Abstract
An increased prevalence of microdeletions at the 22q11 locus has been reported in samples of patients with schizophrenia. 22q11 microdeletions represent the highest known genetic risk factor for the development of schizophrenia, second only to that of the monozygotic cotwin of an affected individual or the offspring of two schizophrenic parents. It is therefore clear that a schizophrenia susceptibility locus maps to chromosome 22q11. In light of evidence for suggestive linkage for schizophrenia in this region, we hypothesized that, whereas deletions of chromosome 22q11 may account for only a small proportion of schizophrenia cases in the general population (up to ≈2%), nondeletion variants of individual genes within the 22q11 region may make a larger contribution to susceptibility to schizophrenia in the wider population. By studying a dense collection of markers (average one single nucleotide polymorphism/20 kb over 1.5 Mb) in the vicinity of the 22q11 locus, in both family- and population-based samples, we present here results consistent with this assumption. Moreover, our results are consistent with contribution from more than one gene to the strikingly increased disease risk associated with this locus. Finer-scale haplotype mapping has identified two subregions within the 1.5-Mb locus that are likely to harbor candidate schizophrenia susceptibility genes.
Microdeletions of chromosome 22q11 are associated with variable phenotypic expression (1) that includes relatively high frequency of severe mental illness (2, 3). Approximately one-third to one-fourth of patients with the 22q11 microdeletion develop schizophrenia or schizoaffective disorder, as defined in the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) (4). Although the microdeletion occurs in the population at a rate of 0.025% (5), it has been found in up to 2% of adult schizophrenic patients (6) and up to 6% of cases with childhood-onset schizophrenia (7). Taken together, these studies suggest that the risk of schizophrenia for a patient with a 22q11 microdeletion may be ≈25–30 times the general population risk of 1% and that the rate of 22q11 microdeletions in schizophrenia, although relatively low, may be up to 12–80 times the estimated general population rate. In addition, at least two independent studies reported suggestive linkage results in the 22q11 region (8, 9). Finally, a series of recent studies suggest that schizophrenic patients carrying the 22q11 deletion bear the hallmark neuropsychological and neuroanatomical features of classical schizophrenia (10–12). It therefore seems likely that the 22q11 region harbors genes that, alone or in combinations, are causally implicated in schizophrenia in a subset of patients.
The overwhelming majority of the 22q11 deletions (≈87%) are 3 Mb in size, whereas a smaller percentage of them (≈8%) involve the same proximal breakpoint but a different distal breakpoint, resulting in a smaller 1.5 Mb deletion. All deletions are mediated by low copy repeat (LCR) sequences (13, 14). At least one schizophrenic patient has been described to carry the smaller 22q11 microdeletion (6), and therefore the “schizophrenia critical region” has been defined to 1.5 Mb (LCR-A to -B). The majority of the genes in the region are known (http://genome.ucsc.edu), making this locus amenable to a molecular genetic analysis.
The magnitude of the risk attributed to this deletion is unprecedented for a single genetic lesion and is comparable only to the risk among children of two schizophrenic parents or monozygotic cotwins of an affected individual. In both of these cases, the increased risk is due to the contribution of more than one susceptibility gene. It is therefore possible that the increased risk associated with this microdeletion is due to the contribution of more than one physically linked gene at the 22q11 locus. To address the role of individual genes from this chromosomal region, we have undertaken linkage disequilibrium (LD) studies in family samples (trios) that test for preferential transmission of common (>10% frequency) variants and multivariant haplotypes from parents to affected individuals. These studies are based on (and therefore test) the assumptions that, whereas deletions of chromosome 22q11 may account for only a small proportion of schizophrenia cases in the general population, nondeletion common variants of individual genes within the 22q11 region may make a larger contribution to susceptibility to schizophrenia in the wider population. Using this approach, we analyzed initially 13 of the 23 known genes residing in the deleted locus, delineated a region of significant LD within a segment in which PRODH2 and DGCR6 are the only known genes, and identified a relatively common haplotype where the schizophrenia susceptibility variant(s) likely reside (15). One additional result of this analysis was that the observed haplotypic association was particularly pronounced among children with schizophrenia and adults with early onset features of the disease. Further evidence implicating the PRODH2 gene itself was provided by the identification of rare structural variants of this gene, especially in cases with early onset of the disease (15), as well as by behavioral and neurochemical studies of a mouse model (16). However, comparison of the increase in the morbid risk of schizophrenia associated with the identified PRODH2 variation to the risk associated with the 22q11 microdeletion could not exclude contribution from other genes in the region. We therefore extended our analysis to include all of the known genes in the 1.5-Mb 22q11 locus.
Methods
Patient Samples.
