Abstract
Mice heterozygous for the retinoblastoma (Rb) tumor suppressor gene develop pituitary and thyroid tumors with high penetrance. We demonstrate here that loss of the ARF tumor suppressor strongly accelerates intermediate lobe pituitary tumorigenesis in Rb heterozygous mice. These effects in the pituitary are greater than those conferred by p53 loss in that Rb+/−;ARF−/− mice display significantly more early atypical lesions than Rb+/−; p53−/− mice. Also, Rb+/−;ARF−/− compound mutants do not develop many of the novel tumors or precancerous lesions seen in Rb+/−;p53−/− compound mutants. Although complete loss of ARF expression is not obligatory for pituitary tumorigenesis in Rb+/− mice, alterations of the ARF locus are observed in tumors from Rb+/−;ARF+/− mice, consistent with a selective advantage of ARF inactivation in this context. We conclude that inactivation of ARF acts more broadly than that of p53 in connecting abrogation of the Rb pathway to tumorigenesis.
The retinoblastoma (RB) tumor suppressor gene is critical for control of the G1–S cell cycle transition and tumor suppression (1). Individuals heterozygous for an RB mutation are predisposed to retinoblastoma and osteosarcoma, and RB is also inactivated in a wide variety of spontaneously arising human cancers (2). In addition, the protein product, pRB, is regulated by a number of factors that are also mutated or otherwise inactivated in other familial cancer syndromes and sporadic tumors (2). These include amplification and overexpression of cyclin D1 (CCND1) in carcinomas, amplification and dominant activating mutation of CDK4 in melanomas, and mutation or deletion of p16INK4A (CDKN2A) in multiple tumor types (3–5). Because this pathway appears to be a central regulator of tumor suppression, elucidation of the downstream consequences of its inactivation is critical for understanding the molecular bases of cellular transformation and tumor development.
pRB interacts with a number of cellular proteins to regulate a multitude of cellular events. One such role is in the regulation of the late G1 to S transition of the cell cycle through its interactions with the E2F family of transcription factors. In addition, pRB is known to regulate other cellular functions, including differentiation through other cellular factors, and is known to be important in the development of multiple cell lineages (6, 7).
The effects of Rb mutation have been studied extensively in mice (8–10). Homozygous Rb mutant (Rb−/−) embryos die in midgestation because of a defect in hematopoiesis. Rb heterozygous mice (Rb+/−) develop pituitary tumors and thyroid tumors with high penetrance as well as a number of neuroendocrine tumors (9, 11–13), representing a syndrome of multiple endocrine neoplasia (13). These tumors exhibit loss of the WT allele of Rb consistent with a requirement for this event in tumorigenesis (11, 12, 14). By generating mice mutant for combinations of genes, we and others have been able to use this system to probe downstream components of the Rb pathway and establish genetic interactions that impinge on the consequences of Rb inactivation and subsequent tumor formation (15–18).
The analysis of chimeras composed partly of Rb homozygous mutant embryonic stem cells showed that inactivation of both alleles of Rb is a required, rate-limiting step for pituitary tumorigenesis (19, 20). Pituitary-specific ablation of homozygous conditional alleles of Rb by a rat pro-opiomelanocortin promoter-driven Flp enzyme resulted in mice with significantly shorter tumor latency than that of Rbfrt/− controls lacking the enzyme or Rb+/− mice (21), confirming this result. Other genes, notably upstream regulators of pRB, also contribute to pituitary tumorigenesis in the mouse and further highlight the centrality of the Rb pathway in this process. Mutation of members of the Ink4 and Cip/Kip families of cyclin-dependent kinase inhibitors predispose to pituitary hyperplasia. Mice lacking p27Kip1 (Cdkn1b) or p18Ink4c (Cdkn2c) develop pituitary hyperplasia, and the compound mutants for p21Cip1 (Cdkn1a) demonstrate enhanced tumorigenesis (22–25).
A number of tumor models based on selective inactivation of Rb family function have been developed as well, including strains that express a truncated simian virus 40 large T (TgT121) antigen in the choroid plexus (26) or human papillomavirus (HPV-16) E7 in photoreceptors (27) or ocular lens (28). These strains have been used to examine the genetic interactions between inactivation of Rb and other tumor suppressor genes in tumorigenesis.
By crossing the TgT121 strain into the p53 null background, it was shown that the proliferative advantage of tumor cells achieved through abrogation of Rb family function resulted in high levels of p53-dependent apoptosis contributing to the long tumor latency. Removal of this latter mechanism of tumor suppression resulted in rapid growth of tumors (27, 29, 30). Subsequently, the study of Rb+/−;p53−/− germ-line mutant mice revealed clear cooperative roles for loss of both tumor suppressor genes in the development of pinealoblastoma, bronchial epithelial hyperplasia, and pancreatic islet cell hyperplasia, all of which were lesions not found in high frequency in Rb+/− or p53−/− mice (15, 16), although the predisposition to bronchial and islet cell hyperplasias differs for mice harboring a different targeted Rb allele (13). Furthermore, the pinealoblastomas exhibited loss of heterozygosity (LOH) for both Rb and p53 in Rb+/−;p53+/− mice, indicating that the inactivation of both genes are obligate genetic events in the development of these lesions (15). Surprisingly, pituitary tumorigenesis in Rb+/− mice was not significantly affected by inactivation of p53, and LOH at this locus was observed very infrequently (1/16) in pituitary tumors isolated from Rb+/−;p53+/− mice (15).
