Abstract
The extensive infraabdominal (iab) region contains a number of cis-regulatory elements, including enhancers, silencers, and insulators responsible for directing the developmental expression of the abdominal-A and Abdominal-B homeotic genes at the Drosophila bithorax complex. It is unclear how these regulatory elements are primed for activity early in embryogenesis, but the 100-kb intergenic region is subject to a complex transcriptional program. Here, we use molecular and genetic methods to examine the functional activity of the RNAs produced from this region and their role in cis regulation. We show that a subset of these transcripts demonstrates a distinct pattern of cellular localization. Furthermore, the transcripts from each iab region are discrete and the transcripts do not spread across the insulator elements that delineate the iab regions. In embryos carrying a Mcp deletion, the intergenic transcription pattern is disrupted in the iab4 region and the fourth abdominal segment is transformed into the fifth. We propose that intergenic transcription is required early in embryogenesis to initiate the activation of the Drosophila bithorax complex and define the domains of activity for the iab cis-regulatory elements. We also discuss a possible mechanism by which this may occur.
Expression of the abdominal-A (abdA) and Abdominal-B (AbdB) homeotic genes from the bithorax complex (BX-C) in Drosophila is regulated by cis-regulatory elements in a 100-kb intergenic region (1–3). This region harbors an organized series of infraabdominal (iab) regions (Fig. 1A). Genetic, transgenic, and molecular studies have identified a number of classes of distinct cis-regulatory elements within the iab regions capable of directing abdA and AbdB expression. Insulator DNAs have been identified at the boundaries of the iab8-iab7 domains (Frontabdominal-8, Fab8) (4), iab7-iab6 domains (Fab7) (5–10), and iab5-iab4 domains (Miscadastral pigmentation, Mcp) (Fig. 1A) (7, 11). Although not all insulators are thought to act in the same way (12, 13), it has been suggested that the BX-C insulators organize the different iab regions into discrete chromatin domains (Fig. 1A) (8, 14, 15). In this way, cis-regulatory elements within the separate domains are able to direct expression of abdA and AbdB in specific abdominal segments of the Drosophila embryo. Indeed, transgenic studies have identified enhancers from the iab8 (IAB8) (4), iab7 (IAB7) (4), and iab5 (IAB5) (16, 17) regions that drive expression in specific abdominal segments. Furthermore, elements capable of silencing gene transcription, Polycomb response elements, have been discovered in the iab8 (4), iab7 (10, 18), and iab5 regions (17, 19). It is likely that analogous cis-regulatory elements, capable of directing segment-specific gene expression, are present in the other iab regions but remain to be characterized. Although many of the transcription factors that are involved in regulation of abdA and AbdB expression have been identified (1, 18, 20–24), it is not known how the iab cis-regulatory elements are activated in specific domains along the anteroposterior axis early in embryogenesis and ultimately are able to define unique segmental patterns of expression for the abdA and AbdB genes.
Fig 1.
Cellular localization of BX-C transcripts. (A) Summary of abdA-AbdB locus. The abdA and AbdB transcription start sites are indicated by leftward arrows. The intergenic region is ≈100 kb in length. The iab regions that control expression of the two Hox genes are indicated (IAB2 to IAB8). IAB2, IAB3, and IAB4 (shown in green) regulate expression of abdA. IAB5, IAB6, IAB7, and IAB8 (shown in yellow) direct AbdB expression. The insulator DNAs (black ellipses) separate the different iab regions. The presumptive Fab6 insulator has yet to be identified (gray ellipse). Characterized enhancers within the iab regions are shown as blue rectangles. The location of probes used for in situ hybridization analysis in this study are shown as black bars under the locus. Sense transcripts (s), relative to the direction of AbdB transcription, are detected by antisense probes, and antisense transcripts (as) are detected by sense probes. (B and C) iab6as expression pattern at the surface of blastoderm stage 5 embryos. Hybridization to the transcripts appears uniform throughout the cells in which they are expressed. (D and E) Sense transcripts from the IAB8 enhancer sequence are visible as discrete foci in the cells at the surface of blastoderm stage 5 embryos (D). At higher magnification, two foci can be seen in each expressing cell at the focal plane (E). (F and G) AbdB expression in stage 9 embryos visualized by confocal microscopy. The transcript (red) is excluded predominantly from the nucleus (green) and distributed throughout the cellular cytoplasm (G). (H and I) IAB8 sense transcripts (red) are localized at the nuclear periphery and not distributed in the cytoplasm of the cell (I).
Previous studies have shown that a complex transcriptional program exists within the iab regions themselves (4, 22, 25, 26). However, the function of these transcripts is elusive. In this study, we have used molecular and genetic methods to examine the role of the intergenic transcription in regulating the activation of the iab cis elements in the early embryo. In blastoderm-stage embryos, the intergenic RNAs exhibit a distinct pattern of cellular localization. Transcription is restricted to individual iab regions as the transcripts do not cross the insulator elements at the boundaries that separate the regulatory regions. Mutant embryos with a deletion of the Mcp sequence have a disruption in intergenic transcription in the iab4 region, and, subsequently, their fourth abdominal segment is transformed into the fifth. We discuss these findings in relation to regulation of homeotic gene expression at the BX-C and propose a model by which the intergenic transcripts function.
