Abstract
With an ever-increasing resource of validated single-nucleotide polymorphisms (SNPs), the limiting factors in genome-wide association analysis have become genotyping capacity and the availability of DNA. We provide a proof of concept of the use of pooled DNA as a means of efficiently screening SNPs and prioritizing them for further study. This approach reduces the final number of SNPs that undergo full, sample-by-sample genotyping as well as the quantity of DNA used overall. We have examined 15 SNPs in the cholesteryl ester transfer protein (CETP) gene, a gene previously demonstrated to be associated with serum high-density lipoprotein cholesterol levels. The SNPs were amplified in two pools of DNA derived from groups of individuals with extremely high and extremely low serum high-density lipoprotein cholesterol levels, respectively. P values <0.05 were obtained for 14 SNPs, supporting the described association. Genotyping of the individual samples showed that the average margin of error in frequency estimate was ≈4% when pools were used. These findings clearly demonstrate the potential of pooling techniques and their associated technologies as an initial screen in the search for genetic associations.
Association testing may be regarded as a comparison of allele frequency between cases and controls, in which a statistically significant difference is used to implicate a locus in a disease etiology. The methodology relies on the investigator testing either the causal variant itself or a marker in linkage disequilibrium (LD) with the causal variant. It is now well known that the distance over which LD extends is highly variable, not only between populations (1–3) but also across genomic regions (4, 5). A comprehensive genome scan by association analysis is therefore likely to require tens or hundreds of thousands of markers (6, 7).
To date, we have performed validation experiments on ≈200,000 putative single-nucleotide polymorphisms (SNPs) from the public domain, and the confirmation rate is ≈60% in a Caucasian sample (unpublished results). The theoretical advantages of analyzing these SNPs by an association rather than by a linkage approach are clear (8). The reality, however, is hindered by the expense of generating the necessarily large number of genotypes. The use of pooled DNA has been offered as an alternative in the case of qualitative traits (9–11), but practical extensions to quantitative traits have not been presented thus far.
To evaluate the applicability of pooled samples, SNPs in the cholesteryl ester transfer protein (CETP) gene were tested for association by using a pair of pools containing DNA from individuals with low and high serum high-density lipoprotein cholesterol (HDL-C) levels, respectively. CETP is known to play an essential role in reverse cholesterol metabolism. It is believed to mediate the exchange of cholesteryl ester in HDL-C for triglyceride in low-density lipoprotein (LDL) or very low-density lipoprotein (VLDL) (12–14). A number of polymorphisms in the gene have been shown to be associated with serum HDL-C levels (15–18).
Materials and Methods
Study Population.
Participants were female, white subjects from the St. Thomas' Hospital, London, adult-twin registry, which is a voluntary registry of >4,000 twin pairs ranging from 18 to 76 years of age at first interview. They were recruited through media campaigns asking for United Kingdom twins and were unaware of specific hypotheses to be tested. Informed consent was obtained for all subjects. For current purposes, one member of each pair was selected randomly, and of these, the following were excluded: males, those on lipid-lowering drugs or diuretics, and those with no or insufficient DNA available. The subset derived was 1,393 unrelated female, white English individuals. Ages ranged from 18 to 76 years, with a mean age of 47.56 years. Serum levels of HDL-C ranged from 0.24 to 3.760 mmol/liter. To accommodate the correlation between age and HDL-C, quadratic least-squares regression was applied to derive residual HDL-C values, corrected for age. DNA pools were designed by placing these corrected values in order from smallest to largest and selecting the extremes. In this instance, DNA from individuals in the lowest 400 was selected and mixed in equimolar amounts to form a low-HDL-C pool, and likewise a high-HDL-C pool was formed from individuals in the highest 400. There were 1.17 SDs between the highest trait value in the low-HDL-C pool and the lowest trait in the high HDL-C pool; standardized means for the two pools were −1.08 and +1.09, respectively.
HDL-C Measurement.
Quantitative determination of HDL-C from human serum was performed by using a RANDOX kit (19). Normal serum values for women lie in the range of 1.26–1.94 mmol/liter.
SNP Selection.