A previously unexplored candidate interactor is the ARF tumor suppressor, which like the cyclin-dependent kinase inhibitor has growth-suppressive properties, but is not known to directly affect cyclin-dependent kinase activity or components (31). ARF was identified as an alternative transcript of the Ink4a locus possessing a unique first exon (1β) and promoter (31). ARF functions as a tumor suppressor gene in mice (32, 33) and there is evidence to suggest such a role in humans as well (34–36). Biochemically, ARF inhibits multiple functions of MDM2, leading to stabilization of p53 (37–42). ARF has been shown genetically to be required for efficient p53-dependent responses to cellular stresses including overexpression of oncogenes such as RAS, MYC, and E2F-1 as well as DNA damage (43–47). Furthermore, ARF is up-regulated in Rb-deficient cells (47) and has been shown to be a transcriptional target of E2F-1 (44, 48, 49). Therefore, as a potential bridge between the Rb and p53 pathways, ARF would be expected to play a prominent role in a number of processes, including tumorigenesis. In at least one strain of Rb+/− mice, the development of pinealoblastoma, bronchial epithelial hyperplasia, and pancreatic islet cell hyperplasia occurred in appreciable frequency only in Rb+/−; p53−/− mice (15). Given the role of ARF in activating p53 in a variety of contexts, one might expect the inactivation of ARF to have similar effects on the Rb+/− background.
ARF also has functions that do not depend on p53. For example, ARF−/− mice have eye abnormalities associated with failed hyaloid vascular system regression that does not depend on p53 (50). Also, overexpression of ARF has been demonstrated to impair S-phase progression in p53-deficient tumor cell lines (51). It is not clear, however, if p53-independent functions of ARF contribute to tumor suppression. The lack of a clear cooperative effect between loss of p53 and loss of Rb in promoting intermediate lobe pituitary tumorigenesis (15, 16) raises the interesting question of whether loss of ARF can affect this process. Such an interaction might represent effects much broader than those conferred by p53 inactivation, perhaps demonstrating roles for ARF in tumorigenesis beyond that of a simple link between Rb inactivation and p53.
Materials and Methods
Mice.
Rb+/−;ARF−/− animals were generated by breeding Rb+/− mice to ARF−/− mice on a mixed 129 × B6 background to generate compound mutants. Rb+/−;ARF−/−, Rb+/−; ARF+/−, and ARF−/− mice were intercrossed to generate the Rb+/−;ARF−/− compound mutant mice. A similar strategy was used to generate mixed 129 × B6 Rb+/−; p53−/− compound mutant controls. Survival curves were compiled from mice that were killed when moribund. The logrank (Mantel–Haenszel) test was performed to establish statistical significance (PRISM 3, GraphPad, San Diego).
Tumor and Early Lesion Analysis.
For tumor analysis, mice were killed, heads were cut along the midline, fixed in 10% formalin overnight, and processed, and midsagittal sections were cut and stained for BrdUrd and terminal deoxynucleotidyltransferase-mediated dUTP end labeling (see below). For early atypical proliferates (EAPs), mice were anesthetized and perfused with 4% paraformaldehyde (14) or pituitaries were microdissected immediately after death, fixed in Bouin's fixative at room temperature for 16 h, and rinsed in 70% ethanol. Tissues were processed and paraffin serial sections were cut at 4 μm. Sections were stained with hematoxylin (Mayer's) and eosin and screened at ×600–1,000 for early lesions. The proliferation index was calculated by counting the percentage of BrdUrd-positive nuclei out of fields containing ≈500 nuclei, and significance was tested by using a paired t test. Volumetric analysis of EAPs was conducted by measuring the diameters of each lesion at the widest point in each dimension, assuming an ellipsoid shape, and calculating the volume as (π/6 × d1 × d2 × d3).
Terminal Deoxynucleotidyltransferase-Mediated dUTP End Labeling (TUNEL) and Immunochemistry.
Apoptosis was assayed by using the TUNEL assay (52). For BrdUrd analysis, a mixture of BrdUrd (Sigma) and 5-fluoro-2′-deoxyuridine (Sigma) was injected i.p. (100 μg and 10 μg/g body weight, respectively) 1 h before death. Sections were rehydrated, blocked in 3% H2O2, processed in pepsin and HCl, and incubated with a mouse monoclonal anti-BrdUrd antibody (Becton Dickinson). All immunohistochemistry used the ABC peroxidase detection system (Vector Laboratories).
ARF Expression.
Whole pituitary tumor samples were lysed in 100 mM Tris (pH 8), 100 mM NaCl, 1% Nonidet P-40, with Complete protease inhibitor tablets (Roche Molecular Biochemicals). A total of 450 μg of total protein was electrophoretically separated on 12.5% PAGE, blotted to poly(vinylidene difluoride) (Millipore) membrane, and probed with anti-ARF antibodies (Novus, 1:2,000).
Southern and Northern Blot Analysis.
Pituitary tumors were microdissected immediately after death and frozen in liquid nitrogen. Southern blot was performed by using standard protocols with DNA digested with PstI/Asp718i for Rb southern or AflII for the ARF southern. Blots were hybridized with radiolabeled probes spanning exon 3 of the Rb locus and exon 1β of the ARF locus. Northern blot analysis was performed by using standard protocols with 10 μg total RNA isolated from pituitary tumors. Probe was made to exon 1β of the ARF locus or to GAPDH cDNA.
Laser Capture Microdissection–PCR.
Samples processed for laser capture analysis were fixed in 4% paraformaldehyde, paraffin-embedded, sectioned, and stained with hematoxylin and eosin. Early abnormal proliferates or surrounding normal pituitary tissue was captured by using a PixCell II laser capture microdissection system (Arcturus, Mountain View, CA). Laser capture microdissection-captured samples were digested in proteinase K overnight at 42°C, inactivated for 10 min at 95°C, and used in 50 μl [2 mM MgCl2, 50 mM KCl, 10 mM Tris⋅HCl (pH 8.3), 200 μM dNTPs, primers 0.4 μM (RI3, RbpA) or 0.8 μM (Rbint3f), 0.001% gelatin, 1 μl Taq Polymerase (Amplitaq Gold)] PCRs [94°C for 10 min, 40 times (94°C for 30 s, 61°C for 1 min, and 72°C for 1 min), 72°C for 10 min]. Primers used were: Rbint3f (common), 5′-CACCATGTGCAATGCTTGA-3′; RI3 (WT), 5′-CCCATGTTCGGTCCCTAG-3′; and RbpA (mutant), 5′-ACGAGATCAGCAGCCTCTGT-3′. PCR products were resolved on 2% agarose gels, with ≈130-bp WT and 160-bp mutant bands.