The Adult Schizophrenia (AS) Sample, the Childhood Onset Schizophrenia (COS) Sample, and the South African Afrikaner (SAA) case/control sample have been previously described (15). The SAA family sample included 93 patients of Afrikaner origin (age 19 or older) who met lifetime criteria for DSM-IV (4) schizophrenia or schizoaffective disorder and both their biological parents (trios). Participants were interviewed by specially trained clinicians using the Diagnostic Interview for Genetic Studies (17). Close genetic relatedness among the patients included in this analysis was excluded up to the fifth degree by family history information.
All participants provided written informed consent before participation in the study. The protocol and the consent forms were approved by the Rockefeller University Institutional Review Board and the Institutional Review Boards of all participating sites.
Single Nucleotide Polymorphism (SNP) Identification and Genotyping/PCR Primers and Conditions.
See Appendix and Table 6, which are published as supporting information on the PNAS web site, www.pnas.org.
Statistical Analysis
Pairwise Background LD.
See Appendix, which is published as supporting information on the PNAS web site (18–22).
Association Analysis.
Transmissions of single SNPs, as well as multilocus SNP haplotypes, were analyzed by using the haplotype-based haplotype relative risk (HHRR) test (23), as described (15). Tightly linked markers that are in LD provide redundant information, making Bonferroni significance levels conservative. To estimate a global significance level, we randomly permuted transmitted and nontransmitted chromosomes for each trio (24). We generated 1,000,000 such permutations of the data by using the qtdt computer program (25, 26) and calculated the maximum observed allelic transmission score for each permutation.
Results
Pairwise Background LD.
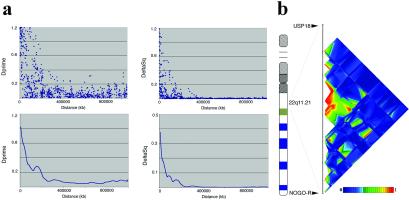
The measurement, extent, and variability of marker-to-marker disequilibrium are important factors in deciding the number of markers required for association scans and for interpreting marker-disease association results. We have therefore assessed the extent and characteristics of LD within the 1.5-Mb 22q11 locus. We calculated pairwise disequilibrium coefficients for all marker pairs within the region. The two distributions are summarized in Fig. 1a. Strong disequilibrium between a marker and a nearby disease allele can allow marker alleles to serve as surrogates for nearby markers. Multiple definitions of strong or “useful” disequilibrium have been proposed. For example, Δ2 > 0.10 corresponds to a 10-fold increase in the sample size required to detect an association compared with testing disease alleles directly. Δ2 takes maximum values only for markers that have identical allele frequencies; at any particular distance, markers with similar allele frequencies can exhibit higher values of Δ2 than those with divergent allele frequencies. Alternatively, D′ > 0.33 also corresponds to a 10-fold increase in sample size, provided marker and trait allele frequencies are well matched. At 10, 20, and 50 kb, ≈97, 91, and 65% of markers in our sample exhibit D′ > 0.33, respectively. In addition, 90, 68, and 41% of these also show Δ2 > 0.10, indicating they are well matched in allele frequency (Table 1). We estimate the half-length of D′ in the region to 48 kb, which compares well with the pattern observed in a recent study of LD along chromosome 22, which overlaps with our region (19). We identified 17 sets of three or more markers that define haplotypes with limited diversity. Some of these sets are overlapping, reflecting the difficulties in placing ancestral recombination events (data not shown; see also below). Finally, we examined the regional distribution of LD (Fig. 1b). The distribution of LD was found to be highly irregular, with significant LD concentrated in the middle of the locus over a distance of ≈250 kb (Fig. 1b).
Fig. 1.
(a) Decay of LD at the 22q11 locus. (Upper) Scatter plot of D′ and Δ2 values vs. physical distance. Values were calculated on the basis of the haplotype frequencies estimated by using partial phase information in the 106 unrelated AS trios. (Lower) Sliding window averages of disequilibrium coefficients. (b) Distribution of pair-wise disequilibrium (D′) across the 22q11 locus. Multiple pair-wise comparisons are shown with D′ statistics color coded (bright red and bright blue are opposite ends of the scale) and plotted at the marker locations.
Table 1.
Extent of significant LD in the 22q11 locus
| Distance
|
Δ2 | D′ | ||||
|---|---|---|---|---|---|---|
| Avrg. | >0.80 | >0.10 | Avrg. | >0.90 | >0.33 | |
| <10 kb | 0.45 | 0.19 | 0.90 | 0.88 | 0.61 | 0.97 |
| 10–20 kb | 0.25 | 0.06 | 0.68 | 0.70 | 0.24 | 0.91 |
| 20–50 kb | 0.15 | 0.00 | 0.41 | 0.55 | 0.16 | 0.65 |
| 50–100 kb | 0.09 | 0.00 | 0.28 | 0.36 | 0.10 | 0.35 |
Average (Avrg.) Δ2 and D′
and the percentage of SNP pairs showing LD coefficients
above specific thresholds at different inter-SNP intervals are shown.
