Abstract
Gene duplication has been a major mechanism for increasing genomic complexity and variation during evolution. The evolutionary history of duplicated genes has been poorly studied along the vertebrate lineage. Here, we attempt to study that history by analyzing the expression of two members of the Snail family, Snail and Slug, in representatives of the major vertebrate groups. We find a surprising degree of variability in a subset of the expression sites for both genes in different species. Although some of the changes can be explained by neofunctionalization or subfunctionalization, others imply reciprocal changes in the expression of the two genes and the reappearance of expression in sites lost earlier in evolution. Because these changes do not fit easily into current models, we need to invoke additional mechanisms acting on enhancer elements to distribute expression domains and functions of duplicated genes unequally during evolution.
Keywords: DDC model, EMT, gene duplication
Multiple gene families contain several members in vertebrates when compared with a single gene in their closest invertebrate relative (amphioxus), which is indicative of large-scale duplications after the protochordate/craniate divergence (1–3). Under a simple adaptationist scenario, the preservation of duplicates would be based on the acquisition of a new function (neofunctionalization) or the existence of a dosage-dependent advantage. If the latter were the case, only a small fraction of duplicated genes would be present in two copies, with the vast majority of them being inactivated by the random accumulation of deleterious mutations. However, there is a high rate of retention of duplicated copies in vertebrate genomes.
In addition to neofunctionalization, a further mechanism has been proposed recently to account for the maintenance of gene duplicates. It implies the occurrence of complementary degenerative mutations causing the differential loss of independent regulatory elements in either duplicate. In this way, each copy will carry out unique nonredundant roles, with the original functions being distributed between the two daughter copies. This mechanism has been called subfunctionalization or partitioning of ancestral functions and has been formulated under the “duplication–degeneration–complementation” model (DDC) that justifies the high rate of duplicated gene preservation during vertebrate evolution (4, 5). A central requirement for this model is the modularity and dissociability of gene expression by means of discrete and separable enhancer elements responsible for spatially and temporally restricted transcription. The unequal distribution of active enhancers in the daughter copies of a given gene explains their differential expression.
The DDC model has been extensively examined in the recent extra-duplication events that occurred in the teleost lineage (6, 7) by comparing expression patterns of duplicated genes in zebrafish to single-copy genes in other vertebrates (8–11). However, in the case of duplications at the base of vertebrates (1–3), the comparison and study of how duplicated genes diverged is more complex because of the evolutionary distances that separate extant vertebrate and invertebrate species. To examine divergences following these ancient duplications, we have analyzed and compared two members of the Snail family, Snail and Slug, in different species representative of the major vertebrate groups.
Snail genes constitute an evolutionarily conserved superfamily of zinc-finger transcription factors composed of the Snail and Scratch families with central roles during embryonic development (12). Members of the Snail family are involved in mesoderm formation and neural development from Drosophila to mammals (13). In vertebrates, they are fundamental for the triggering of the epithelial–mesenchymal transition (EMT), a process that occurs in mesoderm and neural crest development and confers epithelial cells with migratory and invasive properties (13). In addition, Snail has been coopted for the triggering of EMT during the acquisition of malignancy in tumor progression (13, 14). Robust phylogenies for this gene family are available, as well as distinctive peptide motifs for each family member, making the assignment of any given gene straightforward (12).
Only one Snail gene is present in protochordates (15, 16), whereas at least two genes, Snail and Slug, have been found in all vertebrate species analyzed so far (12). After duplication, these genes distribute some of their functions. Only one gene of the pair is expressed in the dorsal neural tube or in the tail bud mesenchyme of chicken and mouse embryos (17), which are two sites of expression of the single ancestor Snail gene in amphioxus and ascidians (15, 16). However, this distribution of functions did not occur at a single point in vertebrates, as it has been shown that a subset of the early sites of expression and function of these two genes are interchanged between mouse and chick (17–20). Snail is the gene expressed in the mouse primitive streak and premigratory neural crest (pnc) cells, whereas in the chicken embryo, the gene expressed is Slug. Accordingly, Snail-null mutant mice, but not Slug mutants, die early at gastrulation because of defects in mesoderm formation and migration (19, 20). On the other hand, interfering with Slug expression in chicken embryos results in severe defects in neural crest and mesoderm development (18).