Results
Loss of ARF Dramatically Accelerates Pituitary Tumorigenesis in Rb+/− Mice.
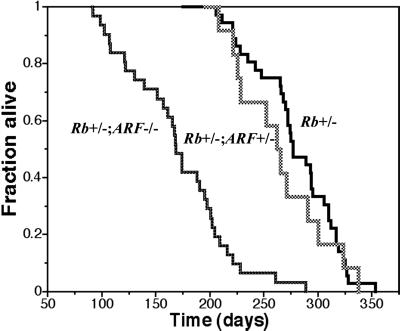
A comparison of the survival curves of Rb+/− mice versus Rb+/−;ARF−/− compound mutant mice on a mixed 129 × B6 background revealed a strong interaction between the inactivation of these two tumor suppressor genes. Whereas Rb+/− mice (n = 37) in this cohort survived for 276 ± 41 days (median = 277 days), the Rb+/−;ARF−/− mice (n = 32) survived for 168 ± 49 days (median = 168.5 days; P < 0.0001) (Fig. 1). Clinical observation and necropsy of these mice revealed that virtually all of these mice died of intermediate lobe pituitary tumors. Although ARF−/− mice have not been observed to have pituitary or thyroid lesions (ref. 33 and data not shown), Rb+/−;ARF−/− mice were highly predisposed to development of pituitary tumors as well as thyroid C cell carcinomas.
Fig 1.
Rb+/−;ARF−/− compound mutant mice exhibit significantly decreased survival relative to Rb+/− and Rb+/−;ARF+/− mice. Survival curves of Rb+/− (n = 37), Rb+/−;ARF−/− (n = 32), and Rb+/−;ARF+/− (n = 13) mice show that Rb+/−;ARF−/− mice had a significantly decreased median survival of 168.5 days vs. 277 days for Rb+/− controls (P < 0.0001). The Rb+/−; ARF+/− mice (n = 13) exhibited a median survival of 262 days, comparable to that of Rb+/− mice (P = 0.34) and significantly longer than that of Rb+/−; ARF−/− mice (P < 0.0001).
Importantly, Rb+/−;ARF−/− mice did not exhibit the pancreatic islet cell hyperplasia (0/10), pinealoblastoma (0/14), or bronchial epithelial hyperplasia with appreciable frequency (0/10), as reported in Rb+/−;p53−/− mice (refs. 15 and 16; data not shown). Therefore, in these tissues, ARF loss is not equivalent to p53 loss, implying that p53 is regulated by other means. Similar findings have been demonstrated in a mouse model for medulloblastoma in which loss of p53 but not ARF accelerates tumor development in animals heterozygous for Patched (Ptch) (53).
Pituitary Tumors of Rb+/−;ARF−/− Mice Have Higher Proliferative Capacity.
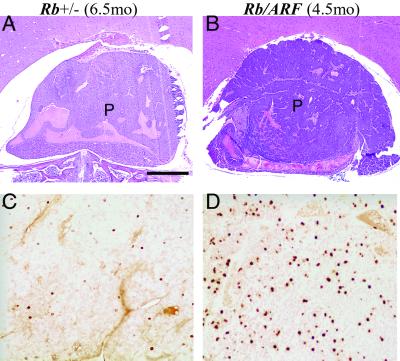
Examination of tumors isolated from Rb+/−; ARF−/− mice and Rb+/− controls revealed that those derived from the compound mutants developed much faster even though end-stage tumors from both populations were histologically indistinguishable. Tumors in 3- to 5-month-old Rb+/−; ARF−/− mice appeared similar to 5- to 7-month-old Rb+/− controls and were obvious on histological examination. We examined comparably sized tumors from these two populations to establish whether they were different with respect to their cellular content or growth characteristics. Although grossly similar (compare Fig. 2 A to B), BrdUrd labeling of cells in S phase was significantly greater in the samples of Rb+/−; ARF−/− mice (Fig. 2D) relative to Rb+/− controls (Fig. 2C), indicating that these tumors had higher proportions of proliferating cells. Quantitation of the proliferation index (see Materials and Methods) for three matched pairs of tumor samples demonstrated that 15.3 ± 4% of nuclei in Rb+/−;ARF−/− tumors were labeled as compared with 2.3 ± 0.6% (P < 0.05) of nuclei for Rb+/− controls. This finding indicates that loss of ARF facilitates the proliferation of tumor cells that have lost Rb.
Fig 2.
Comparison of tumors of similar stage from Rb+/−;ARF−/− and Rb+/− mice shows higher levels of proliferation in Rb+/−;ARF−/− samples. Midsagittal sections of pituitary tumors (P) at comparable stages of development were stained with hematoxylin and eosin (A and B) or used for BrdUrd immunohistochemistry (C and D). Samples from a Rb+/− (6.5 months) mouse (A) and a Rb+/−;ARF−/− (4.5 months) compound mutant mouse (B) are shown. These animals were injected with BrdUrd (10 μg/kg body weight) 1 h before death, and analysis of BrdUrd incorporation demonstrated a significantly higher S-phase fraction (P < 0.05 for three matched pairs) in the Rb+/−; ARF−/− sample (D) relative to the Rb+/− control (C). (Calibration bar: 100 μm for A and B; 50 μm for C and D.)
Alternatively, ARF mutation could accelerate tumorigenesis by abrogating an apoptotic response in cells that might otherwise have been eliminated. However, terminal deoxynucleotidyltransferase-mediated dUTP end labeling of tumor sections isolated in this group of samples did not reveal any differences (data not shown). We also examined hematoxylin/eosin-stained sections of these tumors to identify cells exhibiting hallmarks of apoptosis, including marginated chromatin and fragmented nuclei. Low levels of apoptosis were observed in all samples examined with no distinctive differences observed (data not shown). This evidence indicates that at this stage in tumor development, examining comparatively sized lesions, simultaneous inactivation of Rb and ARF provides tumor cells with a proliferation advantage without obvious effects on apoptosis.
Rb+/−;ARF−/− Mice Develop Early, Aggressive Focal Lesions.