Association Studies in Patient Samples.
We used a three-step strategy to search for a signal of LD from the 22q11 locus. Our goals were: (i) to detect a significant and reproducible LD signal; (ii) to bind the critical region within the limits imposed by the density of our genetic map; and (iii) to identify the causal genetic variant. We first studied 106 trios from the AS sample and 26 trios from the COS sample. A primary scan included SNPs distributed throughout the 1.5-Mb region. Only relatively common (minor allele >10%), informative SNPs were further pursued at this stage. The average marker density is ≈1 SNP/20 kb, although the distribution of intermarker distances follows the distribution of genes and shows considerable variation.
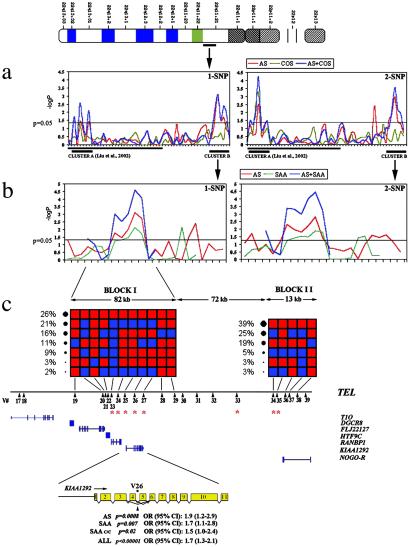
Phase I of our association studies focused primarily in the region between the DGCR6 and CLDN5 genes, whereas Phase II focused in the region between the CLDN5 and NOGO-R genes. We examined each allele of each marker or a combination of two adjacent markers for evidence of transmission disequilibrium using the HHRR test (23). The results of the primary association screen identified two clusters of neighboring SNPs having alleles with nominally significant HHRR results (Fig. 2a): Cluster A at the proximal end of the locus included three SNPs (15), and Cluster B at the distal end of the locus included four SNPs (V24, -26, -34, -35) (Table 2). Notably, the two clusters are separated by ≈1 Mb, and they are not in LD. We have reported separately our results from the detailed analysis of the proximal (centromeric) Cluster A (15).
Fig. 2.
(a) Single SNP and 2-SNP HHRR P values (−log10) across the 22q11 locus. The solid line underneath the x axis indicates the SNPs presented in Liu et al. (15). The remaining SNPs are detailed in Table 2 (blue shading). The two clusters of significant SNP association across the entire 22q11 locus (Clusters A and B) are also underlined. The AS and COS samples were analyzed both separately and as a combined sample, because they originated from the same geographical area (U.S.). Distances are not to scale. (b) Detailed view of HHRR significance levels at Cluster B when additional SNPs were genotyped in the vicinity of the initial association shown in a. Detailed description can be found in Table 2. The family samples used for this analysis were the AS and SAA, as well as a combination of the two (see text). Distances are not to scale. (c) Haplotypic structure of Cluster B. Two haplotypic BLOCKs (I and II) are shown. Red squares indicate major and blue squares minor alleles. Observed haplotype frequencies are shown on the side of each block. The variant number (V#) is indicated at the bottom, and each variant's position relative to the genes residing in this region is also approximately indicated. Asterisks indicate SNPs showing significant association with schizophrenia in the AS and/or SAA samples. The position of the most significantly associated SNP (V26) within intron 4 of the KIAA1292 candidate gene is indicated along with the associated P values and odds ratios (approximate relative risks). Blue bars represent the approximate distribution of exons.
Table 2.
HHRR P values
Significance levels (P values) <0.05 are highlighted. Location within genes or intergenic regions is indicated. SNPs from public databases are indicated by reference SNP identifications. For simplicity, abbreviated Variant numbers (V#) are assigned and used in the text. Inter-SNP distances and PCR primers and conditions are shown in the Appendix.
Delineation of the Associated Region.
To investigate further the primary genetic evidence at Cluster B, we studied a denser collection of markers in its vicinity (Table 2, yellow shading). Unlike Cluster A (15), there was no evidence for preferential overtransmission of the associated alleles in the patient group with early onset features (Table 2/COS sample column and data not shown). We therefore restricted our subsequent analysis in the 106 adult trios. Additional SNPs were typed, and significant HHRR results occurred at SNPs V23 (P = 0.017), V25 (P = 0.017), and V27 (P = 0.002) surrounding the previous significant SNPs (Fig. 2b; Table 2). In all, seven neighboring SNPs having alleles with nominally significant HHRR results were identified in both primary and secondary screen, distributed over a region of ≈250 kb from the HTF9C gene to the NOGO-R gene supporting the existence of LD with the disease over the investigated region.