We have extended these observations to cartilaginous and bony fishes, reptiles, and turtles, and found a much higher degree of variability in domains of expression for this gene pair than expected. Distribution of functions between Snail and Slug has occurred at multiple points in vertebrate phylogeny, and evolutionary reversals are present. Previously uncharacterized mechanisms involving enhancer function must be invoked to explain this situation. Rare genomic rearrangements or epigenetic modification of enhancer function could be the causal explanations for our observations.
Materials and Methods
Embryos.
Scyliorhinus canicula embryos were kindly provided by Ramón Muñoz-Chápuli (University of Malaga, Malaga, Spain), who obtained them from local fishermen and raised them in the lab under running seawater to the desired stages. Freshly laid Mauremys caspica eggs were provided by Gervasio Martín-Partido (University of Badajoz, Badajoz, Spain; with permission for recollection from the Junta de Extremadura) and incubated at 28°C in the lab to the desired stages. Chelydra serpentina eggs were obtained from a commercial provider and incubated at 37°C. Embryos for both turtles were staged according to tables of normal development by Yntema (21). Eublepharis macularius embryos were dissected from freshly laid eggs provided by Oscar Campos (Madrid Zoo, Madrid), and zebrafish (Danio rerio) embryos were kindly provided by Qiling Xu [National Institute of Medical Research (London)].
PCR Amplifications.
PolyA+ RNA from S. canicula, E. macularius, or M. caspica embryos was isolated with Microfast Track isolation kit (Invitrogen), and Snail and Slug coding fragments were amplified by degenerate RT-PCR. PCRs were performed by two rounds of amplification of 30 cycles each at an annealing temperature of 50–55°C with the following primers: forward, ATGCCI(A/C)GI(A/T)(C/G)ITT(C/T)(C/T)TIGTIAA(A/G); reverse, GCIC(G/T)IA(A/G)(A/G)TTI(C/G)(A/T)IC(G/T)(A/G)TCIGC(A/G)AAAGC; nested forward, ATGCCI(A/C)GI(A/T)(G/C)ITT(C/T)TIGT; and nested reverse, AIGG(T/C)TT(T/C)TCICCIGT(A/G)TGIGT.
Sequence Analysis.
PCR products were cloned in the pGem-T easy (Promega) or the PCRII-TOPO (Invitrogen) vectors, sequenced, and compared by using the BLAST family of programs (www.ncbi.nlm.nih.gov/blast/; ref. 22). Sequence alignments were carried out by using CLUSTAL (23) and corrected by visual inspection. Sequence alignments of vertebrate Snail and Slug proteins and zebrafish EST, and pufferfish genomic DNA accession numbers are available in Fig. 8, which is published as supporting information on the PNAS web site, www.pnas.org.
In Situ Hybridization.
Snail and Slug amplified fragments were used as templates for in vitro transcription of DIG-11-UTP-labeled antisense RNA probes. In situ hybridizations were carried out on whole mounted embryos as described (24). After hybridization, embryos were fixed in 4% (wt/vol) paraformaldehyde in PBS, washed in PBS, and photographed in whole mount under a Leica M10 dissecting scope. Subsequently, they were gelatin-embedded, sectioned at 50 μm on a vibratome, cleared in 50% glycerol in PBS, mounted in the same solution, and photographed by using a Leica DMR microscope with Nomarski optics using an Olympus DP-10 digital camera. Snapping turtle (C. serpentina) embryos were used for expression studies because of their easier availability. Although scarce, the expression data obtained in M. caspica are identical to those shown for C. serpentina.
Results
Cloning and Identification of Snail and Slug Genes from Vertebrates.