Materials and Methods
Whole-Mount in Situ Hybridization.
Probes from the BX-C were PCR-amplified by using Drosophila yw67 adult genomic DNA as a template. The DNA probes were cloned into pGEMT-Easy (Promega). Sense and antisense riboprobes (relative to the direction of abdA and AbdB transcription; see Fig. 1) were prepared by using a digoxigenin (DIG) RNA-labeling kit (Roche, Gipf-Oberfrick, Switzerland). PCR primer sequences and positions in BX-C (27) were as described previously (26).
In situ hybridizations were carried out on 2- to 4-h and 0- to 10-h yw67 and homozygous Mcp mutant embryos (28) as described previously (16). Fluorescent in situ hybridizations were performed as described previously (29), using an anti-DIG-rhodamine Fab fragments antibody (Roche). In situ hybridizations were repeated at least three times.
Transgenic Drosophila Assay.
A modified pCasPer vector (16, 30) was used to construct the abdA-lacZ-IAB5 (pA5) transgene. The previously described 1-kb genomic IAB5 enhancer was inserted into the PstI site, which is located 3′ of the lacZ coding region in pCasPer (16). The abdA promoter was inserted into the AscI–BamHI sites of pCasPer-IAB5 as a 1-kb genomic fragment (S. Ohtsuki, personal communication). The promoter sequence extends ≈1 kb 3′ from nucleotide position 152853 in the published BX-C sequence (27), across the abdA transcriptional start site (−538 to approximately +462). The pA5 vector was introduced into the Drosophila germ line by injecting yw67 embryos as described previously (30).
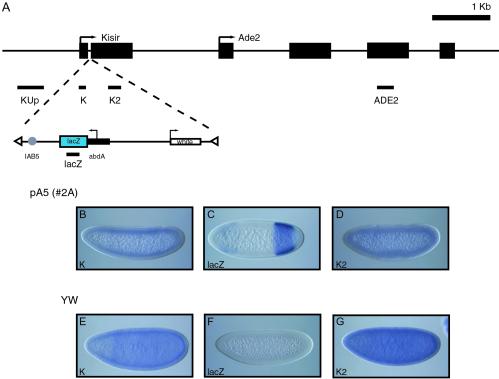
Inverse PCR mapping of the genomic insertion site for a number of independent pA5 transgenic lines was carried out essentially as described previously [Berkeley Drosophila Genome Project (BDGP), www.fruitfly.org/about/methods/inverse.pcr.html]. PCR products were cloned into pGEMT-Easy (Promega) and sequenced by using T7 and SP6 primers. The insertion site for line pA5 2A was identified as nucleotide position 102422 on a sequenced BAC clone (BDGP, BAC clone BACR10M11, EM:AC092216) in the first intron of the Kisir transcriptional unit, located on chromosome 2L. The Kisir predicted transcriptional start is at 101910 (512 bp 5′ of the transgene insertion site), and the transcriptional start site of the neighboring ade2 gene is at 104463 (2,041 bp 3′ of the insertion site) (see Fig. 2A).
Fig 2.
IAB5 enhancer activity at an ectopic locus. (A) Summary of transgene insertion locus on chromosome 2L. The direction of transcription for the endogenous Kisir transcription unit and ade2 gene (arrows) and exon structure (black boxes) are shown. The pA5 transgene is inserted in the Kisir intron. The orientation of the transgene results in the abdA promoter (black rectangle) directing transcription of the lacZ coding sequence (blue rectangle) toward the 5′ exon of Kisir. The IAB5 enhancer (gray circle) is located 512 bp 3′ of the predicted Kisir transcription start site and ≈3 kb 5′ of the abdA promoter. The insertion site was mapped by using inverse PCR with primers from the P element ends (white triangles). The miniwhite reporter gene (white rectangle) is transcribed in the opposite direction from lacZ and is not activated by the IAB5 enhancer. The probes used for in situ hybridization analysis are shown as black bars below the insertion locus or transgene. (B–G) Transgenic and yw67 blastoderm-stage embryos hybridized with antisense RNA probes. Probes against Kisir exon 1 detect a weak expression pattern in transgenic embryos (B and D) that is indistinguishable from the pattern in nontransgenic embryos (E and G). A probe against lacZ exhibits a strong posterior band typical of IAB5 enhancer-directed expression in transgenic embryos (D) (16), which is absent in nontransgenic embryos (F). No expression is detectable from the region upstream of Kisir (Kup) or from exon 3 of ade2 (ADE2) in transgenic or nontransgenic blastoderm-stage embryos (data not shown).