Twenty-two putative SNPs were selected from dbSNP (www.ncbi.nlm.nih.gov/SNP) based on location in the CETP gene. They were amplified and analyzed in triplicate by using DNA pools composed of 94 unrelated individuals of European ancestry from the Centre d'Etude du Polymorphisme Humaine (Paris) panel (20). Seven of these SNPs (rs291046, rs158480, rs289716, rs289717, rs289741, rs1800776, and rs1800777) showed no evidence of polymorphism in our study population. The remaining 15 were confirmed as polymorphic and are given in Table 1.
Table 1.
Results of association testing of 15 SNPs in the CETP gene using pools of DNA
| Assay | Position | Plow | SD | Phigh | SD | P value |
|---|---|---|---|---|---|---|
| Rs1800775 | Promoter | 0.74 | 0.014 | 0.50 | 0.012 | 2.71E-24 |
| (0.58) | (0.42) | (1.07E-10) | ||||
| Rs708272 | Intron 1 | 0.32 | 0.007 | 0.62 | 0.009 | 8.41E-32 |
| (0.38) | (0.53) | (7.59E-10) | ||||
| Rs5883 | Exon 9 | 0.91 | 0.011 | 0.91 | 0.014 | 0.83 |
| (0.96) | (0.95) | (0.59) | ||||
| Rs158477 | Intron 9 | 0.54 | 0.008 | 0.62 | 0.015 | 0.002 |
| (0.48) | (0.50) | (0.43) | ||||
| Rs158478 | Intron 9 | 0.51 | 0.014 | 0.57 | 0.014 | 0.02 |
| (0.57) | (0.61) | (0.11) | ||||
| Rs158479 | Intron 9 | 0.61 | 0.010 | 0.52 | 0.008 | 0.0004 |
| (0.54) | (0.52) | (0.44) | ||||
| Rs158617 | Intron 9 | 0.10 | 0.011 | 0.19 | 0.021 | 1.83E-06 |
| (0.12) | (0.16) | (0.03) | ||||
| Rs289715 | Intron 9 | 0.09 | 0.006 | 0.15 | 0.024 | 2.67E-05 |
| (0.12) | (0.14) | (0.17) | ||||
| Rs289718 | Intron 10 | 0.24 | 0.016 | 0.38 | 0.015 | 2.73E-10 |
| (0.26) | (0.36) | (6.42E-05) | ||||
| Rs289719 | Intron 10 | 0.27 | 0.027 | 0.38 | 0.022 | 3.72E-06 |
| (0.27) | (0.35) | (0.0003) | ||||
| Rs291044 | Intron 10 | 0.64 | 0.021 | 0.76 | 0.008 | 1.13E-07 |
| (0.63) | (0.70) | (0.003) | ||||
| Rs5880 | Exon 12 | 0.13 | 0.061 | 0.03 | 0.026 | 7.72E-10 |
| (0.07) | (0.03) | (0.0005) | ||||
| Rs5882 | Exon 14 | 0.78 | 0.004 | 0.63 | 0.014 | 2.07E-12 |
| (0.72) | (0.65) | (0.002) | ||||
| Rs1801706 | Exon 16 | 0.11 | 0.011 | 0.19 | 0.026 | 5.51E-05 |
| (0.15) | (0.19) | (0.03) | ||||
| Rs289742 | Exon 16 | 0.91 | 0.010 | 0.84 | 0.015 | 4.37E-05 |
| (0.88) | (0.85) | (0.06) |
Results from individual genotyping are given in parentheses. E-n, ×10−n.
Allele frequency derived from the low-HDL-C set.
SD of five estimates of allele frequency derived from five PCRs.
Allele frequency derived from the high-HDL-C set.
Pool Construction.
DNA was measured by using Fluoroskan Ascent (Thermo Labsystems, Franklin, MA) together with Pico green reagents and kits (Molecular Probes, P-7589). The selected DNAs were diluted to a standard concentration, and individual aliquots of DNA were transferred into a single tube by using the Hamilton ML2200 and Vivace automated pipetting stations to ensure that a constant amount of each DNA sample was transferred to the pool. The pool was then mixed gently and requantitated before further dilution to a working concentration of 5 ng/μl.
PCR.