To investigate the basis for the marked acceleration of tumor development in Rb+/−;ARF−/− mice, we analyzed serial sections of pituitary glands for EAPs (14), the first morphologically distinct lesions that can be identified in Rb+/− animals. The abnormal cells that comprise EAPs often cluster at the border between the intermediate and posterior (neural) lobes and are characterized by high nuclear-cytoplasmic ratios, irregularly shaped nuclei, and coarse chromatin (14). Rb+/− mice develop the first EAPs between postnatal day (PND) 35 and PND 60 (14).
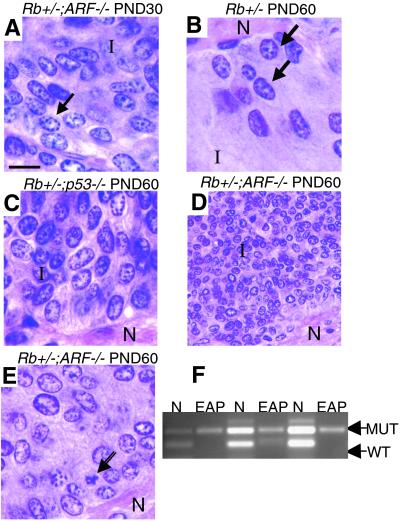
We dissected pituitary glands from PND 30 and PND 60 Rb+/−, Rb+/−;ARF−/−, and Rb+/−;p53−/− mice and used hematoxylin/eosin-stained serial sections for our analysis. In our sample, none of the PND 30 Rb+/− or Rb+/−;p53−/− samples had any lesions, consistent with the interpretation that loss of p53 does not accelerate the onset of pituitary tumorigenesis. Surprisingly, a number of PND 30 Rb+/−;ARF−/− samples had EAPs that were larger than the EAPs found in PND 60 Rb+/− controls (compare Fig. 3 A to B; Table 1). Importantly, PCR analysis of microdissected EAPs demonstrated LOH at Rb, showing that loss of Rb is still required for formation of tumors in Rb+/−;ARF−/− mice (Fig. 3F). The cells from EAPs show presence of the mutant allele only with loss of the WT allele (Fig. 3F). Whereas all of the Rb+/− PND 60 EAPs were of grade 1 or 2, 40% of the Rb+/−;ARF−/− PND 30 lesions were of grade 4 (Table 1). Furthermore, by PND 60, the Rb+/−;ARF−/− mice had about three times as many independent EAPs on average (Fig. 3 D and E; Table 2) as the Rb+/− (Fig. 3B) or Rb+/−;p53−/− (Fig. 3C) controls, and some were so large they had begun to fuse (these were counted as single lesions). Whereas PND 60 Rb+/−;p53−/− samples and Rb+/− controls had comparable numbers of lesions (Table 2), the Rb+/−;p53−/− pituitaries contained nodules that were larger than the lesions found in Rb+/− controls (Fig. 3C; Table 1). Interestingly, whereas the PND 60 Rb+/−;ARF−/− mice had a significantly greater number of lesions than the PND 60 Rb+/−;p53−/− mice, the grade distributions of the EAPs overlapped for these two groups, indicating that loss of p53 contributes to pituitary tumor development in Rb+/− mice, but that this effect appears to be mechanistically distinct from the effects observed with loss of ARF.
Fig 3.
EAPs appear in the pituitaries of Rb+/−;ARF−/− compound mutants earlier than those in Rb+/− and Rb+/−; p53−/− compound mutant mice and are larger by PND 30. Comparison of hematoxylin-stained EAPs from Rb+/−, Rb+/−;ARF−/−, and Rb+/−; p53−/− compound mutant mice. (A) EAP of a PND 30 Rb+/−;ARF−/− compound mutant mouse (×1,000) showing dense groups of abnormal cells in the intermediate lobe (I) containing coarse chromatin, and large, irregular nuclei at the border with the posterior (neural) lobe. (B) EAP of a PND 60 Rb+/− mouse at the intermediate-neural lobe border (×1,000). Arrows indicate abnormal cells. Note the size of this EAP relative to the lesion in A. (C) EAP of a PND 60 Rb+/−;p53−/− compound mutant mouse showing a large lesion with densely packed abnormal cells in the intermediate lobe (×1,000). (D) Low-power view of an EAP of a PND 60 Rb+/−;ARF−/− compound mutant mouse (×400) showing typical large cluster of intermediate lobe (I) tumor cells near the neural lobe (N). (E) Higher-power (×1,000) view of one of the same lesions (D) demonstrating typical morphology of tumor cells. Note the presence of an apoptotic cell (arrow). (F) LOH at the Rb locus is observed in PND 60 EAPs from Rb+/−;ARF−/− mice. PCR analysis of DNA from laser capture-dissected samples shows retention of mutant allele with absence of WT allele in captured EAPs, while both alleles are intact in adjacent normal pituitary (N). (Calibration bar: 10 μm for A–C and E; 25 μm for D.)
Table 1.
Size distribution of EAPs in PND 30 and PND 60 mice
| Genotype and age | Grade 1 | Grade 2 | Grade 3 | Grade 4 |
|---|---|---|---|---|
| Rb+/− PND 60 | 40% (2) | 60% (3) | 0 | 0 |
| Rb+/−;ARF−/− PND 30 | 40% (2) | 20% (1) | 0 | 40% (2) |
| Rb+/−;ARF−/− PND 60 | 22.9% (8) | 51.4% (18) | 2.9% (1) | 22.9% (8) |
| Rb+/−;p53−/− PND 60 | 0 | 54.5% (6) | 9.1% (1) | 36.4% (4) |
All EAPs from all affected mice were measured and classified according to tumor volume: grade 1, <103 μm3; grade 2, 103–5×104 μm3; grade 3, 5×104–1×105 μm3; and grade 4, >105 μm3. Listed are the percentages and numbers in parentheses, for each tumor grade for mice of each genotype.
Table 2.