Probes from the chromosomal regions adjacent to the insertion locus were PCR-amplified by using Drosophila yw67 adult genomic DNA as a template. The DNA probes were cloned into pGEMT-Easy (Promega). Sense and antisense riboprobes (relative to the direction of Kisir and ade2 transcription; see Fig. 2A) were prepared by using a DIG RNA-labeling kit (Roche). PCR primer sequences and positions on the BAC clone described above were as follows: KUp s, 5′-GAA GAG CAG GTT GCC TTT CAG C-3′ (101330); KUp as, 5′-GGT GTC GTA GAA CTT GTA GCC C-3′ (101833); K s, 5′-ATA CCC GAG TCA CGC TAT GC-3′ (102014); K as, 5′-GAG AAA ATG AAA GTG CCG CAG G-3′ (102419); K2 s, 5′-ATG AAA ATC TGG ACA TCG GAG C-3′ (102717); K2 as, 5′-TTC CTT CCA GTT CAG CCA TTC G-3′ (103628); ADE2 s, 5′-ATC TGG AAC AGC CGT TGA ATG C-3′ (106825); and ADE2 as, 5′-TCT TGT AGA GCA CAC GGA GTG G-3′ (108072).
Embryos were collected, fixed, and hybridized with these DIG-labeled RNA probes and with a lacZ antisense RNA probe as described previously (16).
RNA Isolation and RT-PCR.
Total RNA was isolated from 0- to 4-h yw67 wild-type and homozygous Mcp mutant (28) Drosophila embryos by using Trizol (Life Technologies) according to the manufacturer's instructions. RT-PCR was performed with the Qiagen One-Step RT-PCR kit, using primers described above, and the PCR products were electrophoresed on 1% agarose gels.
Results
Cellular Localization of Transcripts from the BX-C.
Comparison of the cellular distribution of the sense and antisense transcripts from the iab regions at the BX-C revealed a significant difference between the two. The early antisense transcripts from iab4 and iab6 are distributed uniformly throughout the cytoplasm of the cells in which they are expressed, as shown for iab6as (Fig. 1 B and C). In contrast, all of the iab sense transcripts appear as discrete foci within each cell. This localization is particularly clear when the in situ hybridization signal is photographed at the surface of the blastoderm embryo, as shown for iab8s (Fig. 1 D and E). The nuclear foci appear in duplicate within single cells, presumably because transcripts are originating from both of the diploid chromosomes. We also examined localization of the transcripts by confocal microscopy using fluorescent in situ hybridization. At stage 9 of development, AbdB is expressed predominantly in abdominal segments 7, 8, and 9 (Fig. 1F). At higher magnification, the AbdB mRNA appears to be widely distributed in the cytoplasm of cells in which it is expressed (Fig. 1G). In contrast, the iab8 sense transcript is visualized as discrete foci that appear to be restricted to a predominantly perinuclear location (Fig. 1I). Earlier studies also noted this nuclear localization of the transcripts within the cell (25). It is conceivable that these different cellular localization patterns are indicative of different functional roles for the sense and antisense orientation transcripts.
IAB5 Enhancer Selectively Activates a Hox Gene Promoter.
It is a formal possibility that the intergenic iab RNAs are transcribed because of enhancer-mediated activation of cryptic promoters within the iab regions and, therefore, may not serve a functional role in vivo. To test this idea, we generated several independent transgenic lines carrying an IAB5 enhancer-containing transgene (pA5) (Fig. 2A) to test promoter specificity. We identified the exact genomic insertion site in these lines by inverse PCR and identified one potentially informative line (Fig. 2A). In this line, the pA5 transgene is inserted in a small intron between the two exons of a short noncoding RNA, named Kisir, on chromosome 2L (Fig. 2A) (Berkeley Drosophila Genome Project, www.flybase.org). In blastoderm-stage embryos, we assayed for IAB5-driven transcription of lacZ, Kisir, the genomic region 5′ of Kisir, and the neighboring ade2 gene (see Fig. 2A). The probes against the Kisir upstream region, Kisir and ade2, all detect transcription patterns indistinguishable from those in nontransgenic embryos (Fig. 2). Conversely, the lacZ probe detects a posterior band of expression typical of IAB5 enhancer-driven transcription (Fig. 2C). Therefore, it appears that the IAB5 enhancer can selectively direct expression from the abdA promoter in a complex chromosomal environment early in embryogenesis, rather than promiscuously activating adjacent cis promoters. Consequently, it might be expected that at the endogenous locus, the only targets for IAB5 and the other iab enhancers are the promoters of the abdA and AbdB genes and not the promoters from which intergenic transcription is fired. This idea is supported by our observation that intergenic transcription occurs before the time when the iab enhancers begin to direct expression of the protein-coding Hox genes in the embryo.
Intergenic Transcription Is Blocked at Insulators.