Using the relatively high-density SNP map, it was possible to determine the fine structure of the haplotypes in this region. Specifically, the analysis shows that the region can be parsed into two major haplotype BLOCKs (I and II), with each block having limited diversity (≤5 haplotypes account for >90% of all chromosomes) with only a small number of SNPs between blocks. SNPs V23–27 reside in BLOCK I, whereas SNPs V34 and V35, at the 3′ end of the NOGO-R gene, reside in BLOCK II. There is no strong evidence for LD between the significant SNPs in BLOCK I and the significant SNPs in BLOCK II (Table 3). On the other hand, within-block SNPs demonstrated highly significant LD with each other (data not shown), indicating that the observed associations in Cluster B could represent two independent disease susceptibility-conferring polymorphisms. Consistent with this prediction, when families were divided into two groups according to the presence or absence in the probands of the risk allele of SNP V26 (the most significant SNP in BLOCK I), significance of the schizophrenia associations with BLOCK I SNPs V23–27 increased dramatically in families with the V26 risk allele present. By contrast, associations with BLOCK II SNPs V34 and V35 remained unchanged or even decreased modestly in the same families (data not shown), indicating a minimal contribution of the V26 risk allele to these latter associations.
Table 3.
LD (D′) between SNPs in BLOCKs I and II
| BLOCK I
|
BLOCK II | |
|---|---|---|
| V34 | V35 | |
| V23 | 0.22 | 0.04 |
| V24 | 0.07 | 0.09 |
| V25 | 0.05 | 0.07 |
| V26 | 0.01 | 0.04 |
| V27 | 0.02 | 0.07 |
In conclusion, the strongest observed association at this stage (P < 0.0008) occurs at SNP V26, with four nearby SNPs (V23, -24, -25, -27) also exhibiting nominal evidence for association. All five SNPs fall in one haplotypic block (Fig. 2c), and thus the evidence for association that they provide is not independent. In these circumstances, a Bonferroni correction is conservative. In fact, the haplotype and disequilibrium structure of this region implies that the 72 markers investigated here correspond to much less than 72 independent tests. We therefore generated 100,000 permutations of the data (24) and recorded the most significant association for each one. Overall, the probability of observing an association as strong as the one presented here is P < 0.043, after adjusting for multiple testing at 63 markers (nine markers with low minor allele frequencies and a correspondingly small number of informative trios were not included in this analysis). Similarly with the Cluster A variation (15), no association was detected with the representative SNP V26 in a sample of 80 North American families not afflicted with schizophrenia (not shown), arguing against a generalized transmission distortion at the 22q11 locus.
Replication of the Association in Two Independent Samples.
We sought additional confirmation of our findings by examining both the primary and secondary set of SNPs in another family sample derived from an independently collected population (Fig. 2b; Table 2) consisting of 93 trios from the Tshwane (formerly known as Pretoria) area in South Africa (SAA sample). Five of the eight SNPs demonstrating significant association in the AS sample also showed significant association in the SAA sample (Fig. 2b; Table 2). All significant SNPs are located in BLOCK I. Moreover, single and multilocus analyses within BLOCK I showed that all of the same alleles and the same multiallelic haplotypes were overtransmitted in the two independent family data sets (Fig. 2b; Tables 2 and 4). Finally, we sought confirmation of the associations observed in both family samples in an independent case-control sample consisting of 93 unrelated schizophrenics of Afrikaner origin and 75 ethnically matched controls. We observed a similar pattern of association, consistent with involvement of an identical common disease variant in all three samples (Table 2). Combined analysis of all three samples at SNPs residing in BLOCK I resulted in a P < 10−5 and revealed an odds ratio (approximate relative risk associated with the causative variant) of ≈1.7 at SNP V26. We did not replicate the association with SNPs V34 and V35, from the 3′ end of the NOGO-R gene located in BLOCK II, in the SAA family set, although significant one- and two-SNP associations were observed ≈15 kb proximal, in the vicinity of SNP V33 (Fig. 2b; Table 2). Because different causal variants may be involved in this case in different populations, confirmation or exclusion of the NOGO-R gene (27) (an excellent candidate gene from this region) from association with schizophrenia will require the identification of all polymorphisms in the region and the assaying of most of these polymorphisms in both populations.
Table 4.