In an effort to characterize Snail and Slug genes from a range of vertebrate species as broad as possible, we have cloned both genes by RT-PCR with degenerate primers from a cartilaginous fish (dogfish, Scyliorhinus canicula), a turtle (caspian turtle, Mauremys caspica), and a lizard (leopard gecko, Eublepharis macularius; Fig. 8). Furthermore, we have exhaustively analyzed zebrafish (D. rerio) and pufferfish (Takifugu rubripes) sequence databases to identify the full set of Snail genes in teleost fishes. Two Snail genes (snail1 and snail2) have been described in both fish species (25–28), and sequence and syntheny relationships clearly show that they are more closely related to the single Snail gene present in other vertebrates than to Slug. Data mining allowed us to identify ESTs for a single Slug gene in zebrafish and a genomic fragment containing its pufferfish homologue (Fig. 8). These searches, combined with cDNA library screenings in zebrafish, failed to identify a presumptive second Slug homologue. Therefore, we suggest that teleost fishes have two Snail genes and a single Slug gene. The comparison of the expression of these genes to that in mouse, chick, and Xenopus provides a phylogenetic range covering all mayor extant vertebrate groups.
Embryonic Expression of Snail and Slug in Vertebrates.
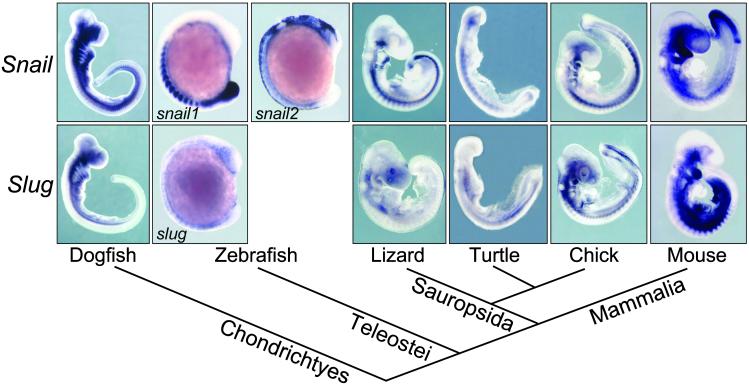
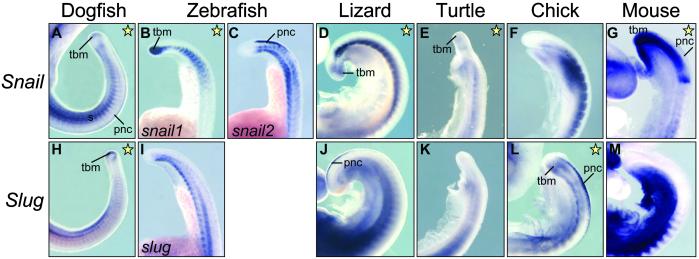
We analyzed by in situ hybridization Snail and Slug expression patterns in dogfish, zebrafish, turtle, and lizard embryos at different developmental stages and compared their distribution with that of the previously characterized mouse and chicken Snail and Slug (17, 18, 29, 30) and zebrafish snail1 and snail2 genes (Fig. 1; refs. 25 and 26). These genes are expressed at multiple sites during embryogenesis in all species analyzed. Among them are the premigratory and migratory neural crest, cranial mesenchyme, branchial arches, mesoderm, limb buds, and tail bud mesenchyme. In some territories, such as in the migratory neural crest and the branchial arches, there is a significant overlap between the expressions of the two genes. Other domains of expression have been unequally distributed among them, such as the pnc, the paraxial mesoderm, or the tail bud (see below). However, as shown in Fig. 1, the sum of Snail and Slug expression sites is conserved among all vertebrates. If we accept that the duplication event occurred at or very close to the origin of vertebrates (1), and we do not consider cases of neofunctionalization, we can assume that the single Snail family gene present in the predecessor of all vertebrates was expressed in these locations or their corresponding evolutionary precursors. Later, expression sites could be distributed between Snail and Slug after duplication and divergence in accordance with the DDC model.
Fig 1.
Phylogenetic relationships and whole mount in situ hybridization of Snail and Slug genes in different vertebrates at mid-embryonic stages. Vertebrate phylogeny is based on recent studies that place turtles as a sister group of avians and not as basal sauropsids (43, 44).