EAP analysis in PND 30 and PND 60 mice
| Mice | No. of samples | No. of mice with lesions | Average no. of lesions per affected mouse |
|---|---|---|---|
| PND 30 | |||
| Rb+/− | 8 | 0 | N/A |
| Rb+/−;ARF−/− | 12 | 3 | 1.7 ± 0.4 |
| Rb+/−;p53−/− | 9 | 0 | N/A |
| PND 60 | |||
| Rb+/− | 5 | 3 | 1.7 ± 0.7 |
| Rb+/−;ARF−/− | 5 | 5 | 7.0 ± 1.0 |
| Rb+/−;p53−/− | 5 | 4 | 2.8 ± 1.0 |
EAPs were identified in hematoxylin/eosin-stained serial sections of pituitaries isolated from Rb+/−, Rb+/−;ARF−/−, and Rb+/−;p53−/− mice at PND 30 and PND 60. Only EAPs that had distinct borders were counted as individual lesions. N/A, not available.
The great difference between the sizes of the PND 60 lesions makes it impractical to compare their apoptotic or proliferative indices. Careful examination of the large nodules in Rb+/−;ARF−/− samples revealed the presence of obvious apoptotic nuclei. Virtually all of the sections through each nodule contain apoptotic figures, indicating that apoptosis is not completely compromised in these developing tumors in the absence of ARF. We cannot, however, rule out the possibility that the absence of ARF may protect cells from apoptosis before the development of early lesions that can be identified histologically.
The ARF Locus Is Altered in Tumors of Rb+/−;ARF+/− Mice.
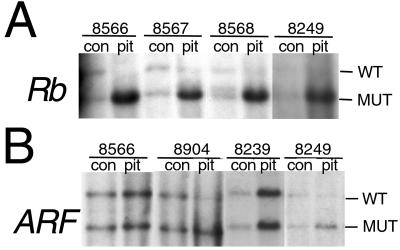
Acquired proliferative advantage in developing tumors might provide a selection pressure to inactivate ARF in tumors of Rb+/− mice, perhaps making ARF loss an obligatory event in pituitary tumorigenesis. We addressed this possibility by examining whether ARF expression is affected in tumors of Rb+/− mice. Because ARF can behave as a classic tumor suppressor gene, reduction to homozygosity for the mutant allele may be selected for in lesions of ARF+/− mice as reported (32). Despite the fact that Rb+/−;ARF+/− mice have a survival curve that is similar to that for Rb+/− mice (Fig. 1; P = 0.34), two of six tumors from double heterozygotes demonstrated alterations at the ARF locus (Fig. 4). Southern blot analysis showed that all pituitary tumor samples exhibited LOH for Rb with only the band associated with the mutant allele remaining (Fig. 4A). DNA isolated from normal brain tissue confirmed heterozygosity for the Rb mutation (9). Similarly, the same control samples, when analyzed with a probe specific for ARF, confirmed heterozygosity for the ARF mutation (Fig. 4B) (33). Although four pituitary samples (Fig. 4B; pit 8566 and 8239, and data not shown) maintained equivalent levels of hybridization to both alleles of ARF, the mutant allele was enriched in two pituitary samples (Fig. 4B; pit 8904 and 8249). Western and Northern blot analyses indicated that ARF is expressed in pituitary tumors from Rb+/− mice (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). These results, showing alterations in the ARF locus in two of six samples, indicate that although ARF loss may not be a requisite event in pituitary tumorigenesis, there is selective advantage for inactivating ARF in developing tumors.
Fig 4.
Tumors isolated from Rb+/−;ARF+/− animals exhibit alterations at the ARF locus. (A) DNA from normal adjacent cerebellum (con) confirms that the animals were heterozygous for Rb, and all samples of pituitary (pit) tumors showed LOH for Rb with only the 5.2-kB lower band associated with the mutant allele remaining. (B) DNA from normal adjacent cerebellum (con) confirmed that all of the animals were heterozygous for ARF. Of four pituitary samples that also showed this pattern of equivalent hybridization, two are shown here (pit 8566 and 8239). Two pituitary samples exhibited alterations at the ARF locus. These samples demonstrated enrichment for the lower 6.0-kB band associated with mutant allele (pit 8904 and 8249).
Discussion
The Rb pathway is critically important in pituitary tumorigenesis in mice. In addition to inactivation of Rb, mutations in upstream cyclin-dependent kinase inhibitors such as p21Cip1, p27Kip1, and p18Ink4c can contribute to this process (17, 18, 22–25). Unlike mutants lacking genes that cooperate with loss of Rb in this process, ARF–deficient mice do not sustain pituitary lesions (32, 33), and evidence obtained in other systems suggested that ARF might play an important role downstream of Rb inactivation in tumorigenesis (44, 47–49). We probed this possibility directly by generating mice containing mutations in both Rb and ARF.
Our results demonstrate a marked acceleration of pituitary tumorigenesis in Rb+/− mice lacking ARF. Based on the analysis of early lesions, it is clear that the tumor suppressor function of ARF, although important, does not appear to act solely in a typical p53-dependent fashion. Although it has been proposed that ARF connects the Rb and p53 pathways through E2F activity and MDM2, respectively (3, 54, 55), other functions of ARF appear to be critical in pituitary tumorigenesis.
Rb+/−;ARF−/− compound mutants have significantly more early lesions at PND 30 and PND 60 than do Rb+/− or Rb+/−;p53−/− mice (Table 2). This early lesion analysis extends previous work in Rb+/−;p53−/− mice (15, 16). Nevertheless, p53 loss has a measurable effect manifested in the significantly larger size of the lesions noted at that time point (Table 1). In addition, Rb+/−;ARF−/− compound mutants do not exhibit the pancreatic islet cell hyperplasia, pinealoblastoma, or bronchial epithelial hyperplasia reported in Rb+/−;p53−/− mice (15, 16), and at lower frequency, in Rb+/− mice (13, 15). In these tissues, ARF mutation does not recapitulate p53 inactivation, indicating that p53 may be regulated through ARF-independent mechanisms in these contexts.
