Abstract
High-throughput expression profiling enables the global study of gene activities. Genes with positively correlated expression profiles are likely to encode functionally related proteins. However, all biological processes are interlocked, and each protein may play multiple cellular roles. Thus the coexpression of any two functionally related genes may depend on the constantly varying, yet often-unknown cellular state. To initiate a systematic study on this issue, a theory of coexpression dynamics is presented. This theory is used to rationalize a strategy of conducting a genome-wide search for the most critical cellular players that may affect the coexpression pattern of any two genes. In one example, using a yeast data set, our method reveals how the enzymes associated with the urea cycle are expressed to ensure proper mass flow of the involved metabolites. The correlation between ARG2 and CAR2 is found to change from positive to negative as the expression level of CPA2 increases. This delicate interplay in correlation signifies a remarkable control on the influx and efflux of ornithine and reflects well the intrinsic cellular demand for arginine. In addition to the urea cycle, our examples include SCH9 and CYR1 (both implicated in a recent longevity study), cytochrome c1 (mitochondrial electron transport), calmodulin (main calcium-binding protein), PFK1 and PFK2 (glycolysis), and two genes, ECM1 and YNL101W, the functions of which are newly revealed. The complexity in computation is eased by a new result from mathematical statistics.
Keywords: gene expression, microarray, urea cycle, correlation, glycolysis
Microarrays have generated an enormous amount of gene-expression data from a variety of biological studies (1–5). After proper preprocessing, the data can be stored as a matrix of real numbers with N rows and m columns. Rows represent the gene-expression profiles, and columns represent the cell types, time points, or environmental or other experimental conditions under which the mRNA samples are taken. To elucidate microarray data, most methods (6–10) rely on the notion of profile similarity as described for Fig. 1. It is argued that coexpressed genes are likely to encode proteins that participate in a common structural complex, metabolic pathway, or biological process (6, 10).
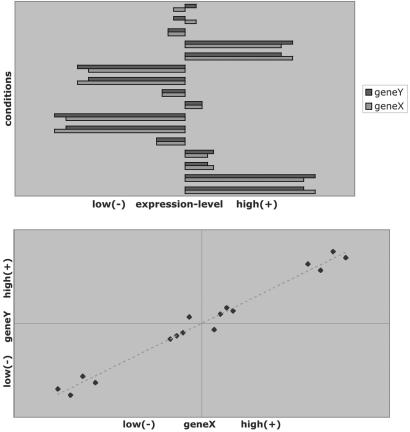
Fig 1.
Profile similarity. (Upper) Profiles of genes X and Y under 16 conditions. (Lower) The same data is shown in a scatter plot: one point for one condition. Coexpressed genes have most points on either the first (coactivated) or third (co-inactivated) quadrant. The strength of the coexpression pattern can be measured by correlation coefficient, which equals (X1Y1 +⋯+ XmYm)/m after standardization of each profile. On the other hand, X and Y are contraexpressed if most points are on the other two quadrants, meaning that when one gene is up-regulated, the other gene is down-regulated; the correlation coefficient is negative. However, contraexpression is rarely discussed in the literature.
Despite the many successful applications reported in the literature, there is an important issue that is hard to address by profile-similarity analysis. As is known, all biological processes are interlocked, and many proteins have multiple cellular roles. Two proteins engaged in a common process under certain conditions may disengage and embark on activities of their own under other conditions, which implies that both the strength and pattern of association between two gene profiles may vary as the intrinsic cellular-state changes. Weaver (11) discussed two transcription factors, Max and thyroid hormone receptor (TR). They can serve either as activators or repressors depending on other molecules bound to them. Max can bind to Myc and form a Myc–Max dimer that acts as a transcription activator. But when bound to Mad, the Mad–Max dimer serves as a repressor. For the case of TR, it associates with retinoid X receptor (RXR) to form a TR–RXR dimer that serves as a repressor in the absence of thyroid hormone. In the presence of thyroid hormone, the TR–RXR dimer is converted into an activator. Histone deacetylation is involved in both repressing events. Thus for example, if we take X to be the expression profile of the gene encoding TR and take Y to be the profile of one of its target genes, then X and Y may be either positively or negatively correlated depending on the hormone level. If the hormone level fluctuates in an unspecific manner, the opposing directions of correlation may cancel out each other, and no similarity-based analysis may succeed in detecting the functional association between X and Y.