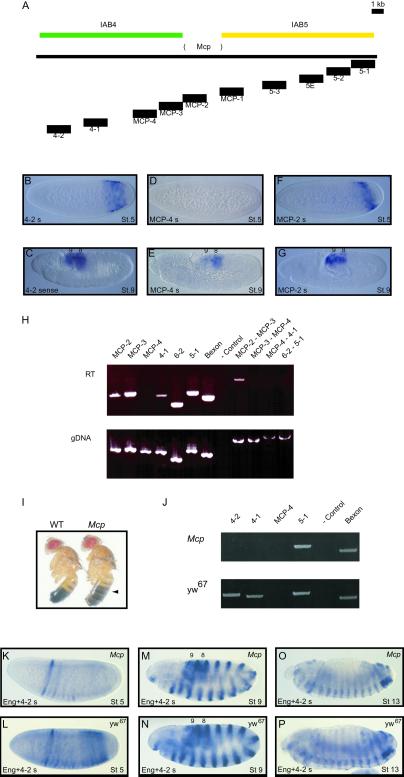
The different embryonic domains of sense transcription that have been characterized from the iab regions led us to hypothesize that each iab region may contain a discrete transcription unit. One prediction of this hypothesis is that a transcript originating in one iab region should not spread to the neighboring iab region. To test this prediction, we examined transcription across the boundaries of the iab5-iab4 and iab6-iab5 regions. The iab5 and iab4 domains are separated by the presumptive Mcp insulator element, which has been defined by chromosomal lesions (28) and molecular analysis (7, 11, 19) (Fig. 3A). In blastoderm-stage embryos, probes from chromosomal regions within the Mcp sequence demonstrate the characteristic iab5 pattern of transcription (Fig. 3F, MCP-2). However, the MCP-4 probe fails to detect transcription (Fig. 3D), whereas probes mapping closer to the abdA gene detect an iab4 transcription pattern (Fig. 3B, 4–2). These observations suggest that early transcription is blocked on the abdA side of the Mcp insulator element and does not spread from iab5 into iab4. Later in development, transcription is detected by all three probes in abdominal segments 8 and 9 (Fig. 3 C, E, and G).
Fig 3.
Mcp genomic region regulates transcription. (A–H) Intergenic transcription is blocked at insulator DNAs. (A) Genomic Mcp region. The 3-kb deletion in the Mcp mutant (28) is shown between the iab4 (green) and iab5 (yellow) regulatory regions. The locations of expected RT-PCR products and in situ hybridization probes are shown below the line. (B–G) Transcription across the Mcp insulator sequence detected by in situ hybridization. In blastoderm stage 5 embryos, sense transcripts can be detected in the iab4 region (4-2; B) and, in the characterized Mcp sequence (MCP-2; F), with the transcription pattern at MCP-2 restricted farther toward the posterior of the embryo. No transcription is detected from probe MCP-4 (D), which maps adjacent to the characterized Mcp sequence early in development. All three probes detect expression in the posterior abdominal segments at stage 9 (C, E, and G). (H) RT-PCR detection of transcription at insulator sequences. PCR amplifications were performed in parallel on reverse-transcribed total RNA (RT) and genomic DNA (gDNA) from 0- to 4-h embryos. Transcription could be detected with primer pairs within the characterized Mcp element (MCP-2 and MCP-3) and the iab4 region (4-1), but not at the intervening MCP-4 sequence. No product was detected from the RT cDNA sample when amplified from MCP-3 to MCP-4 or from MCP-4 to 4-1, suggesting that the iab4 (4-1) and Mcp (MCP2/3) transcripts are not joined. However, transcription could be detected with primers from MCP-2 into MCP-3, suggesting that they are part of the same transcription unit. No product was detected from 6-2 to 5-1, although these sequences are transcribed independently, indicating that transcription is blocked at the intervening region. The expected size products were obtained with all primer pairs from gDNA. Control lanes are RT− (absence of reverse transcriptase) for RT and water for gDNA (−Control). (I–P) Mcp deletion results in disrupted transcription in the iab4 region. (I) Mcp mutant phenotype. In contrast to wild-type adult males, the A4 segment is pigmented in homozygous Mcp mutants (black arrow), indicating that the fourth abdominal segment is transformed into a more posterior abdominal segment. (J) PCR amplifications were performed in parallel on reverse-transcribed total RNA from homozygous Mcp mutant (Mcp) and wild-type yw67 0- to 4-h embryos. No transcription could be detected in the Mcp mutant with primer pairs from the iab4 region (4-1 and 4-2), although the iab5 (5-1) and AbdB (Bexon) transcripts are present. In contrast, transcription in the iab4 region was readily detectable in wild-type embryos. Negative control lane is RT−. (K–P) Transcription in Mcp mutant and yw67 embryos detected by in situ hybridization with engrailed and iab4s probes. In blastoderm stage 5 embryos, sense transcripts can be detected in the iab4 region and in the anterior engrailed stripe pattern in yw67 embryos (L). The iab4s transcript is absent in the Mcp mutant embryos (K). The transcription patterns detected at stages 9 and 13 in Mcp (M and O) and yw67 (N and P) are indistinguishable from each other.
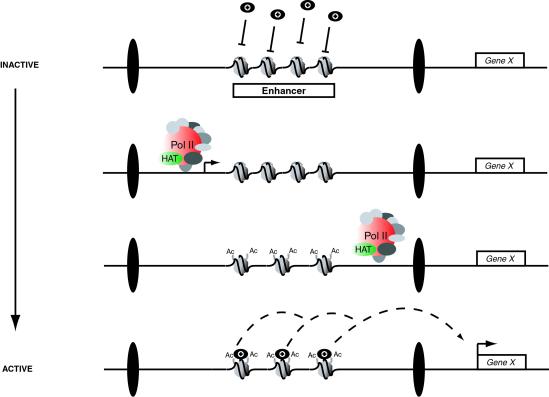
Fig 4.