The early lesion analysis has also demonstrated that EAPs can be detected as early as PND 30 in the Rb+/−;ARF−/− compound mutants but are absent in PND 30 Rb+/− and Rb+/−;p53−/− controls (14). By PND 60, Rb+/−;ARF−/− compound mutants have approximately three times as many EAPS that are also larger than those identified in the control populations (Table 2). Furthermore, PCR analysis of microdissected PND 60 EAPs from Rb+/−;ARF−/− animals demonstrate that loss of Rb is still required for tumor formation (Fig. 3F), even in the absence of ARF.
We propose this finding can be explained by at least three possible mechanisms. First, ARF could directly regulate pituitary development, for example, by enforcing cell cycle arrest in differentiated melanotrophs; however, we have not observed any gross differences in the structures of pituitaries from mice lacking ARF, nor have we identified any lesions in adult ARF-deficient mice that would suggest such a role. Second, ARF deficiency may increase the proportion of individual cells that eliminate the remaining WT allele of Rb in development or accelerate the timing of the loss of the WT allele of Rb. Conditional inactivation of Rb specifically in the pituitary resulted in mice with significantly shorter tumor latency than that of Rb+/− mice (21), as was observed with chimeric mice partly composed of Rb homozygous mutant embryonic stem cells (19, 20). Because the conditional Rb alleles were inactivated by a rat pro-opiomelanocortin-driven Flp enzyme (21), and pro-opiomelanocortin is expressed in midgestation (56), these results indicate that inactivation of both alleles of Rb earlier in development is sufficient to accelerate tumorigenesis. However, there is no evidence that ARF regulates the frequency of LOH events in tumorigenesis, for example, through increased frequency of sister-chromatid exchange or through a more general role in genomic instability independent of p53. Finally, ARF deficiency may enhance the survival or outgrowth of incipient tumor cells after the LOH event at the Rb locus. This proposal explains the findings here most efficiently in that it accounts for the fact that there are more individual lesions in Rb+/−;ARF−/− compound mutants as early as PND 30. If ARF deficiency can enhance the ability of these first Rb-deficient cells to survive or proliferate, then the number of histologically apparent lesions that emerge would be increased.
Importantly, this hypothesis can also explain why there is incomplete selection against ARF in the Rb+/−;ARF+/− mice as LOH was detected by Southern blot in only 2/6 samples. Perhaps the temporal window within which ARF function is critical is quite limited and therefore incompatible with temporal requirements for LOH. The advantage conferred by the absence of ARF may be restricted to such a short period early in the development of tumors that selection is not maintained long enough to result in LOH. Alternatively, ARF loss would not be selected for in tumor cells if ARF acted in a noncell-autonomous fashion to suppress tumorigenesis. The incomplete ARF LOH in Rb+/−;ARF+/− animals, and our findings that ARF can be detected at the RNA and protein level in tumors from Rb+/− animals suggests that complete loss of ARF is not required for pituitary tumor formation in Rb+/− animals. However, we were technically limited in our ability to obtain an appropriate amount of normal pituitary intermediate lobe at an appropriate time in pituitary development to compare ARF levels to those in Rb+/− tumors. Therefore it remains possible that ARF levels may be reduced in tumors from Rb+/− animals and that ARF reduction may be important for pituitary tumorigenesis in Rb+/− mice.
Our evidence in more advanced tumors of 4- to 7-month-old mice indicates that the tumors from Rb+/−;ARF−/− mice have higher proliferation indices than do tumors at similar stages in Rb+/− controls (Fig. 2). Thus ARF loss likely contributes to tumor development by also enhancing the ability of cells within well-developed tumors to proliferate.
We have shown that mutation of ARF significantly accelerates pituitary tumor development in Rb+/− mice. Because ARF loss has such a dramatic effect, particularly on the number of early lesions that is not observed with p53 loss, ARF may be regulating a p53-independent mode of tumor suppression. Given the role of ARF in inhibiting MDM2 (55), it is possible that the p53-independent functions of MDM2 are being regulated by ARF. For example, MDM2 can inhibit the transactivation function of the p53-related protein p73, which has been shown to be important in a variety of p53-independent apoptotic pathways (57–59). In addition, MDM2 has been shown to potentiate E2F-1-mediated transactivation and cellular proliferation (60, 61). Even in the absence of Mdm2, it has been shown that loss of both ARF and p53 can cooperate in the development of novel tumors (62), and prolonged exposure to MYC in B cell lymphomas provides selection for overexpression of Mdm2 in the absence of ARF (63). Therefore, it is likely that even in the absence of a requirement to inactivate p53 in pituitary tumorigenesis, ARF may be restraining some other activity of MDM2 or acting independently of the Mdm2-p53 pathway to inhibit tumor development. The finding that ARF expression can enforce a cell cycle arrest in Mdm2/p53 double mutant mouse embryo fibroblasts (MEFs) but not in p53−/− MEFs is evidence for novel targets of ARF (62). There is also evidence that ARF can induce apoptosis in the absence of p53 (64). In vivo, ARF−/− mice have defects in eye development that are not observed in p53−/− mice (50).
Our analysis is an in vivo demonstration that ARF plays an important role in tumor suppression in the context of Rb inactivation. The early lesion analysis suggests that ARF loss does not completely compromise apoptosis because dying cells can be readily detected in PND 60 Rb+/−;ARF−/− nodules. However, it is not possible to know whether ARF promotes apoptosis immediately after loss of the WT allele of Rb as this event occurs before the appearance of histologically recognizable lesions. Later in tumor development, it is clear that loss of ARF can increase the proportion of tumor cells that are proliferating. These data help to define a broad scope of effects of the ARF tumor suppressor in the context of Rb inactivation that extends beyond functional inactivation of p53.