A general issue arising from the above discussion is how to systematically study the coexpression patterns between functionally related genes as the cellular-state changes. The issue is compounded by the numerous intracellular and intercellular conditions that can alter the cellular state. A direct approach would be to specify a number of them and conduct more profiling accordingly, but this depends on the resource availability and the knowledge about what conditions are relevant.
We approach the problem in a reverse manner. For any two given genes, we attempt to delineate the cellular-state changes that may affect their coexpression pattern. This is made possible via a theory of coexpression dynamics. We use Saccharomyces cerevisiae as a model to illustrate our method. Our data come from four cell-cycle experiments accessible at http://genome-www.stanford.edu/cellcycle. We use all of them to construct gene profiles with a total of 73 conditions. Genes with too many missing values are excluded, leaving a total of 5,878 genes under study.
Methods
Our theory is illustrated schematically in Fig. 2. The term liquid association (LA) is used to conceptualize the internal evolution of coexpression pattern for a pair of genes (X,Y). Because the relevant cellular states typically are unknown, it is difficult to detect LA from the profiles of X and Y alone. However, if the state change turns out to be associated with the differential expression of a third gene Z, then the profile of Z can be used to screen the scatter plot of (X,Y) for the LA activity. Specifically, if an increase in Z is associated with a decrease in the correlation of (X,Y), then gene Z is a negative LA-scouting gene for (X,Y), and a negative score is assigned to quantify the strength of LA. The pair (X,Y) is called a negative LA pair (LAP) of Z. Likewise, a positive LA-scouting gene can be defined if an increase in Z is associated with an increase in the correlation of (X,Y), and the LA score is positive. Thus when comparing the low with the high expression levels of a positive LA-scouting gene, the scouted LAP is likely to change from being contraexpressed to being coexpressed. For a negative LA-scouting gene, the change goes in the opposite direction: from coexpression to contraexpression. In general, an LA-scouting gene serves only as a whistle blower, a surrogate for the intrinsic-state variable that facilitates the LA activity. The protein encoded by an LA-scouting gene may not have the direct physical contact with its LAP or the proteins encoded. The word “scouting” may be replaced by “monitoring,” “mediating,” or their synonyms with the above note in mind.
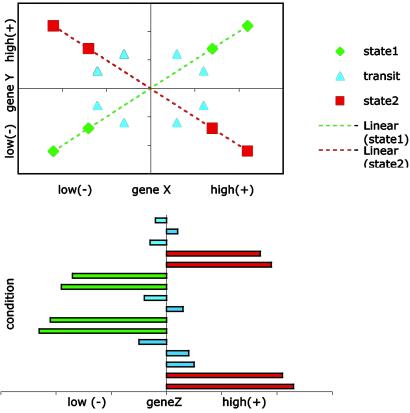
Fig 2.
Coexpression dynamics. (Upper) Profiles of genes X and Y are displayed in a scatter plot. The four green points represent four conditions for cellular state 1 wherein X and Y are coregulated. Likewise, the four red points represent four conditions for cellular state 2 wherein X and Y are contraexpressed. To depict this kind of internal evolution in the association pattern, we say (X,Y) forms a LAP. Because the relevant cellular states usually are unknown, it is hard to detect LAP directly from the profiles of X and Y alone. However, if the cellular states are correlated with the differential expression of a third gene Z, then we can use Z to scout (X,Y) for information about their LA activity. (Lower) The four green bars represent the expression of Z for the same four green-colored conditions as shown in Upper. Likewise, the four red bars correspond to the four red-colored conditions shown in Upper. We see when Z is down-regulated (green), X and Y are coexpressed; when Z is up-regulated (red), X and Y become contraexpressed. We assign a score to quantify the strength of LA. The LA score for this illustration is a negative value. On the other hand, if the low expressions of Z correspond to the red points shown in Upper and the high expressions of Z correspond to the green points shown in Upper, then the LA score will be positive.
For the genome-wide study, there is still a computational hurdle to overcome, because there are too many combinations for choosing three genes from N genes. With N = 5,878, the number is already hovering above 33 billion. We surely cannot afford to inspect every scatter plot to find all triplets with LA patterns, which is why an LA score is needed and must be easy to compute. Aided by a statistical theory as outlined next, this turns out to be possible. In fact, only two simple steps are required.