Model of intergenic transcription-mediated enhancer activation. At the very early stages of embryonic development, the cis-regulatory elements (shown here as an enhancer, but also possible for Polycomb response elements) at the intergenic iab regions are inactive. A closed chromatin configuration prevents access to the trans factors (black circles), such as the products of the gap or segmentation genes, which are necessary to direct expression of the abdA and AbdB protein-coding genes (Gene X). Within each iab region, discrete anteroposterior patterns of transcription (black arrow) are initiated at around late stage 4/early stage 5 in development. RNA polymerase II (Pol II) and its associated remodeling complex (HAT, histone acetyltransferase) pass through the enhancer and epigenetically modify the histone tails (shown here as acetylation, but also may involve deacetylation, methylation, or demethylation of histone tail residues), creating an open chromatin configuration. Spreading of transcription from one iab region to the next is prevented by insulator DNAs (black ellipses) at the boundaries of each iab region. The newly modified open histones now permit the binding of trans factors to the enhancer sequence, and this stable interaction is able to direct segment-specific expression of the neighboring Hox genes from midstage 5 of development onward.
To confirm these results, we examined transcription in 0- to 4-h embryos by using RT-PCR. Transcription is readily detected by using primer pairs within iab5 or iab4 (Fig. 3H). However, primers mapping within the MCP-4 sequence do not detect any transcription (Fig. 3H, MCP-4). Furthermore, PCR amplification with primer pairs consisting of one primer within the MCP-4 sequence and one from the iab5 or iab4 region also fail to produce amplification products (Fig. 3H). In contrast, larger PCR products contained solely within the iab5 region are obtained (Fig. 3H). Therefore, the sense-orientation transcription detected in the iab4 region must be initiated within the iab4 region, and the transcription in the iab5 region terminates near the Mcp element. Similar results were obtained with primer pairs spanning the iab6 and iab5 regions. The transcripts within each of these domains appear to be discrete and do not form part of a larger transcription unit (Fig. 3H). This result, in fact, may indicate the location of the putative Fab6 insulator (Fig. 1A) (15).
Mcp Deletion Causes Disrupted Transcription in iab4.
Mcp is a spontaneous dominant mutation that deletes ≈3 kb of DNA at the Mcp insulator region (Fig. 3A). The Mcp phenotype is a posterior transformation of the fourth abdominal segment into the fifth (11, 28) (Fig. 3I). This transformation of segmental identity is due to ectopic activation of AbdB in the fourth abdominal segment, anterior to its normal expression domain (1, 3). We reasoned that deletion of the Mcp sequence may disrupt expression of the sense transcripts in the iab4 region. Consequently, we compared transcription of the iab4 and iab5 regions in 0- to 4-h yw67 (BX-C+) and Mcp mutant embryos by RT-PCR. Although transcription is readily observed in the iab4 regions of yw67 embryos, iab4 transcripts are not detectable in Mcp mutant embryos (Fig. 3J). In contrast, expression from the iab5 region and the AbdB gene is comparable in yw67 and Mcp mutant embryos (Fig. 3J). It therefore appears that deletion of the Mcp sequence results in an absence of transcription in the iab4 domain and a subsequent failure to correctly establish the identity of the fourth abdominal segment (Fig. 3I). To confirm this observation, we also examined transcription by in situ hybridization. Embryos were cohybridized with probes against the iab4s transcript and the segment polarity gene engrailed. Early in development, the distinct stripes of the engrailed pattern are seen initially in the anterior of the embryo (Fig. 3 K and L). In yw67 embryos, iab4s transcription also is detectable in posterior regions (Fig. 3L) but is absent in Mcp mutant embryos (Fig. 3K). Later in development, the transcription patterns in Mcp mutant and yw67 embryos are indistinguishable, suggesting that the Mcp deletion disrupts iab4 transcription only early in development.
Discussion
The iab region between the abdA and AbdB homeotic genes in the BX-C has been studied extensively in recent years. A number of different cis-regulatory elements from the iab regions have been identified and characterized at the molecular level. Nonetheless, a complete molecular understanding of how these regulatory elements function in directing homeotic gene expression has yet to be achieved. In particular, it has remained unclear how these elements are activated in specific segmental domains during embryogenesis. Previous studies have shown the existence of a highly ordered developmental transcription program at the iab regions of the BX-C, although they were unable to assign any function to the transcripts (4, 22, 25, 26). This study provides evidence that transcription through the intergenic region early in development may act to define the embryonic domains of activity for the cis-regulatory elements at the iab regions.
Insulator Sequences Regulate Intergenic Transcription.
If the initial activation of the cis-regulatory elements in the iab regions is dependent on the intergenic transcripts, then it would be necessary not only to have unique transcription patterns within each region but also to ensure that transcription does not spread from one region to the next. Our study provides evidence that the insulators that organize the different iab regions into discrete chromatin domains (8, 14, 15) act to regulate the intergenic transcription program in this way.