Supplementary Material
Acknowledgments
We thank C. Sherr, M. Roussel, F. Zindy, and E. van de Kamp (St. Jude, Memphis, TN) for helpful discussions, technical advice, and the generous gifts of the ARF−/− mice (33) and exon 1β probe. Additionally, we thank J. Sage and E. Flores for helpful discussions and technical advice. This work was supported in part by the National Institutes of Health, the Medical Scientist Training Program (K.Y.T. and D.A.R.), and a fellowship from the Koch Foundation (to K.Y.T.). T.J. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
RB, retinoblastoma
EAP, early atypical proliferate
PND, postnatal day
LOH, loss of heterozygosity
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Weinberg R. A. (1995) Cell 81, 323-330. [DOI] [PubMed] [Google Scholar]
- 2.Fearon E. R. (1997) Science 278, 1043-1050. [DOI] [PubMed] [Google Scholar]
- 3.Sherr C. J. (2000) Cancer Res. 60, 3689-3695. [PubMed] [Google Scholar]
- 4.Sherr C. J. (1996) Science 274, 1672-1677. [DOI] [PubMed] [Google Scholar]
- 5.Mulligan G. & Jacks, T. (1998) Trends Genet. 14, 223-229. [DOI] [PubMed] [Google Scholar]
- 6.Dyson N. (1998) Genes Dev. 12, 2245-2262. [DOI] [PubMed] [Google Scholar]
- 7.Lipinski M. M. & Jacks, T. (1999) Oncogene 18, 7873-7882. [DOI] [PubMed] [Google Scholar]
- 8.Clarke A. R., Maandag, E. R., van Roon, M., van der Lugt, N. M., van der Valk, M., Hooper, M. L., Berns, A. & te Riele, H. (1992) Nature 359, 328-330. [DOI] [PubMed] [Google Scholar]
- 9.Jacks T., Fazeli, A., Schmitt, E. M., Bronson, R. T., Goodell, M. A. & Weinberg, R. A. (1992) Nature 359, 295-300. [DOI] [PubMed] [Google Scholar]
- 10.Lee E. Y., Chang, C. Y., Hu, N., Wang, Y. C., Lai, C. C., Herrup, K., Lee, W. H. & Bradley, A. (1992) Nature 359, 288-294. [DOI] [PubMed] [Google Scholar]
- 11.Harrison D. J., Hooper, M. L., Armstrong, J. F. & Clarke, A. R. (1995) Oncogene 10, 1615-1620. [PubMed] [Google Scholar]
- 12.Hu N., Gutsmann, A., Herbert, D. C., Bradley, A., Lee, W. H. & Lee, E. Y. (1994) Oncogene 9, 1021-1027. [PubMed] [Google Scholar]
- 13.Nikitin A. Yu., Juarez-Perez, M. I., Li, S., Huang, L. & Lee, W. H. (1999) Proc. Natl. Acad. Sci. USA 96, 3916-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikitin A. Yu. & Lee, W. H. (1996) Genes Dev. 10, 1870-1879. [DOI] [PubMed] [Google Scholar]
- 15.Williams B. O., Remington, L., Albert, D. M., Mukai, S., Bronson, R. T. & Jacks, T. (1994) Nat. Genet. 7, 480-484. [DOI] [PubMed] [Google Scholar]
- 16.Harvey M., Vogel, H., Lee, E. Y., Bradley, A. & Donehower, L. A. (1995) Cancer Res. 55, 1146-1151. [PubMed] [Google Scholar]
- 17.Brugarolas J., Bronson, R. T. & Jacks, T. (1998) J. Cell Biol. 141, 503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park M. S., Rosai, J., Nguyen, H. T., Capodieci, P., Cordon-Cardo, C. & Koff, A. (1999) Proc. Natl. Acad. Sci. USA 96, 6382-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams B. O., Schmitt, E. M., Remington, L., Bronson, R. T., Albert, D. M., Weinberg, R. A. & Jacks, T. (1994) EMBO J. 13, 4251-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maandag E. C., van der Valk, M., Vlaar, M., Feltkamp, C., O'Brien, J., van Roon, M., van der Lugt, N., Berns, A. & te Riele, H. (1994) EMBO J. 13, 4260-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vooijs M., van der Valk, M., te Riele, H. & Berns, A. (1998) Oncogene 17, 1-12. [DOI] [PubMed] [Google Scholar]
- 22.Kiyokawa H., Kineman, R. D., Manova-Todorova, K. O., Soares, V. C., Hoffman, E. S., Ono, M., Khanam, D., Hayday, A. C., Frohman, L. A. & Koff, A. (1996) Cell 85, 721-732. [DOI] [PubMed] [Google Scholar]
- 23.Fero M. L., Rivkin, M., Tasch, M., Porter, P., Carow, C. E., Firpo, E., Polyak, K., Tsai, L. H., Broudy, V., Perlmutter, R. M., et al. (1996) Cell 85, 733-744. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama K., Ishida, N., Shirane, M., Inomata, A., Inoue, T., Shishido, N., Horii, I. & Loh, D. Y. (1996) Cell 85, 707-720. [DOI] [PubMed] [Google Scholar]
- 25.Franklin D. S., Godfrey, V. L., O'Brien, D. A., Deng, C. & Xiong, Y. (2000) Mol. Cell. Biol. 20, 6147-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saenz Robles M. T., Symonds, H., Chen, J. & Van Dyke, T. (1994) Mol. Cell. Biol. 14, 2686-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howes K. A., Ransom, N., Papermaster, D. S., Lasudry, J. G., Albert, D. M. & Windle, J. J. (1994) Genes Dev. 8, 1300-1310. [DOI] [PubMed] [Google Scholar]
- 28.Griep A. E., Herber, R., Jeon, S., Lohse, J. K., Dubielzig, R. R. & Lambert, P. F. (1993) J. Virol. 67, 1373-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Symonds H., Krall, L., Remington, L., Saenz-Robles, M., Lowe, S., Jacks, T. & Van Dyke, T. (1994) Cell 78, 703-711. [DOI] [PubMed] [Google Scholar]
- 30.Pan H. & Griep, A. E. (1994) Genes Dev. 8, 1285-1299. [DOI] [PubMed] [Google Scholar]