(i) Standardize each gene-expression profile with a normal score transformation. Specifically, the m values in the profile are compared with each other and their ranks R1, … , Rm are recorded. The ranks are then used to obtain the transformed profile, Φ−1(R1/(m + 1)), Φ−1(R2/(m + 1)), … , Φ−1(Rm/(m + 1)), where Φ(.) is the cumulative normal distribution.
- (ii) Compute the average product of the three transformed profiles,
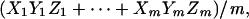
which is the LA score that we need.
As illustrateard in the flow chart (Fig. 3), there are several ways of using the LA approach. Two of them are given in this article: (i) screen the genome to find the LA-scouting genes for a given pair of genes (an order of N comparisons is required), or (ii) screen the genome to find the LAPs of a given gene (an order of N2 comparisons is required).
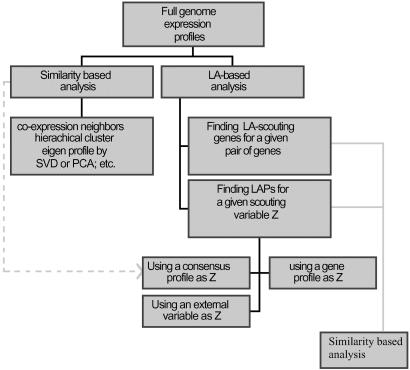
Fig 3.
Organization chart for incorporating LA with similarity-based methods. In this article, we only consider the use of a third gene to detect the LA activities. Coexpressed genes found by profile-similarity analysis can be pooled to obtain a consensus profile for LA scouting. Likewise, the genes identified through the LA system can be analyzed further for patterns of clustering. For some applications, the scouting variable may come from external sources related to the expression profiles. SVD, singular value decomposition; PCA, principal component analysis.
Statistical Development of LA Score.
Change of the cellular state is most likely a continuous process. It is convenient to present the theory in terms of continuous random variables. Suppose X, Y, and Z are already standardized to have mean 0 and variance 1. Then the correlation coefficient between X and Y is equal to E(XY). By conditioning, E(XY) = E (E(XY|Z)) = Eg(Z), where g(z) = E(XY|Z = z) denotes the conditional expectation of XY given Z = z. The notion of LA now arises from a straightforward dynamic perspective. We regard g(z) as the coexpression measure between gene X and gene Y when Z is at level z and ask how g(z) varies as z increases. Denote the derivative of g(z) with respect to z by g′(z), which leads to the following definition for measuring the average amount of coexpression change.
Definition:
Suppose X, Y, and Z are random variables with mean 0 and variance 1. The LA of X and Y with respect to Z, denoted by LA(X,Y|Z), is
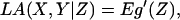 |
where
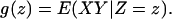 |
This definition is fairly general. It can be estimated by applying one of the many existing nonparametric regression techniques to estimate the curve g′(z) first, but here is a shortcut. We assume that Z follows a normal distribution and obtain the following.
Theorem.
If Z is standard normal, LA(X,Y|Z) = E(XYZ).
Using the celebrated Stein Lemma (12), the theorem follows from
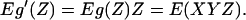 |
This theorem justifies why we need the normal score transformation as the first step in computing the LA score. Another advantage is that the interference of abnormally large values is tempered, because only the ranks of the expression levels are used in the transformation. More statistical discussion is given in Supporting Text, which is published on the PNAS web site, www.pnas.org.
We use a permutation test to tell whether the LA score is statistically significant. The procedure generates as many as 105 or 106 artificial profiles Z* of Z by randomly permuting (Z1, … , Zm) and computes their LA scores. The P value is obtained by counting how often the LA score of (X,Y|Z*) exceeds the LA score of (X,Y|Z).
Results
Urea Cycle.