For all assays processed in this study, the same conditions were used. Genomic DNA (25 ng, either pooled or individual), 1 unit of Taq polymerase (HotStarTaq, Qiagen, Valencia, CA), 200 μmol of each dNTP, 25 pmol of nonbiotinylated gene-specific PCR primer 1, 4 pmol of gene-specific PCR primer 2 carrying a universal sequence tag, and 10 pmol biotinylated universal sequence primer (5′-bio-AGCGGATAACAATTTCACACAGG-3′) were subjected, in 50 μl total volume, to the following temperature profile: initial denaturation at 95°C for 15 min followed by 45 cycles of 95°C for 20 s, 56°C for 30 s, 72°C for 30 s, and a final extension of 3 min at 72°C.
Preparation of Single-Stranded Template for MassEXTEND (SEQUENOM, San Diego) Reactions.
PCR products were immobilized onto streptavidin-coated paramagnetic particles (Dynal, Oslo) via the biotinylated universal PCR primer. After immobilization, the double strand was denatured by using 50 μl of 0.1 M NaOH at room temperature. After removal of the NaOH and neutralization with 10 mM Tris⋅HCl solution, the beads carrying single-stranded PCR product were used for primer extension by MassEXTEND reactions.
MassEXTEND Reaction.
Primer extension reactions were performed by using triple terminator mixes. Assays were grouped according to the SNP-specific requirements on the termination mixes (ddACG, ddACT, ddAGT, and ddCGT, respectively). The final reaction volume of 15 μl comprised 1 unit of Thermosequenase (Amersham Pharmacia), 50 μM of the respective termination mix, and 20 pmol of the assay-specific extension primer. All assays were run with the same temperature profile, comprising an initial denaturation at 80°C for 30 s followed by three cycles of 45°C for 15 s and 72°C for 1 min.
Primer extension products were resolubilized from the solid support by applying an ammonium hydroxide solution to the streptavidin beads. For the subsequent matrix-assisted laser desorption ionization/time-of-flight MS analysis, 15 nl of sample were transferred from a 96-well microtiter plate serially onto four patches of a 384-element silicon chip preloaded with 3-hydroxy-picolinic-acid (3-HPA) (SpectroCHIP, SEQUENOM) by means of a piezoelectric pipette.
Matrix-Assisted Laser Desorption Ionization/Time-of-Flight MS.
Application of the analyte solution onto the preloaded matrix patches of the silicon chip dissolved the matrix. After evaporation of the solvent, homogenous crystals of matrix and analyte were formed. Automated analysis of these samples was performed on a SEQUENOM–Bruker MassARRAY mass spectrometer by scanning the 384 elements with a 337-nm laser pulse. Twenty shots were summed per element to yield the final spectra.
Allele Frequency Determination.
Mass spectra were processed by proprietary software (SPECTROTYPER, SEQUENOM) using baseline correction, peak identification, and peak area calculation algorithms. Normalized peak areas were computed as individual peak areas divided by the sum of total peak area.
Results and Discussion
SNP Confirmation and Validation.
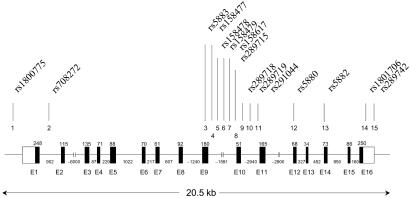
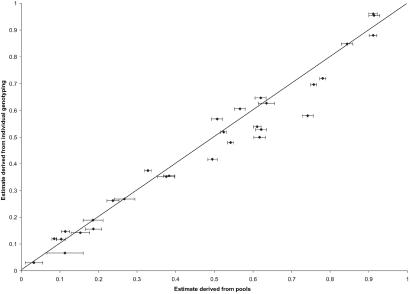
Fifteen SNPs from dbSNP were confirmed to be polymorphic in a pool of 94 Centre d'Etude du Polymorphisme Humaine DNA samples as described (21). The relative locations of these SNPs are shown in Fig. 1. For each HDL-C-based pool and each SNP, five allele frequency estimates, each derived from separate PCR amplifications, were calculated. The mean and SD of these estimates are listed in Table 1. Formal tests of association were performed by a one degree-of-freedom χ2 test of equality of frequency. By this test, a significant difference in allele frequency between pools provides evidence to support the hypothesis that the SNP is in LD with a genetic variant influencing trait value. Finally, all 800 DNA samples were genotyped individually for the 15 SNPs to allow a comparison of the two approaches. Fig. 2 shows a scatter plot of allele frequency estimates by testing pools and genotyping individual samples.