The Mcp element (28) has been shown to harbor a number of regulatory activities, including the ability to act as an insulator (7, 19). We have discovered two activities of the Mcp element that are related to the regulation of intergenic transcription. First, the Mcp element prevents the spreading of transcription from the iab5 region to the iab4 region. The characterized Mcp element (28) itself is transcribed, but the immediately adjacent sequences in the iab4 region are not (Fig. 3). The Fab8 insulator also is transcribed early in development (4), but we failed to detect transcripts spanning across the insulator from the iab8 region to the iab7 region (data not shown) and also from the iab6 region to iab5 region (Fig. 3). It therefore is possible that while the insulator elements at the BX-C are transcribed, they also act to terminate transcription from one iab region to the next. Second, the Mcp element appears to positively regulate transcription in the iab4 region, because its deletion results in the absence of the iab4 sense transcript. If the iab4 transcript is required to activate the iab4 cis-regulatory elements that normally direct abdA expression in the fourth abdominal segment (31), then it would be predicted that the loss of transcription may result in a failure to produce the fourth abdominal segment. The phenotype of Mcp deletion mutant flies supports this idea, because they have a posterior transformation of the fourth abdominal segment into the fifth (Fig. 3I) (11, 28). It therefore is conceivable that the Mcp deletion removes some part of the promoter or initiation elements required for iab4 transcription and, ultimately, results in a failure of the embryo to establish the correct identity of the fourth abdominal segment. When the iab4s transcript is fully mapped, it should be possible to make a small, targeted deletion of the iab4 promoter sequence by homologous recombination to analyze its role in regulating expression. It appears that the sense transcripts are functionally required only early in development, because their activation in later stages of development in the Mcp embryos (Fig. 3 M and O) is not sufficient to rescue the mutant phenotype.
Our RT-PCR analysis indicates that the transcripts within each iab region are part of the same transcription unit (Fig. 3). We can detect high molecular weight smears in the 10- to 20-kb range on Northern blots with probes from various iab regions (data not shown). This finding is consistent with the notion that large RNAs are synthesized across entire individual iab regions and are restricted not only to the characterized cis-regulatory elements within the regions. In this way, the iab RNAs share some similarities with the bithoraxoid transcripts of the neighboring Ubx region in the BX-C (32, 33). The bithoraxoid transcription unit is ≈25 kb in length, is expressed earlier in development than the adjacent homeotic genes, and is transcribed through characterized cis-regulatory elements (34). These observations may indicate that all of the intergenic sense transcripts at the BX-C serve a similar function.
It is a formal possibility that the early intergenic transcripts are simply products of spurious activation of cryptic promoters by the enhancers in the iab regions. However, our transgenic assay suggests that the IAB5 enhancer may not be capable of activating neighboring promoters (Fig. 2). It could be argued that in this assay, the IAB5 enhancer may not be able to activate the Kisir or ADE2 genes because of enhancer–promoter incompatibility. This is unlikely because the IAB5 enhancer has been shown to be relatively promiscuous when directing transcription from a range of different promoters. At the endogenous locus, IAB5 directs expression of AbdB. The promoter for this gene does not contain a recognizable TATA element or a consensus Inr or DPE element (35). However, in transgenic assays, IAB5 will preferentially activate TATA-containing promoters when challenged with linked TATA-less promoters (16). The IAB5 enhancer also will interact strongly with Inr- and DPE-containing promoters. Overall, these observations indicate that IAB5 has the potential to interact with many different classes of core promoters. In this case, any IAB5-directed transcription from the Kisir or ADE2 promoters should be detectable against the weak levels of endogenous transcription we can detect from these two genes in early embryogenesis. We have evidence that, at the endogenous locus, the IAB5 enhancer in fact is recruited selectively only to the AbdB promoter by a tethering mechanism (R.A.D., unpublished data). In addition, all of the existing data indicate that the iab enhancers are active only later in development, when they begin to direct expression of the neighboring homeotic genes (4, 16, 17, 36). However, we cannot discount the possibility that the intergenic transcription is directed by existing or as yet unknown enhancers in the BX-C. It is conceivable that, early in development at the endogenous locus, these enhancers regulate localized intergenic transcription, before the initiation of long-range interactions with the Hox gene promoters.
Functional Role for Intergenic Transcription.
Intriguingly, the iab sense transcripts demonstrate specific localization patterns in the cells in which they are expressed. Their expression can be detected as two foci that appear to correspond to localized nuclear expression (Fig. 1 B–I), because the transcripts are not distributed throughout the cytoplasm. It is possible that the RNA transcripts have some function in the nucleus. Alternatively, their nuclear localization simply may indicate that they have no cellular function but it is the act of their transcription that is important. The tightly regulated temporal and spatial expression patterns of the transcripts in the embryo, along with their nuclear localization, point toward a molecular function of the sense transcription itself. It is possible that the passage of an RNA polymerase II complex during transcription through the iab regions permits the recruitment of trans factors to otherwise inaccessible cis-regulatory elements (Fig. 4). The RNA polymerase II complex is known to include a histone acetyltransferase (37) that could modify the histone tails in nucleosomes across the transcribed region. It is also conceivable that the RNA polymerase II complex may recruit other chromatin-modifying enzymes, such as histone methyltransferases or deacetylases, capable of contributing to the remodeling process. In this way, transcription would facilitate the “opening” of chromatin in the iab regions, corresponding to the activation of the cis-regulatory elements in specific domains of the embryo (Fig. 4). We cannot discount the possibility that intergenic transcription also may affect “higher-order” chromosome structure or simply transiently displace nucleosomes, allowing access to the DNA sequences of cis-regulatory elements. It is also possible that transcription from the iab regions may use a different polymerase complex with similar, associated activities. Our analysis of the Mcp deletion reveals that the insulator elements flanking each iab region may contain sequences that act to initiate the recruitment of the RNA polymerase complex and prevent the spreading of transcription from one iab region to the next (Fig. 4).