Five-SNP risk haplotypes in BLOCK I
| V23
|
V24
|
V25
|
V26
|
V27
|
T/nT ratio | |
|---|---|---|---|---|---|---|
| AS | SAA | |||||
| 2 | 2 | 2 | 2 | 1 | 1.5 | 1.4 |
| 1 | 2 | 2 | 2 | 1 | 1.3 | 1.3 |
| 1 | 1 | 1 | 1 | 2 | 0.6 | 0.6 |
| 1 | 2 | 2 | 1 | 2 | 0.6 | 0.8 |
| Other | 0.5 | 0.5 | ||||
T, transmitted; nT, non-transmitted.
Genetic Interactions and Parent-of-Origin Analysis.
Having established a reproducible pattern of association represented by SNP V26, which serves as a tag of the risk haplotypes, we then checked whether the proximal and distal disease risk-associated variation contributes to distinct or overlapping sets of patients. We stratified the AS families according to the presence or absence of the Cluster A (PRODH2/DGCR6) risk haplotype (15) in the proband (carriers vs. noncarriers). This stratified analysis provided significant evidence for association with SNP V26 only in the subset of noncarrier AS families (HHRR χ2 = 15.5, P < 0.001; transmitted (T)/nontransmitted (nT): allele 1, 36/68; allele 2, 106/74). In this subset, transmission of the V26 risk allele was associated with an odds ratio of 1.9 [95% confidence interval: 1.3–2.6], statistically indistinguishable when compared with the entire AS sample. By contrast, there was no evidence of association with SNP V26 in the carrier subset (HHRR χ2 = 0, P = 1; T/nT: allele 1, 27/27; allele 2, 36/36). As expected, further analysis of the distribution of the V26 alleles in the two proband groups provided similar results (alleles 1/2: noncarriers, 36/106; carriers, 27/27; χ2 = 10.9, P < 0.001). This observation is consistent with a disease-risk contribution from the proximal or distal subregions to distinct subsets of patients, possibly with distinct syndromic features (such as early onset features). However, the significance of this finding is, at present, unclear, because it could not be replicated in the South African samples (data not shown). A more conclusive analysis of interaction will require considerable increase in the sample size. We also conducted a parent-of-origin analysis, which did not provide any significant evidence for a parent-of origin effect. For example, when marker V26 is considered, parental origin can be discerned in 74 of 96 transmissions/nontransmissions. In this case, maternal and paternal transmissions appear to contribute equally to the V26 association (8 vs. 24 paternal and 13 vs. 29 maternal).
Candidate Genes from the Associated Region.
The replication of the association between BLOCK I SNPs and schizophrenia in an independent family sample and an independent case-control sample, in an identical pattern, adds considerable support to the genetic evidence associating genes at this region with schizophrenia. We analyzed the preferential transmission of specific haplotypes using genotypes from the five BLOCK I SNPs that gave the lowest P values in the association study (Table 4). We identified two overtransmitted risk haplotypes that are distinguished by the presence of allele 2 of SNP V26 and allele 1 of SNP V27 (Table 4). All of the other common haplotypes in this region are undertransmitted. Although there may be separate mutations in the two risk haplotypes, the simplest explanation is that a putative causative allele is strongly linked to these two SNPs rather than to a specific haplotype. On the basis of this latter assumption, we attempted to identify a linked causal missense allele, examining the coding region of all known genes within BLOCK I (as well as that of the T10 gene located immediately adjacent to it) (Fig. 2c) for missense or nonsense allelic variants, by sequencing and comparing a panel of five homozygous carriers and five homozygous noncarriers of the V26 risk allele 2. We identified four missense variants in three of the seven tested genes (Table 5). Because of their relatively low frequency, it is unclear whether they can explain the full effect associated with the very common SNP V26 risk allele. The causal SNP or SNPs may, therefore, affect regulation of one or more of the resident genes, many of which are plausible candidates. Four known genes found in the RefSeq database are located in BLOCK I (Fig. 2c): the DGCR8 gene (which encodes for a protein of undetermined function), the FLJ22127 gene (which encodes for a protein with a double-strand RNA-binding domain), the HTF9C gene (28) (which likely encodes for a methyltransferase), and the RANBP1 gene (29) (which encodes for a protein that interacts with the cell-cycle regulatory protein Ran/TC4). Two more genes predicted by analysis of the syntenic region on mouse chromosome 16 and supported by available EST and mRNA databases are located either within BLOCK I (KIAA1292) (30) or immediately outside it (T10) (31) (Fig. 2c).
Table 5.
Coding region variants in BLOCK I genes
| Gene | Nucleotide position | Nucleotide change | Amino acid change | No. carriers | Associated allele |
|---|---|---|---|---|---|
| DGCR8 | 610 | A → G | I → V | 1 (10) | I |
| HTF9C | 497 | G → T | A → S | 2 (10) | S |
| 683 | C → A | L → I | 1 (10) | I | |
| KIAA1292 | 1707 | G → A | A → T | 5 (25) | A |
DGCR8 : GI:12225241; HTF9C : GI:15426493; KIAA1292 : GI:17484904.