Fig 1.
Structure of the CETP gene and the locations of SNPs tested.
Fig 2.
Allele frequencies derived from individual genotyping plotted against estimates derived from pools for 15 SNPs in CETP. Error bars with a width of 2 SDs are also presented for estimates derived from pools.
As detailed in Table 1, by using the pooled DNA approach 14 of 15 SNPs exhibited significantly different frequencies between low- and high-HDL-C groups at the 5% level. Nine of these associations were confirmed in individual genotyping (P < 0.05) and found to be distributed across the length of the gene. Among them are rs1800775, a promoter SNP, rs708272, an SNP in intron 2 (known as TaqIB), and rs5882, an amino acid substitution in exon 14 (known as I405V), all of which have been reported to be associated with HDL levels (17, 18, 22).
In all cases, the difference in allele frequency was magnified when pooled analysis was applied. This was likely to have been by chance, because the differences in estimates appear randomly distributed (Fig. 2); however, it demonstrates how the type I error rate may be raised by this approach. The mean difference between a pooled estimate of allele frequency and the individual sample estimate was 0.049 and 0.037 for the low- and high-HDL-C pools, respectively.
Factors Influencing Accuracy of Allele Frequency Estimates.
For DNA pools, as with individual samples, assay performance is variable and is of great importance. To mimic a typical high-throughput setting in the current study, all assays were designed in silico and run without any preassessment of amplification efficiency, preferential amplification of alleles, or stability of amplification. The SD values in Table 1 range from 0.006 to 0.061, showing that a substantial portion of the error is introduced at the PCR stage. All other non-PCR factors (MassEXTEND reaction, matrix-assisted laser desorption ionization/time-of-flight MS analysis) showed lower variability (data not shown). The quality of the genomic DNA used, as well as accurate quantification of the DNA concentration, are further factors that can contribute greatly to deviations of the estimated allele frequency in pools from the true allele frequency. These aspects might partially account for the reproducible deviations from the true allele frequencies seen in Fig. 2.
For practical purposes, it would be useful to design a future study to determine how many PCRs it is optimal to perform. The current study covers too few SNPs to answer this question adequately. In our data, successively recalculating the SD as replicates were added led to a stabilization of the estimate. However, this may be easily attributed to a successive improvement in the estimate as the number of data points increased. A similar recalculation of the mean showed varying behavior between assays. There were insufficient data to conclude, for example, a correlation between minor allele frequency and the number of PCRs required to produce a stable estimate of allele frequency.
The correctness of frequency determination could be improved by the following protocol changes that, despite diminishing throughput, would still permit cost savings in comparison to individual genotyping. First, increasing the number of samples incorporated into each pool may decrease the influence of random quantitation effects and statistical sampling error. Second, the genotyping of 10–20 random DNAs would allow an examination of heterozygotes and thus the correction of pool-based allele frequency estimates for allelic PCR bias (unequal peak heights in heterozygotes). Third, the number of PCR replicates could be increased. Taking the arithmetic mean of a larger number may reduce the influence of random PCR effects, a step most likely to be useful for assays with high SD. Future studies may allow a cost-benefit assessment of these changes as well as a formal determination of the type I and type II errors involved.
Haplotype Analysis for 15 Markers in CETP.
By using a custom-built program, maximum-likelihood estimates of haplotype frequency were obtained by the E-M algorithm (23), and a likelihood ratio test was applied to test the null hypothesis of equal haplotype frequencies between low- and high-HDL-C groups, against the alternative that at least one haplotype was at a different frequency in the two groups. This test was applied to a sliding window of five markers, and strong evidence of association was derived for all but two windows. The results are displayed in Table 2.
Table 2.