Implications for Complex Gene Clusters.
The sense iab transcription program shares a number of characteristics with intergenic transcription at mammalian loci. The β-globin locus in humans also is divided into distinct subdomains, and long, intergenic transcripts are responsible for delineating the active domains by association with chromatin-remodeling activities (38). An active enhancer at the H2 hypersensitive site of the β-globin locus control region also was found to be transcribed from transgenes in mice, irrespective of the position and orientation of the enhancer (39). At imprinted loci in mice, a number of cis-regulatory elements, including enhancers and the imprinting control regions at H19/Igf2 and Igf2r, are transcribed (40–42). These observations have led to the suggestion that intergenic transcription may be a common characteristic of active complex loci in eukaryotes. However, we expanded our study to examine transcription at the hairy, even-skipped, fushi tarazu, and gooseberry loci in Drosophila embryos and failed to detect any significant transcription beyond the protein-coding transcription units (data not shown). This result raises the possibility that the intergenic transcripts at the BX-C may be unique in Drosophila.
Finally, the functional characterization of these transcripts provides a plausible reason for the evolutionarily conserved chromosomal arrangement of the iab regions and their large size. In our model, the establishment of autonomous chromatin environments for the iab regions depends on the initiation of individual transcription units within each iab region. The resulting complex colinear transcription program, responsible for defining the domains of activity for the iab regions, may require both the spatially ordered arrangement of cis-regulatory sequences and a large, initially naïve chromatin environment.
Note
After this paper was submitted for publication, similar observations on the functional activity of intergenic transcription in the Drosophila BX-C were reported by three other laboratories (43–45).
Acknowledgments
We thank Sumio Ohtsuki for unpublished DNA constructs, Mark Stapleton for providing the engrailed probe and Cory Olsen for technical assistance. We also thank Katharine Arney, Andrew Dowsett, and Joanne Topol for comments on the manuscript. This work was supported by a grant from the National Institutes of Health to E.B.L. (HD06331). R.A.D. was funded by a Wellcome Trust Prize Traveling Fellowship.
Abbreviations
BX-C, bithorax complex
DIG, digoxigenin
References
- 1.Sanchez-Herrero E. (1991) Development (Cambridge, U.K.) 111, 437-449. [DOI] [PubMed] [Google Scholar]
- 2.Boulet A. M., Lloyd, A. & Sakonju, S. (1991) Development (Cambridge, U.K.) 111, 393-405. [DOI] [PubMed] [Google Scholar]
- 3.Celniker S. E., Sharma, S., Keelan, D. J. & Lewis, E. B. (1990) EMBO J. 9, 4277-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J., Ashe, H., Burks, C. & Levine, M. (1999) Development (Cambridge, U.K.) 126, 3057-3065. [DOI] [PubMed] [Google Scholar]
- 5.Gyurkovics H., Gausz, J., Kummer, J. & Karch, F. (1990) EMBO J. 9, 2579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagstrom K., Muller, M. & Schedl, P. (1996) Genes Dev. 10, 3202-3215. [DOI] [PubMed] [Google Scholar]
- 7.Karch F., Galloni, M., Sipos, L., Gausz, J., Gyurkovics, H. & Schedl, P. (1994) Nucleic Acids Res. 22, 3138-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galloni M., Gyurkovics, H., Schedl, P. & Karch, F. (1993) EMBO J. 12, 1087-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J., Barolo, S., Szymanski, P. & Levine, M. (1996) Genes Dev. 10, 3195-3201. [DOI] [PubMed] [Google Scholar]
- 10.Mihaly J., Hogga, I., Gausz, J., Gyurkovics, H. & Karch, F. (1997) Development (Cambridge, U.K.) 124, 1809-1820. [DOI] [PubMed] [Google Scholar]
- 11.Karch F., Weiffenbach, B., Peifer, M., Bender, W., Duncan, I., Celniker, S., Crosby, M. & Lewis, E. B. (1985) Cell 43, 81-96. [DOI] [PubMed] [Google Scholar]
- 12.Parnell T. J. & Geyer, P. K. (2000) EMBO J. 19, 5864-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geyer P. K. (1997) Curr. Opin. Genet. Dev. 7, 242-248. [DOI] [PubMed] [Google Scholar]