Conserved Unique Expression Domains of Snail and Slug in Vertebrates.
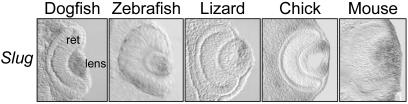
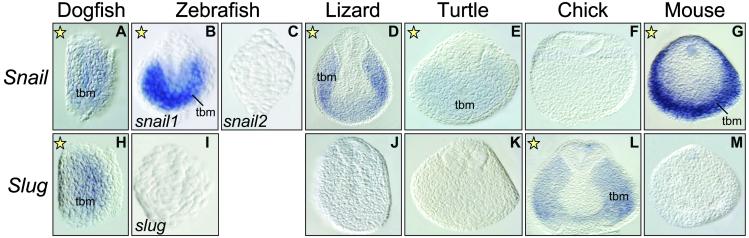
In our expression analysis, we could also detect specific domains for either Slug or Snail. Slug genes from all of the species examined are expressed in the lens at early developmental stages (Fig. 2), whereas Snail genes are not expressed in this tissue at these stages (not shown). The lens can be considered a vertebrate-specific character, derived from a nonneurogenic ectodermal placode and absent in other chordates (31). Therefore, this expression site could have been newly acquired before the divergence of cartilaginous fishes and, very likely, concomitantly with the appearance of this structure.
Fig 2.
Expression of Slug in the developing lens of vertebrates at early optic cup stages. ret, retina.
On the other hand, we could also detect asymmetric Snail expression on the right side of the lateral plate mesoderm in turtle embryos (not shown), similar to what has been described for mouse and chick embryos (17, 32). This expression is a very transient one in all three species and is what precluded us from detecting it in the other vertebrates used in this study. The amphioxus homologue of Ptx, a putative Snail downstream target (13), shows a conserved asymmetric expression (33), but this asymmetry has not been examined for amphioxus Snail itself (16). However, left-right asymmetric expression has been found for one snail homologue in the mollusk Patella vulgata (34). Thus, asymmetric expression might have been present before gene duplication at the base of vertebrates, and later on retained only by the Snail gene.
Variability of Snail and Slug Expression in the Neural Crest and the Tail Bud.
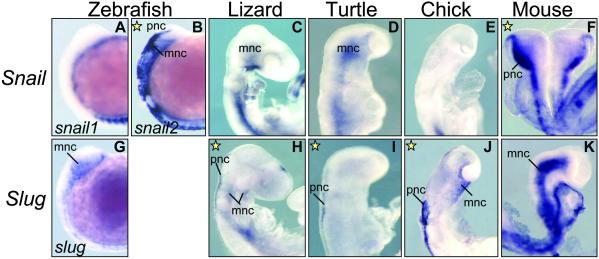
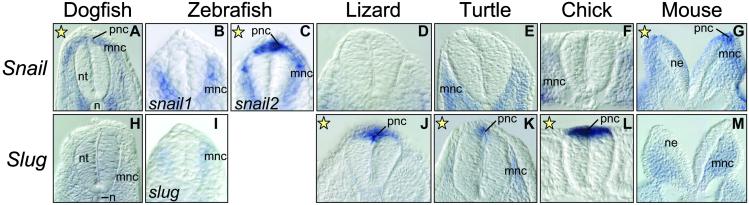
As for other expression sites, we noticed an unusual rate of variability in the expression of Snail and Slug genes in the pnc (Figs. 3 and 4) and in the tail bud mesenchyme (Figs. 5 and 6), which is indicative of a high plasticity and modularity in the regulation of these genes. Snail is expressed in the pnc of zebrafish (snail2) and mouse (Fig. 3 B and F) and in dogfish embryos (Fig. 4A). Slug is expressed in this population in the sauropsid lineage (lizard, turtle, and chick; Fig. 3 H–J). This finding can be better assessed in the sections taken at the trunk level and shown in Fig. 4. This observation suggests that a swapping in Snail and Slug expression occurred after the divergence of sauropsids and mammals. A different situation is observed in the tail bud mesenchyme. Both Snail and Slug are expressed in the undifferentiated tail tip mesenchyme of dogfish embryos (Fig. 5 A and H, and sections in Fig. 6 A and H), whereas nly Snail is expressed in the tail bud of zebrafish (snail1), lizard, turtle, and mouse embryos (Figs. 5 B, D, E, and G, and the corresponding sections in Fig. 6). Conversely, the chicken tail bud only expresses Slug (Figs. 5L and 6L). Thus, in this tissue, the silencing of Snail and activation of Slug is specific to the avian lineage. It is noteworthy that in zebrafish, expression in the pnc and tail bud has been distributed between snail1 and snail2 (Figs. 4 B and C and 6 B and C), whereas the single slug gene is not expressed in either (Figs. 4I and 6I). Therefore, we detect changes in the distribution of expression domains of Snail genes that can be mapped at precise points in the phylogeny of vertebrates.