Haplotypic association results obtained for sliding windows of five SNP markers using maximum-likelihood estimates of haplotype frequency derived by an E-M algorithm
| Window | No. of haplotypes observed | P value |
|---|---|---|
| 1-2-3-4-5 | 16 | 6.45E-07 |
| 2-3-4-5-6 | 17 | 7.70E-06 |
| 3-4-5-6-7 | 13 | 0.09 |
| 4-5-6-7-8 | 13 | 0.09 |
| 5-6-7-8-9 | 16 | 8.87E-06 |
| 6-7-8-9-10 | 17 | 1.47E-21 |
| 7-8-9-10-11 | 15 | 5.28E-27 |
| 8-9-10-11-12 | 12 | 9.54E-31 |
| 9-10-11-12-13 | 15 | 5.31E-30 |
| 10-11-12-13-14 | 16 | 0.00019 |
| 11-12-13-14-15 | 11 | 1.70E-05 |
E-n, ×10−n.
The 3-4-5-6-7 and 4-5-6-7-8 windows gave only weak evidence of association, consistent with the earlier finding that markers 3–6 and 8 provided little single-point evidence of association.
The application of this approach to those seven markers with single-point P values <0.01 (markers rs1800775, rs708272, rs289718, rs289719, rs291044, rs5880, and rs5882) gave a P value of 2.75642 × 10−24. This value was far lower than any pointwise P value, exemplifying the increased power of haplotype analysis in certain situations. Interestingly, the single haplotype test of the set of alleles that, individually, were at elevated frequency in the low-HDL-C group gave a P value of 0.126736, failing to explain the global test result.
A total of 37 haplotypes were estimated to be present, and of these three had frequency >10% in both low- and high-HDL-C groups. Together they accounted for 69% of the frequency in the high-HDL-C group and 50% in the low-HDL-C group. Two of them (TATCCGA and TACTCGG) showed substantially different frequencies between groups (raw differences of 9.3% and 10.9%, respectively), and it was noted that they shared the allelic pattern TA**CG*. Testing this configuration singly, against all others, revealed a highly significant result (P value of 5.21 × 10−21).
In summary, the results obtained for the haplotypic association are consistent with causal variant(s) occurring on multiple haplotypic backgrounds. Possible explanations include the presence of multiple mutations in the population (allelic heterogeneity), elevated rates of recombination in the region, or a single, very old mutation now showing disrupted LD with surrounding markers. Based on the results obtained for the two common haplotypes showing different frequency for low- and high-HDL-C groups, one might speculate that an ancestral haplotype contained the four alleles described above.
The Value of Association Testing in Pools.
At the current level of precision, the greatest value of our pooling approach is likely to be its suitability as an initial screen to rapidly and cost-efficiently identify which SNPs should undergo individual genotyping. Our method required 150 reactions (2 pools, 15 SNPs, and 5 PCR replicates) for initial association testing in contrast to 12,000 reactions made necessary by individual-sample genotyping.
A follow-up by individual genotyping of selected SNPs showing evidence of association has two major benefits. First, it allows confirmation of the pooled estimates of frequency, and second, it permits the reconstruction of the haplotype showing LD with the disease.
In conclusion, our results provide an indication that large-scale association analysis can be accelerated when pools of DNA are used. The measurement of the allele frequency of one SNP in one pool by using our approach took ≈3–5 s, demonstrating the feasibility of thousands of loci undergoing preliminary investigation in this high-throughput manner.
Acknowledgments
We thank the study participants, who generously contributed to our research, as well as Tim Spector and the Twin Research Unit of St. Thomas' Hospital for sample ascertainment and data collection. We are also grateful to Frank Dudbridge for the use of his software and for helpful discussions in the interpretation of haplotypes.