- 14.Mihaly J., Hogga, I., Barges, S., Galloni, M., Mishra, R. K., Hagstrom, K., Muller, M., Schedl, P., Sipos, L., Gausz, J., et al. (1998) Cell. Mol. Life Sci. 54, 60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vazquez J., Farkas, G., Gaszner, M., Udvardy, A., Muller, M., Hagstrom, K., Gyurkovics, H., Sipos, L., Gausz, J. & Galloni, M. (1993) Cold Spring Harbor Symp. Quant. Biol. 58, 45-54. [DOI] [PubMed] [Google Scholar]
- 16.Ohtsuki S., Levine, M. & Cai, H. N. (1998) Genes Dev. 12, 547-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Busturia A. & Bienz, M. (1993) EMBO J. 12, 1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagstrom K., Muller, M. & Schedl, P. (1997) Genetics 146, 1365-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller M., Hagstrom, K., Gyurkovics, H., Pirrotta, V. & Schedl, P. (1999) Genetics 153, 1333-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding K., Wedeen, C., McGinnis, W. & Levine, M. (1985) Science 229, 1236-1242. [DOI] [PubMed] [Google Scholar]
- 21.Harding K. & Levine, M. (1988) EMBO J. 7, 205-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casares F. & Sanchez-Herrero, E. (1995) Development (Cambridge, U.K.) 121, 1855-1866. [DOI] [PubMed] [Google Scholar]
- 23.Ingham P. W. & Martinez-Arias, A. (1986) Nature 324, 592-597. [DOI] [PubMed] [Google Scholar]
- 24.Pirrotta V. (1998) Cell 93, 333-336. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Herrero E. & Akam, M. (1989) Development (Cambridge, U.K.) 107, 321-329. [DOI] [PubMed] [Google Scholar]
- 26.Bae E., Calhoun, V. C., Levine, M., Lewis, E. B. & Drewell, R. A. (2002) Proc. Natl. Acad. Sci. USA 99, 16847-16852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin C. H., Mayeda, C. A., Davis, C. A., Ericsson, C. L., Knafels, J. D., Mathog, D. R., Celniker, S. E., Lewis, E. B. & Palazzolo, M. J. (1995) Proc. Natl. Acad. Sci. USA 92, 8398-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis E. B. (1978) Nature 276, 565-570. [DOI] [PubMed] [Google Scholar]
- 29.Lundell M. J. & Hirsh, J. (1992) Dev. Biol. 154, 84-94. [DOI] [PubMed] [Google Scholar]
- 30.Small S., Blair, A. & Levine, M. (1992) EMBO J. 11, 4047-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karch F., Bender, W. & Weiffenbach, B. (1990) Genes Dev. 4, 1573-1587. [DOI] [PubMed] [Google Scholar]
- 32.Lipshitz H. D., Peattie, D. A. & Hogness, D. S. (1987) Genes Dev. 1, 307-322. [DOI] [PubMed] [Google Scholar]
- 33.Hogness D. S., Lipshitz, H. D., Beachy, P. A., Peattie, D. A., Saint, R. B., Goldschmidt-Clermont, M., Harte, P. J., Gavis, E. R. & Helfand, S. L. (1985) Cold Spring Harbor Symp. Quant. Biol. 50, 181-194. [DOI] [PubMed] [Google Scholar]
- 34.Horard B., Tatout, C., Poux, S. & Pirrotta, V. (2000) Mol. Cell. Biol. 20, 3187-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones K. A. & Kadonaga, J. T. (2000) Genes Dev. 14, 1992-1996. [PubMed] [Google Scholar]
- 36.Shimell M. J., Simon, J., Bender, W. & O'Connor, M. B. (1994) Science 264, 968-971. [DOI] [PubMed] [Google Scholar]
- 37.Wittschieben B. O., Otero, G., de Bizemont, T., Fellows, J., Erdjument-Bromage, H., Ohba, R., Li, Y., Allis, C. D., Tempst, P. & Svejstrup, J. Q. (1999) Mol. Cell 4, 123-128. [DOI] [PubMed] [Google Scholar]
- 38.Gribnau J., Diderich, K., Pruzina, S., Calzolari, R. & Fraser, P. (2000) Mol. Cell 5, 377-386. [DOI] [PubMed] [Google Scholar]
- 39.Kong S., Bohl, D., Li, C. & Tuan, D. (1997) Mol. Cell. Biol. 17, 3955-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyle R., Watanabe, D., te Vruchte, D., Lerchner, W., Smrzka, O. W., Wutz, A., Schageman, J., Hahner, L., Davies, C. & Barlow, D. P. (2000) Nat. Gen. 25, 19-21. [DOI] [PubMed] [Google Scholar]
- 41.Moore T., Constancia, M., Zubair, M., Bailleul, B., Feil, R., Sasaki, H. & Reik, W. (1997) Proc. Natl. Acad. Sci. USA 94, 12509-12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drewell R. A., Arney, K. L., Arima, T., Barton, S. C., Brenton, J. D. & Surani, M. A. (2002) Development (Cambridge, U.K.) 129, 1205-1213. [DOI] [PubMed] [Google Scholar]
- 43.Rank G., Prestel, M. & Paro, R. (2002) Mol. Cell. Biol. 22, 8026-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hogga I. & Karch, F. (2002) Development (Cambridge, U.K.) 129, 4915-4922. [DOI] [PubMed] [Google Scholar]
- 45.Bender W. & Fitzgerald, D. P. (2002) Development (Cambridge, U.K.) 129, 4923-4930. [DOI] [PubMed] [Google Scholar]