Fig 3.
Whole-mounted embryos showing Snail and Slug expression in the cranial premigratory neural crest (pnc). Snail is the gene expressed in the pnc of zebrafish (snail2) and mouse embryos, whereas in lizard, turtle, and chicken embryos, Slug is the gene present in this tissue (yellow stars). mnc, migratory neural crest.
Fig 4.
Sections through the trunk region showing expression of Snail: (A) dogfish; (C) zebrafish snail2; (G) mouse; and expression of Slug: (J) lizard; (K) turtle; and (L) chick in the pnc. Yellow stars indicate those genes expressed in this population in each species. n, notochord; ne, neural epithelium; nt, neural tube.
Fig 5.
Close view of Snail and Slug expression patterns in the tails of whole-mounted embryos. Yellow stars indicate which gene is present in the tail bud mesenchyme in each species. tbm, tailbud mesenchyme.
Fig 6.
Sections through the tail bud showing expression of both Snail and Slug in dogfish (A and H), but only Snail in zebrafish (snail1, B), lizard (D), turtle (E), and mouse (G), or Slug in chick (L) in the mesenchyme. Yellow stars indicate those genes expressed in this population in each species.
Discussion
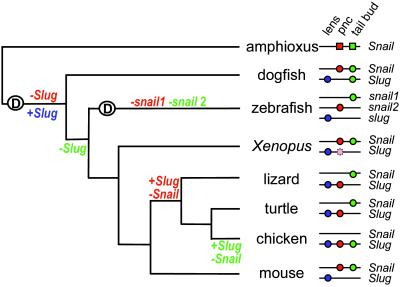
A model to explain the large-scale retention of duplicated genes has been proposed wherein quick divergence accompanied by the novel acquisition of an adaptive advantage (neofunctionalization) and/or the partitioning of the ancestral gene functions between copies (subfunctionalization) leads to positive selection and preservation of the duplicates (DDC model; refs. 4 and 5). Degenerative mutations have been regarded as the main mechanism behind this model (5), and experimental evidence proves so for both exons (11, 35) and cis-regulatory elements (8) in duplicated zebrafish genes. Specific expression of Slug in the lens and the distribution of expression domains between zebrafish snail1 and snail2 (Fig. 7) would fit within this scenario.
Fig 7.
Evolutionary history and schematic representation of the distribution of functional enhancers for Snail and Slug expression in the lens (blue), pnc (red), and tail bud mesenchyme (green) across vertebrate species. Xenopus Slug (light red) is expressed in pnc but at a later stage than Snail (45); Snail is the gene expressed in the tail bud (R. Mayor, personal communication). Amphioxus is shown as an outgroup and is considered to be the ancestral condition before Snail duplication; squares and circles denote enhancers driving expression to the lens, the dorsal neural tube, and the tail bud.
The observed variation in the expression of the two genes in the pnc and tail bud mesenchyme indicates that (i) an event that must have occurred shortly after gene duplication selected Snail expression in the pnc; (ii) a second event led to the interchange between Snail and Slug expression in this territory in sauropsids; (iii) after an initial phase of coexpression in cartilaginous fishes, and before the divergence of bony fishes, Slug lost its expression in the tail bud; and (iv) a subsequent event, specific to the avian lineage, led to Snail disappearance and Slug activation in the tail bud (Fig. 7).