Abbreviations
LD, linkage disequilibrium
SNP, single-nucleotide polymorphism
CETP, cholesteryl ester transfer protein
HDL-C, high-density lipoprotein cholesterol
References
- 1.Goddard K. A., Hopkins, P. J., Hall, J. M. & Witte, J. S. (2000) Am. J. Hum. Genet. 66, 216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kidd J. R., Pakstis, A. J., Zhao, H., Lu, R.-B., Okanofua, F. E., Odunsi, A., Grigorenko, E., Bonne-Tamir, B., Friedlaender, J., et al. (2000) Am. J. Hum. Genet. 66, 1882-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reich D. E., Cargill, M., Bolk, S., Ireland, J., Sabeti, P. C., Richter, D. J., Lavery, T., Kouyoumjian, R., Farhadian, S. F., Ward, R. & Lander, E. S. (2001) Nature 411, 199-204. [DOI] [PubMed] [Google Scholar]
- 4.Clark A. G., Weiss, K. M., Nickerson, D. A., Taylor, S. L., Buchanan, A., Stengård, J., Salomaa, V., Vartiainen, E., Perola, M., Boerwinkle, E. & Sing, C. F. (1998) Am. J. Hum. Genet. 63, 595-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abecasis G. R., Cookson, W. O. & Cardon, L. R. (2001) Am. J. Hum. Genet. 68, 191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruglyak L. (1999) Nat. Genet. 22, 139-144. [DOI] [PubMed] [Google Scholar]
- 7.Collins A., Lonjou, C. & Morton, N. E. (1999) Proc. Natl. Acad. Sci. USA 96, 15173-15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Risch N. & Merikangas, K. (1996) Science 273, 1516-1517. [DOI] [PubMed] [Google Scholar]
- 9.Shaw S. H., Carrasquillo, M. M., Kashuk, C., Puffenberger, E. G. & Chakravarti, A. (1988) Genome Res. 8, 111-123. [DOI] [PubMed] [Google Scholar]
- 10.Barcellos L. F., Klitz, W., Field, L. L., Tobias, R., Bowcock, A. M., Wilson, R., Nelson, M. P., Nagatomi, J. & Thomson, G. (1997) Am. J. Hum. Genet. 61, 734-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Risch N. & Teng, J. (1998) Genome Res. 8, 1273-1288. [DOI] [PubMed] [Google Scholar]
- 12.Drayna D., Jarnagin, A. S., McLean, J., Henzel, W., Kohr, W., Fielding, C. & Lawn, R. (1987) Nature 327, 632-634. [DOI] [PubMed] [Google Scholar]
- 13.Koizumi J., Mabuchi, H., Yoshimura, A., Michishita, I., Takeda, M., Itoh, H., Sakai, Y., Sakai, T., Ueda, K., Takeda, R., et al. (1985) Atherosclerosis 58, 175-186. [DOI] [PubMed] [Google Scholar]
- 14.Kurasawa T., Yokoyama, S., Miyake, Y., Yamamura, T. & Yamamoto, A. (1985) J. Biochem. (Tokyo) 98, 1499-1508. [DOI] [PubMed] [Google Scholar]
- 15.Kuivenhoven J. A., de Knijff, P., Boer, J. M. A., Smalheer, H. A., Botma, G.-J., Seidell, J. C., Kastelein, J. J. P. & Pritchard, P. H. (1997) Arterioscler. Thromb. Vasc. Biol. 17, 560-568. [DOI] [PubMed] [Google Scholar]
- 16.Kondo I., Berg, K., Drayna, D. & Lawn, R. (1989) Clin. Genet. 35, 49-56. [DOI] [PubMed] [Google Scholar]
- 17.Freeman D. J., Packard, C. J., Shepherd, J. & Gaffney, D. (1990) Clin. Sci. 79, 575-581. [DOI] [PubMed] [Google Scholar]
- 18.Corbex M., Poirier, D., Fumeron, F., Betoulle, D., Evans, A., Ruidavets, J. B., Arveiler, D., Luc, G., Tiret, L., Cambien, F., et al. (2000) Genet. Epidemiol. 19, 64-80. [DOI] [PubMed] [Google Scholar]
- 19.Sugiuchi H., Uji, Y., Okabe, H., Irie, T., Uekama, K., Kayahara, N. & Miyauchi, K. (1995) Clin. Chem. (Washington, D.C.) 41, 717-723. [PubMed] [Google Scholar]
- 20.Dausset J., Cann, H., Cohen, D., Lathrop, M., Lalouel, J. M. & White, R. (1990) Genomics 6, 575-577. [DOI] [PubMed] [Google Scholar]
- 21.Buetow K. H., Edmonson, M., MacDonald, R., Clifford, R., Yip, P., Kelley, J., Little, D. P., Strausberg, R., Koester, H., Cantor, C. R. & Braun, A. (2001) Proc. Natl. Acad. Sci. USA 98, 581-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dachet C., Porier, O., Cambien, F., Chapman, J. & Rouis, M. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 507-515. [DOI] [PubMed] [Google Scholar]
- 23.Long J. C. (1995) Am. J. Hum. Genet. 56, 799-810. [PMC free article] [PubMed] [Google Scholar]