- 31.Quelle D. E., Zindy, F., Ashmun, R. A. & Sherr, C. J. (1995) Cell 83, 993-1000. [DOI] [PubMed] [Google Scholar]
- 32.Kamijo T., Bodner, S., van de Kamp, E., Randle, D. H. & Sherr, C. J. (1999) Cancer Res. 59, 2217-2222. [PubMed] [Google Scholar]
- 33.Kamijo T., Zindy, F., Roussel, M. F., Quelle, D. E., Downing, J. R., Ashmun, R. A., Grosveld, G. & Sherr, C. J. (1997) Cell 91, 649-659. [DOI] [PubMed] [Google Scholar]
- 34.Kumar R., Sauroja, I., Punnonen, K., Jansen, C. & Hemminki, K. (1998) Genes Chromosomes Cancer 23, 273-277. [DOI] [PubMed] [Google Scholar]
- 35.Newcomb E. W., Alonso, M., Sung, T. & Miller, D. C. (2000) Hum. Pathol. 31, 115-119. [DOI] [PubMed] [Google Scholar]
- 36.Gardie B., Cayuela, J. M., Martini, S. & Sigaux, F. (1998) Blood 91, 1016-1020. [PubMed] [Google Scholar]
- 37.Zhang Y., Xiong, Y. & Yarbrough, W. G. (1998) Cell 92, 725-734. [DOI] [PubMed] [Google Scholar]
- 38.Tao W. & Levine, A. J. (1999) Proc. Natl. Acad. Sci. USA 96, 6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pomerantz J., Schreiber-Agus, N., Liegeois, N. J., Silverman, A., Alland, L., Chin, L., Potes, J., Chen, K., Orlow, I., Lee, H. W., et al. (1998) Cell 92, 713-723. [DOI] [PubMed] [Google Scholar]
- 40.Honda R. & Yasuda, H. (1999) EMBO J. 18, 22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamijo T., Weber, J. D., Zambetti, G., Zindy, F., Roussel, M. F. & Sherr, C. J. (1998) Proc. Natl. Acad. Sci. USA 95, 8292-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber J. D., Taylor, L. J., Roussel, M. F., Sherr, C. J. & Bar-Sagi, D. (1999) Nat. Cell. Biol. 1, 20-26. [DOI] [PubMed] [Google Scholar]
- 43.de Stanchina E., McCurrach, M. E., Zindy, F., Shieh, S. Y., Ferbeyre, G., Samuelson, A. V., Prives, C., Roussel, M. F., Sherr, C. J. & Lowe, S. W. (1998) Genes Dev. 12, 2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bates S., Phillips, A. C., Clark, P. A., Stott, F., Peters, G., Ludwig, R. L. & Vousden, K. H. (1998) Nature 395, 124-125. [DOI] [PubMed] [Google Scholar]
- 45.Khan S. H., Moritsugu, J. & Wahl, G. M. (2000) Proc. Natl. Acad. Sci. USA 97, 3266-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmero I., Pantoja, C. & Serrano, M. (1998) Nature 395, 125-126. [DOI] [PubMed] [Google Scholar]
- 47.Zindy F., Eischen, C. M., Randle, D. H., Kamijo, T., Cleveland, J. L., Sherr, C. J. & Roussel, M. F. (1998) Genes Dev. 12, 2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeGregori J., Leone, G., Miron, A., Jakoi, L. & Nevins, J. R. (1997) Proc. Natl. Acad. Sci. USA 94, 7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inoue K., Roussel, M. F. & Sherr, C. J. (1999) Proc. Natl. Acad. Sci. USA 96, 3993-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKeller R. N., Fowler, J. L., Cunningham, J. J., Warner, N., Smeyne, R. J., Zindy, F. & Skapek, S. X. (2002) Proc. Natl. Acad. Sci. USA 99, 3848-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yarbrough W. G., Bessho, M., Zanation, A., Bisi, J. E. & Xiong, Y. (2002) Cancer Res. 62, 1171-1177. [PubMed] [Google Scholar]
- 52.Gavrieli Y., Sherman, Y. & Ben-Sasson, S. A. (1992) J. Cell Biol. 119, 493-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wetmore C., Eberhart, D. E. & Curran, T. (2001) Cancer Res. 61, 513-516. [PubMed] [Google Scholar]
- 54.Sherr C. J. (1998) Genes Dev. 12, 2984-2991. [DOI] [PubMed] [Google Scholar]
- 55.Sherr C. J. & Weber, J. D. (2000) Curr. Opin. Genet. Dev. 10, 94-99. [DOI] [PubMed] [Google Scholar]
- 56.Japon M. A., Rubinstein, M. & Low, M. J. (1994) J. Histochem. Cytochem. 42, 1117-1125. [DOI] [PubMed] [Google Scholar]
- 57.Stiewe T. & Putzer, B. M. (2000) Nat. Genet. 26, 464-469. [DOI] [PubMed] [Google Scholar]
- 58.Lissy N. A., Davis, P. K., Irwin, M., Kaelin, W. G. & Dowdy, S. F. (2000) Nature 407, 642-645. [DOI] [PubMed] [Google Scholar]
- 59.Irwin M., Marin, M. C., Phillips, A. C., Seelan, R. S., Smith, D. I., Liu, W., Flores, E. R., Tsai, K. Y., Jacks, T., Vousden, K. H. & Kaelin, W. G., Jr. (2000) Nature 407, 645-648. [DOI] [PubMed] [Google Scholar]
- 60.Martin K., Trouche, D., Hagemeier, C., Sorensen, T. S., La Thangue, N. B. & Kouzarides, T. (1995) Nature 375, 691-694. [DOI] [PubMed] [Google Scholar]
- 61.Loughran O. & La Thangue, N. B. (2000) Mol. Cell. Biol. 20, 2186-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weber J. D., Jeffers, J. R., Rehg, J. E., Randle, D. H., Lozano, G., Roussel, M. F., Sherr, C. J. & Zambetti, G. P. (2000) Genes Dev. 14, 2358-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eischen C. M., Weber, J. D., Roussel, M. F., Sherr, C. J. & Cleveland, J. L. (1999) Genes Dev. 13, 2658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hemmati P. G., Gillissen, B., von Haefen, C., Wendt, J., Starck, L., Guner, D., Dorken, B. & Daniel, P. T. (2002) Oncogene 21, 3149-3161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.