This picture cannot be explained simply by selective loss of Slug enhancer(s) responsible for pnc or tail bud expression after gene duplication and divergence, as we need to account for the reappearance of these elements in the sauropsids or avian lineage. In this case, we would need to invoke extremely unlikely events as the reversal of degenerative mutations or “de novo” creation of tissue-specific regulatory elements to account for the reappearance of these expression domains after their loss at earlier stages in evolution. Although such events may occur over short time-scales (36), it has such a low probability in our situation that it could be described as an evolutionary reversal, in the sense prohibited by Dollo's law on the irreversibility of evolutionary changes (36). A possibility we cannot formally rule out is that ancestors of all of the species analyzed retained expression of both genes in pnc and tail bud, and that independent enhancer loss by mutation occurred in each lineage. In this case, the distribution of expression patterns we observe would not reflect common descent, but random variation. Once more, this is an extremely unlikely scenario, where in all ancestors both duplicated genes were necessarily expressed at all sites simultaneously.
Other possible mechanisms to explain our set of observations include changes in the genomic structure of these genes or epigenetic modifications of enhancer activity. In the first case, chromosomal interchange between Snail and Slug loci could have determined a redistribution of the enhancer elements responsible for driving these sites of expression. Mapping information shows that two genes that lie at opposite sides of the Snail locus in humans and mouse (HCK and BMP7; Fig. 9, which is published as supporting information on the PNAS web site) are linked in the chicken genome (37). This observation excludes the possibility of a single translocation or noncanonical recombination event in the avian lineage but does not rule out a double crossing-over, interchange, or gene conversion (38). In this last scenario, these events must have occurred twice in vertebrate evolution and affected different cis-regulatory elements, once on the pnc enhancer(s) in the sauropsid lineage and another on the tail bud enhancer(s) in the avian lineage.
As for epigenetic changes, we can consider that pnc and tail bud enhancers are present and conserved in both Snail and Slug genes of all species but that, in specific contexts, one of them is functionally dormant. To be structurally maintained, they would need to serve additional functions during development or adult life. Methylation, chromatin structure, or the availability of upstream regulators could be causal explanations for this situation. Lifting of these silencing mechanisms on Slug pnc and tail bud enhancers would result in their reappearance in the sauropsid or avian lineage, respectively. Functional equivalence between Snail and Slug (39) would permit the concomitant disappearance of Snail in these territories with no detrimental effect.
The possibilities described above do not enter into conflict with the current DDC model, but extend the range of mechanisms that can act on enhancer elements during evolution to distribute expression domains and functions of duplicated genes unequally. Both hypotheses, changes in genomic structure or epigenetic modifications, can be tested once the nature and structure of the cis-control elements responsible for Snail and Slug expression are characterized. Sequence comparisons such as those recently carried out for the HoxA cluster (40) and cross-species transgenic assays for enhancer activity in mice with genomic sequences from other species (most importantly chicken) can provide answers to the accuracy of the different predictions made by each hypothesis.
The present analysis of Snail/Slug duplicated genes during vertebrate evolution reveals a much higher degree of plasticity and complexity than expected and highlights the risk of using expression or function as phylogenetic characters when studying the evolution of gene families. This analysis is also a clear demonstration that the irreversible loss (or modification) in the activity of cis-regulatory elements is not a fixed feature of duplicated gene history, and that rare and not parsimonious events can be observed more than once in the evolution of a single genetic regulatory system. This work also underscores the importance of comparative studies of gene expression during development, not only between distantly related model systems, but also over a broad range of more closely related species (9, 41, 42).
