] [PubMed] [Google Scholar]
- 32.Doyle J. J. (1992) Syst. Bot. 17, 144-163. [Google Scholar]
- 33.Wendel J. F. & Doyle, J. J. (1998) in Molecular Systematics of Plants II, eds. Soltis, D. E., Soltis, P. S. & Doyle, J. J. (Kluwer, Dordrecht, The Netherlands), pp. 265–296.
- 34.Wendel J. F. (2000) Plant Mol. Biol. 42, 225-249. [PubMed] [Google Scholar]
- 35.Bowman J. L. & Smyth, D. R. (1998) Int. J. Plant Sci. 159, 65-74. [Google Scholar]
- 36.East E. M. (1916) Genetics 1, 164-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright S. (1934) Genetics 19, 537-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stebbins G. L. (1959) Proc. Am. Philos. Soc. 103, 231-251. [Google Scholar]
- 39.Arnold M. L., (1997) Natural Hybridization and Evolution (Oxford Univ. Press, New York).
- 40.Grant V., (1981) Plant Speciation (Columbia Univ. Press, New York).
- 41.Appel O. & Al-Shehbaz, I. A. (2002) in The Families and Genera of Vascular Plants, ed. Kubitzki, K. (Springer, Berlin), Vol. 4, pp. 75–174. [Google Scholar]
- 42.Toledo J., Dehal, P., Jarrin, F., Hu, J., Hermann, M., Al-Shebaz, I. & Quiros, C. F. (1998) Ann. Bot. 82, 523-530. [Google Scholar]
- 43.Rieseberg L. H. (1995) Am. J. Bot. 82, 944-953. [Google Scholar]
- 44.Rieseberg L. H. (1997) Annu. Rev. Ecol. Syst. 28, 359-389. [Google Scholar]
- 45.Lee I., Wolfe, D. S., Nilsson, O. & Weigel, D. (1997) Curr. Biol. 7, 95-104. [DOI] [PubMed] [Google Scholar]
- 46.Busch M. A., Bomblies, K. & Weigel, D. (1999) Science 285, 585-587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.