The number in parentheses indicates the number of patients sequenced. Unless otherwise stated, all the patients are heterozygous for the change.
Allele associated to V26 allele 2.
Information from more than 10 patients was available for this change.
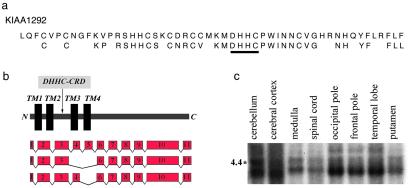
We note that three of the five SNPs that show evidence of association in SAA and AS samples (V25–27) are located within the transcription unit and the putative 5′ control region of KIAA1292 (Fig. 2c). In addition, of all SNPs that show evidence of association, only V26 and V27 are exclusively present in the risk haplotypes (Table 4). SNP V26, located in intron 4 of KIAA1292, shows the strongest and most consistent evidence of association of all SNPs in either primary or secondary panels in all tested samples. For that reason and because all three tested SNPs within the KIAA1292 gene unit demonstrate significant association with schizophrenia in both the AS and the SAA samples, we consider the KIAA1292 gene a prime candidate worthy of further investigation. KIAA1292 is predicted to have four transmembrane domains and a new form of cysteine-rich domain called DHHC-CRD (32) (Fig. 3 a and b). The only member of the family that has been characterized to some extent is yeast Erf2, a component of a previously uncharacterized Ras subcellular localization pathway (33). The KIAA1292 gene is highly expressed in the human brain (Fig. 3c), and in situ hybridization analysis of expression in the mouse brain demonstrated high expression levels especially in the cortex and hippocampus, areas expected to play an important role in the pathophysiology of the disease (Y.-J.C., M.K., and J.A.G., unpublished work). It should be noted that analysis of EST databases as well as RT-PCR experiments on RNA from human and mouse brains identified two major alternatively spliced variants of KIAA1292 involving exons 4 and 5 (Fig. 3b), which encode for protein forms with truncated DHHC-CDR domains and, most likely, reduced or altered function. Of interest, V26 is located in the intron between the alternatively spliced exons 4 and 5, and a potential effect of V26 genotype on the function of putative splicing enhancers and the relative levels of the two splicing variants warrants further investigation.
Fig. 3.
(a) Examination of the deduced KIAA1292 polypeptide sequence shows a new form of cysteine-rich domain called DHHC-CRD that includes a DHHC motif consisting of the amino acids Asp-His-His-Cys (DHHC) and a cysteine-rich domain (CRD) that includes a Cys4 zinc-finger-like metal binding site. The sequence for the KIAA1292 DHHC-CRD as well as a consensus sequence for this domain (32) are indicated. (b) Alternative splice forms of the KIAA1292 gene. Note that the DHHC-CRD is largely part of the loop between adjacent TM2 and TM3 (TM, transmembrane). (c) Brain expression of the KIAA1292 gene.
Discussion
Using a relatively dense genetic map of SNPs across the entire 1.5-Mb 22q11 region, we found strong evidence of LD with schizophrenia. In our study, D′ decayed halfway between maximum and minimum values at 48 kb. The extent of disequilibrium observed in this region of chromosome 22q is similar to that previously described for this (19) and other regions (22, 34). Among the markers investigated, 56 of 72 were in strong LD with surrounding markers and included in a common haplotype block. Nevertheless, a small proportion of the markers we investigated exhibited only low levels of LD with surrounding markers, and in these regions, it would be desirable to investigate additional markers. Given this limitation, our analysis identified two associated subregions (Fig. 2a), suggesting that the 22q11 microdeletion-associated schizophrenia may have the characteristics of a contiguous gene syndrome.
In the proximal part of the locus, we delineated a subregion of significant association within a segment of ≈30 kb in which PRODH2 and DGCR6 are the only known genes, and we identified one relatively common haplotype where the schizophrenia susceptibility variant(s) likely reside (15). In the distal part of the locus, we bound the associated subregion to a haplotypic block that spans at least 80 kb and shows consistent association with the disease in three independent samples from two different populations.
One difference between the two subregions of association is that in the proximal subregion (Cluster A), the association was more pronounced among children with schizophrenia and adults with early onset features of the disease. Similar association was not observed in the distal subregion. These observations seem to suggest that genetic variation at the distal subregion contributes to schizophrenia independently of the onset of the disease, whereas the proximal subregion contains genes (most likely PRODH2) contributing to early onset forms of the disease. It might be relevant that previous studies indicated a relatively wide range of age at onset among 22q11-deleted patients with schizophrenia ranging from childhood onset to up to 26 years of age (2).