Fig. 4 shows the key enzymes and intermediates in the urea cycle. First, we consider two functionally associated genes, GLN3 and CAR1. CAR1 encodes arginase, the enzyme catalyzing the hydrolysis of arginine into urea, whereas GLN3p is a transcription factor known to activate nitrogen-catabolic genes including CAR1 (13). Intuitively we may expect a positive correlation between the two profiles, but this is not the case; very little similarity exists between the two expression profiles. Perhaps this merely reflects the complexity due to many cis elements and trans-acting factors for CAR1 (14), the localization of Gln3p, and its interactions with other factors (15). To apply the LA methodology, we take (GLN3,CAR1) as the pair (X,Y) in Fig. 2, take any gene as Z, and compute the LA score as defined earlier. We then rank all 5,878 genes according to their abilities to serve as a positive or negative LA-scouting gene by ordering their LA scores. It turns out that at the eighth place leading on the negative end is ARG4, the gene immediately ahead of CAR1 in the urea cycle. ARG4 and its two proceeding genes in the flow chart, ARG1 and ARG3, share some mild degree of profile similarity. However, these three genes are not coexpressed with ARG2, the gene encoding acetylglutamate synthase, which carries out the first step in synthesizing ornithine and eventually arginine (16). We speculate that this may be in part because of CAR2p (ornithine aminotransferase). To feed ornithine into the arginine biosynthesis pathway, CAR2 should be inactivated to avoid the immediate degradation of ornithine, which suggests that CAR2 and ARG2 may be contraexpressed. But this prediction is not supported by the profile data; the correlation between ARG2 and CAR2 is nearly zero. We turn to the LA method and proceed as described before by treating (ARG2,CAR2) as the gene pair (X,Y). Again we rank all genes by their LA-scouting abilities for this pair. Among the leading 10 negative LA-scouting genes for (ARG2,CAR2), we find the gene CPA2, which encodes the large subunit of carbamoyl phosphate synthetase. Note that carbamoyl phosphate is needed for ARG3p to synthesize citrulline from ornithine. Thus high expression of CPA2 may reflect the state of cellular demand for arginine. From the LA-activity plot (Fig. 5), we see that under this state, ARG2 and CAR2 are indeed negatively correlated. A similar interpretation can be given to the LA activity between (GLN3,CAR1), with ARG4 being the scouting gene (see Fig. 7, which is published as supporting information on the PNAS web site). Because ARG4p catalyzes the last step of arginine biosynthesis, high expression of ARG4 can be viewed as a cellular signal for arginine demand, too. Thus if CAR1 were also up-regulated, then the newly synthesized arginine would be subject to immediate hydrolysis by CAR1p, leading to a wasteful cycle of metabolism. Such nutrient-wasteful species would be less likely to survive during the course of evolution. Consistent with this argument, from Fig. 7 we do see that up-regulation of GLN3 is concomitant with higher expression of CAR1 when ARG4 is expressed at the low level.
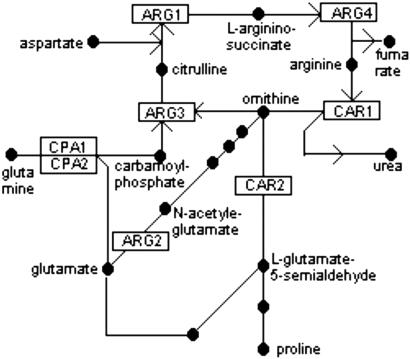
Fig 4.
Urea cycle/arginine biosynthesis pathway. ARG2 encodes acetyl-glutamate synthase, which catalyzes the first step in synthesizing ornithine from glutamate. Ornithine and carbamoyl phosphate are the substrates of the enzyme ornithine transcarbamoylase, encoded by ARG3. Carbamoyl phosphate synthetase is encoded by CPA1 and CPA2. ARG1 encodes argininosuccinate synthetase, ARG4 encodes argininosuccinase, CAR1 encodes arginase, and CAR2 encodes ornithine aminotransferase.
Fig 5.
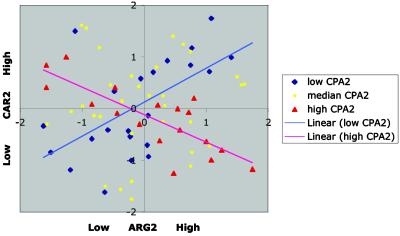
LA between ARG2 and CAR2 as scouted by CPA2. When the expression level of CPA2 is low (conditions represented by blue diamonds), a positive correlation is seen between ARG2 and CAR2. As the level of CPA2 increases, the correlation pattern is gradually weakened. Eventually, when CPA2 is high (red triangle), the association is turned into negative. The LA score is −0.289. For efficient activation of the arginine biosynthesis pathway, up-regulation of ARG2 must be concomitant with down-regulation of CAR2 to prevent ornithine from leaking out of the urea cycle. We see that this occurs only when CPA2 is up-regulated. Because activation of CPA2 provides the influx of carbamoyl phosphate into the urea cycle, a high expression level of CPA2 can be interpreted as a physiological signal for arginine demand. When the demand is relieved and CPA2 is lowered, CAR2 is up-regulated, opening up the channel for ornithine to leave the urea cycle.