Supplementary Material
Acknowledgments
We thank everyone who provided eggs and embryos or suggested where to find them. We thank Christine and Bernard Thisse for the gift of zebrafish snail1 and snail2 probes, Roberto Mayor for communicating unpublished results, Concepción Azuara for excellent technical assistance, Rafael Zardoya and laboratory members for discussion and encouragement, and Michalis Averof, Alberto Ferrús, Jordi García-Fernández, and Pat Simpson for comments on the manuscript. This work was funded by the Spanish Ministries of Education and Culture (Grant PM98-0125), Health (Grant FIS-01/985), and Science and Technology (Grant BMC2002-0383), and by the Comunidad Autónoma de Madrid (Grant CAM 08.1/0044/2000 to M.A.N.). A.L. was supported by a postdoctoral fellowship from the Spanish Secretary of Education and Universities and the European Social Fund.
Abbreviations
DDC, duplication–degeneration–complementation model
pnc, premigratory neural crest
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Furlong R. F. & Holland, P. W. H. (2002) Philos. Trans. R. Soc. London B 357, 531-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLysaght A., Hokamp, K. & Wolfe, K. (2002) Nat. Genet. 31, 200-204. [DOI] [PubMed] [Google Scholar]
- 3.Gu X., Wang, Y. & Gu, J. (2002) Nat. Genet. 31, 205-209. [DOI] [PubMed] [Google Scholar]
- 4.Force A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L. & Postlethwait, J. (1999) Genetics 151, 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch M. & Force, A. (2000) Genetics 154, 459-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Postlethwait J. H., Yan, Y. L., Gates, M. A., Horne, S., Amores, S., Brownlie, A., Donovan, A., Egan, E. S., Force, A., Gong, Z., et al. (1998) Nat. Genet. 18, 345-349. [DOI] [PubMed] [Google Scholar]
- 7.Robinson-Rechavi M., Marchand, O., Escriva, H. & Laudet, V. (2001) Curr. Biol. 11, R458-R459. [DOI] [PubMed] [Google Scholar]
- 8.McClintock J. M., Kheirbek, M. A. & Prince, V. E. (2002) Development (Cambridge, U.K.) 129, 2339-2354. [DOI] [PubMed] [Google Scholar]
- 9.Schilling T. F. & Knight, R. D. (2001) Philos. Trans. R. Soc. London B 356, 1599-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland P. W. H. (1999) Cell Dev. Biol. 10, 541-547. [DOI] [PubMed] [Google Scholar]
- 11.Lister J. A., Close, J. & Raible, D. W. (2001) Dev. Biol. 237, 333-344. [DOI] [PubMed] [Google Scholar]
- 12.Manzanares M., Locascio, A. & Nieto, M. A. (2001) Trends Genet. 17, 178-181. [DOI] [PubMed] [Google Scholar]
- 13.Nieto M. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 155-166. [DOI] [PubMed] [Google Scholar]
- 14.Blanco M. J., Moreno-Bueno, G., Sarrio, D., Locascio, A., Cano, A., Palacios, J. & Nieto, M. A. (2002) Oncogene 21, 3241-3246. [DOI] [PubMed] [Google Scholar]
- 15.Corbo J. C., Erives, A., Di Gregorio, A., Chang, A. & Levine, M. (1997) Development (Cambridge, U.K.) 124, 2335-2344. [DOI] [PubMed] [Google Scholar]
- 16.Langeland J. A., Tomsa, J. M., Jackman, W. R., Jr. & Kimmel, C. B. (1998) Dev. Genes Evol. 208, 569-577. [DOI] [PubMed] [Google Scholar]
- 17.Sefton M., Sanchez, S. & Nieto, M. A. (1998) Development (Cambridge, U.K.) 125, 3111-3121. [DOI] [PubMed] [Google Scholar]
- 18.Nieto M. A., Sargent, M., Wilkinson, D. G. & Cooke, J. (1994) Science 264, 835-839. [DOI] [PubMed] [Google Scholar]