The combination of the study of Liu et al. (15) and the present study represents a comprehensive application of LD mapping across the 22q11 susceptibility locus in relation to schizophrenia. An implication of these studies is that more than one gene from this region may contribute to schizophrenia susceptibility. Functional variants of these genes may independently or synergistically exert modest increases in the disease risk in nondeleted patients. In 22q11 microdeletion carriers, drastic reduction of dosage to half in more than one gene may result in the dramatic 25- to 30-fold increase in disease risk observed. Alternatively, genes in the identified proximal and distal subregions may contribute only partially to the 25- to 30-fold increase in the risk associated with this locus, the largest part of it being due to an unidentified gene that lacks common functional variants and is variably mutated in a small portion of nondeleted schizophrenic patients. Such rare and possibly highly penetrant mutations would be invisible to our present experimental strategy, which focuses on relatively common variants. Nevertheless, our current analysis revealed a limited number of candidate genes (including two membrane-associated proteins, KIAA1292 and NOGO-R, highly expressed in brain regions implicated in schizophrenia), which need to be tested in additional samples and whose biological function needs to be explored in more detail by generation of animal models.
Supplementary Material
Acknowledgments
We thank all of the families who participated in this research. Support for this work was provided in part by National Institute of Mental Health R01 MH61399-01 (to M.K.). Clinical work at the Rockefeller University Hospital General Clinical Research Center was supported in part by National Center for Research Resources at the National Institutes of Health, Grant M01 RR00102.
Abbreviations
LD, linkage disequilibrium
AS, adult schizophrenia
SNP, single nucleotide polymorphism
COS, childhood onset schizophrenia
SAA, South African Afrikaner
HHRR, haplotype-based haplotype relative risk
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Scambler P. J. (2000) Hum. Mol. Genet. 9, 2421-2426. [DOI] [PubMed] [Google Scholar]
- 2.Pulver A. E., Nestadt, G., Goldberg, R., Shprintzen, R. J., Lamacz, M., Wolyniec, P. S., Morrow, B., Karayiorgou, M., Antonarakis, S. E., Housman, D., et al. (1994) J. Nerv. Ment. Dis. 182, 476-478. [DOI] [PubMed] [Google Scholar]
- 3.Murphy K. C., Jones, L. A. & Owen, M. J. (1999) Arch. Gen. Psychiatry 56, 940-945. [DOI] [PubMed] [Google Scholar]
- 4.American Psychiatric Association, (1994) Diagnostic and Statistical Manual of Mental Disorders (Am. Psychiatric Press, Washington, DC).
- 5.du Montcel S. T., Mendizabal, H., Ayme, S., Levy, A. & Philip, N. (1996) J. Med. Genet. 33, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karayiorgou M., Morris, M. A., Morrow, B., Shprintzen, R. J., Goldberg, R., Borrow, J., Gos, A., Nestadt, G., Wolyniec, P. S., Lasseter, V. K., et al. (1995) Proc. Natl. Acad. Sci. USA 92, 7612-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Usiskin S. I., Nicolson, R., Krasnewich, D. M., Yan, W., Lenane, M., Wudarsky, M., Hamburger, S. D. & Rapoport, J. L. (1999) J. Am. Acad. Child Adolesc. Psychiatry 38, 1536-1543. [DOI] [PubMed] [Google Scholar]
- 8.Blouin J. L., Dombroski, B. A., Nath, S. K., Lasseter, V. K., Wolyniec, P. S., Nestadt, G., Thornquist, M., Ullrich, G., McGrath, J., Kasch, L., et al. (1998) Nat. Genet. 20, 70-73. [DOI] [PubMed] [Google Scholar]
- 9.Shaw S. H., Kelly, M., Smith, A. B., Shields, G., Hopkins, P. J., Loftus, J., Laval, S. H., Vita, A., De Hert, M., Cardon, L. R., et al. (1998) Am. J. Med. Genet. 81, 364-376. [DOI] [PubMed] [Google Scholar]
- 10.Bearden C. E., Woodin, M. F., Wang, P. P., Moss, E., McDonald-McGinn, D., Zackai, E., Emannuel, B. & Cannon, T. D. (2001) J. Clin. Exp. Neuropsychol. 23, 447-464. [DOI] [PubMed] [Google Scholar]
- 11.Chow E. W., Mikulis, D. J., Zipursky, R. B., Scutt, L. E., Weksberg, R. & Bassett, A. S. (1999) Biol. Psychiatry 46, 1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow E. W., Zipursky, R. B., Mikulis, D. J. & Bassett, A. S. (2002) Biol. Psychiatry 51, 208-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelmann L., Pandita, R. K., Spiteri, E., Funke, B., Goldberg, R., Palanisamy, N., Chaganti, R. S., Magenis, E., Shprintzen, R. J. & Morrow, B. E. (1999) Hum. Mol. Genet. 8, 1157-1167. [DOI] [PubMed] [Google Scholar]
- 14.Shaikh T. H., Kurahashi, H., Saitta, S. C., O'Hare, A. M., Hu, P., Roe, B. A., Driscoll, D. A., McDonald-McGinn, D. M., Zackai, E. H., Budarf, M. L., et al. (2000) Hum. Mol. Genet. 9, 489-501. [DOI] [PubMed] [Google Scholar]
- 15.Liu H., Heath, S. C., Sobin, C., Roos, J. L., Galke, B. L., Blundell, M. L., Lenane, M., Robertson, B., Wijsman, E. M., Rapoport, J. L., Gogos, J. A. & Karayiorgou, M. (2002) Proc. Natl. Acad. Sci. USA 99, 3717-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gogos J. A., Santha, M., Takacs, Z., Beck, K. D., Luine, V., Lucas, L. R., Nadler, J. V. & Karayiorgou, M. (1999) Nat. Genet. 21, 434-439. [DOI] [PubMed] [Google Scholar]
- 17.Nurnberger J. I., Blehar, M. C., Kaufmann, C. A., York-Cooler, C., Simpson, S. G., Harkavy-Friedman, J., Severe, J. B., Malaspina, D. & Reich, T. (1994) Arch. Gen. Psychiatry 51, 849-859. [DOI] [PubMed] [Google Scholar]
- 18.Abecasis G. R., Cherny, S. S., Cookson, W. O. & Cardon, L. R. (2002) Nat. Genet. 30, 97-101. [DOI] [PubMed] [Google Scholar]
- 19.Dawson E., Abecasis, G. R., Bumpstead, S., Chen, Y., Hunt, S., Beare, D. M., Pabial, J., et al. (2002) Nature 418, 544-548. [DOI] [PubMed] [Google Scholar]
- 20.Hedrick P. W. (1987) Genetics 117, 331-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devlin B. & Risch, N. (1995) Genomics 29, 311-322. [DOI] [PubMed] [Google Scholar]
- 22.Abecasis G. R., Noguchi, E., Heinzmann, A., Traherne, J. A., Bhattacharyya, S., Leaves, N. I., Anderson, G. G., Zhang, Y., Lench, N. J., Carey, A., et al. (2001) Am. J. Hum. Genet. 68, 191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terwilliger J. & Ott, J. (1992) Hum. Hered. 42, 337-346. [DOI] [PubMed] [Google Scholar]
- 24.McIntyre L. M., Martin, E. R., Simonsen, K. L. & Kaplan, N. L. (2000) Genet. Epidemiol. 19, 18-29. [DOI] [PubMed] [Google Scholar]
- 25.Abecasis G. R., Cardon, L. R. & Cookson, W. O. C. (2000) Am. J. Hum. Genet. 66, 279-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abecasis G. R., Cookson, W. O. & Cardon, L. R. (2000) Eur. J. Hum. Genet. 8, 545-551. [DOI] [PubMed] [Google Scholar]
- 27.Fournier A. E., GrandPre, T. & Strittmatter, S. M. (2001) Nature 409, 341-346. [DOI] [PubMed] [Google Scholar]
- 28.Guarguaglini G., Battistoni, A., Pittoggi, C., Di Matteo, G., Di Fiore, B. & Lavia, P. (1997) Biochem. J. 325, 277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coutavas E., Ren, M., Oppenheim, J. D., D'Eustachio, P. & Rush, M. G. (1993) Nature 366, 585-587. [DOI] [PubMed] [Google Scholar]
- 30.Nagase T., Ishikawa, K., Kikuno, R., Hirosawa, M., Nomura, N. & Ohara, O. (1999) DNA Res. 6, 337-345. [DOI] [PubMed] [Google Scholar]
- 31.Halford S., Wilson, D. I., Daw, S. C., Roberts, C., Wadey, R., Kamath, S., Wickremasinghe, A., Burn, J., Goodship, J., Mattei, M. G., et al. (1993) Hum. Mol. Genet. 2, 1577-1582. [DOI] [PubMed] [Google Scholar]
- 32.Putilina T., Wong, P. & Gentleman, S. (1999) Mol. Cell. Biochem. 195, 219-226. [DOI] [PubMed] [Google Scholar]
- 33.Bartels D. J., Mitchell, D. A., Dong, X. & Deschenes, R. J. (1999) Mol. Cell. Biol. 19, 6775-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reich D. E., Cargill, M., Bolk, S., Ireland, J., Sabeti, P. C., Richter, D. J., Lavery, T., Kouyoumjian, R., Farhadian, S. F., Ward, R. & Lander, E. S. (2001) Nature 411, 199-204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.