- 19.Jiang R., Lan, Y., Norton, C. R., Sundberg, J. P. & Gridley, T. (1998) Dev. Biol. 198, 277-285. [PubMed] [Google Scholar]
- 20.Carver E. A., Jiang, R., Lan, Y., Oram, K. F. & Gridley, T. (2001) Mol. Cell. Biol. 21, 8184-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yntema C. L. (1968) J. Morphol. 125, 219-251. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 23.Thompson J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieto M. A., Patel, K. & Wilkinson, D. G. (1996) Methods Cell Biol. 51, 219-235. [DOI] [PubMed] [Google Scholar]
- 25.Thisse C., Thisse, B., Schilling, T. F. & Postlethwait, J. H. (1993) Development (Cambridge, U.K.) 119, 1203-1215. [DOI] [PubMed] [Google Scholar]
- 26.Thisse C., Thisse, B. & Postlethwait, J. H. (1995) Dev. Biol. 172, 86-99. [DOI] [PubMed] [Google Scholar]
- 27.Smith S., Metcalfe, J. A. & Elgar, G. (2000) Gene 247, 119-128. [DOI] [PubMed] [Google Scholar]
- 28.Smith S. F., Snell, P., Gruetzner, F., Bench, A. J., Haaf, T., Metcalfe, J. A., Green, A. R. & Elgar, G. (2002) Genome Res. 12, 776-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith D. E., Del Amo, F. F. & Gridley, T. (1992) Development (Cambridge, U.K.) 116, 1033-1039. [DOI] [PubMed] [Google Scholar]
- 30.Nieto M. A., Bennet, M. F., Sargent, M. G. & Wilkinson, D. G. (1992) Development (Cambridge, U.K.) 116, 227-237. [DOI] [PubMed] [Google Scholar]
- 31.Baker C. V. & Bronner-Fraser, M. (2001) Dev. Biol. 232, 1-61. [DOI] [PubMed] [Google Scholar]
- 32.Isaac A., Sargent, M. G. & Cooke, J. (1997) Science 275, 1301-1304. [DOI] [PubMed] [Google Scholar]
- 33.Yasui K., Zhang, S., Uemura, M. & Saiga, H. (2000) Development (Cambridge, U.K.) 127, 187-195. [DOI] [PubMed] [Google Scholar]
- 34.Lespinet O., Nederbragt, A. J., Cassan, M., Dictus, W. J., Van Loon, A. E. & Adoutte, A. (2002) Dev. Genes Evol. 212, 186-195. [DOI] [PubMed] [Google Scholar]
- 35.Altschmied J., Delfgaauw, J., Wilde, B., Duschl, J., Bouneau, L., Volff, J. N. & Schartl, M. (2002) Genetics 161, 259-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall C. R., Raff, E. C. & Raff, R. A. (1994) Proc. Natl. Acad. Sci. USA 91, 12283-12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groenen M. A., Cheng, H. H., Bumstead, N., Benkel, B. F., Briles, W. E., Burket, T., Burt, D. W., Crittenden, L. B., Dodgson, J., Hillel, J., et al. (2000) Genome Res. 10, 137-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dover G. (2000) BioEssays 22, 1153-1159. [DOI] [PubMed] [Google Scholar]
- 39.del Barrio M. G. & Nieto, M. A. (2002) Development (Cambridge, U.K.) 129, 1583-1593. [DOI] [PubMed] [Google Scholar]
- 40.Chiu C.-H., Amemiya, C., Dewar, K., Kim, C.-B., Ruddle, F. H. & Wagner, G. P. (2002) Proc. Natl. Acad. Sci. USA 99, 5492-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arthur W. (2002) Nature 415, 757-764. [DOI] [PubMed] [Google Scholar]
- 42.Skaer N., Pistillo, D., Gibert, J. M., Lio, P., Wülbeck, C. & Simpson, P. (2002) Trends Genet. 18, 399-405. [DOI] [PubMed] [Google Scholar]
- 43.Zardoya R. & Meyer, A. (1998) Proc. Natl. Acad. Sci. USA 95, 14226-14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hedges S. B. & Poling, L. L. (1999) Science 283, 998-1001. [DOI] [PubMed] [Google Scholar]
- 45.Linker C., Bronner-Fraser, M. & Mayor, R. (2000) Dev. Biol. 224, 215-225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.