The LA scores are statistically significant. For (GLN3,CAR1|ARG4), the score is −0.2589 with a P value 74 of 100,000. For (CAR2,ARG2|CPA2), the score is −0.2894 with a P value 56 of 1 million.
In Table 1, which is published as supporting information on the PNAS web site, we discussed what can be learned from other LA-scouting genes with scores better than ARG4 or CPA2. We are limited by the scanty literature on the gene-regulatory mechanisms even for known genes. Still, there is a high degree of biological relevance and consistency. Among other evidences, the interplay with the tricarboxylic acid (TCA) cycle through the metabolites, fumarate and malate, is especially notable. Fumarate is a byproduct of the enzyme reaction by ARG4p to yield arginine. Malate lies next to fumarate in the TCA cycle.
LAP Database.
For further study, we compile a database that contains the information about the most significant LAPs detected from each of the 5,878 genes under study. After ranking all possible pairs (>17 million pairs in total) by their LA scores, we kept only the top 20 positive LAPs and top 20 negative LAPs. This LAP database can be accessed at www.stat.ucla.edu/∼kcli.
Longevity.
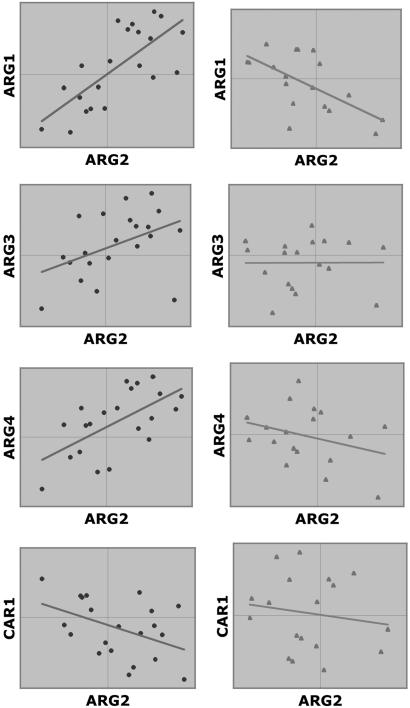
A recent article (17) reveals the role of SCH9 in longevity of yeast. Checking the LA database that we compiled, the pair (ARG1, ARG2) appears as a negative LAP of SCH9 (Fig. 8, which is published as supporting information on the PNAS web site). Indeed, this rather unexpected finding has led us to the above study about yeast's rational control of arginine biosynthesis. To continue the discussion, we take note that ideally, the entire urea cycle genes should be expressed coherently in such a way that not only positive correlation exists among ARG1, ARG2, ARG3, and ARG4, but concomitantly these genes should also be negatively correlated with CAR1. This turns out to be the case when the expression of SCH9 is low (see Fig. 6). SCH9 encodes a serine/threonine protein kinase. It is required for the nitrogen activation of the fermentable growth median-induced pathway (18). Because the sch9-delete mutant has a longer nondividing life span than the wild type (17), this brings out a likely association between the efficient usage of the urea cycle and longevity physiology.
Fig 6.
Coherent expression of urea-cycle genes is mediated by SCH9. When SCH9 is low (Left), all four ARG genes are coexpressed, and CAR1 is contraexpressed with ARG2. Thus, as ARG2 is activated, CAR1 is concomitantly down-regulated, which ensures that the newly synthesized arginine will not be subject to the immediate hydrolysis by arginase. Low SCH9 is concomitant with the down-regulation of CAR2 (Fig. 8 Lower), further shutting down the outlet for ornithine to leave the urea cycle. In contrast, when SCH9 is high (Right), the coherence disappears, some showing no correlation and others displaying negative correlation.
In their longevity study, Fabrizio et al. (17) screened for long-lived mutants after transposon-mutagenizing yeast cells and treating them with heat stress for 1 h and the superoxide-generating agent paraquat for 9 days. In fact, two mutants were isolated and the affected genes were SCH9 and CYR1 (encoding adenylate cyclase). The longevity regulation by SCH9 and CYR1 is consistent with the popular oxidative-damage theory of aging (19). When checking for the LAPs of CYR1, we found an oxidative protection gene, TTR1/GRX2 (glutaredoxin), and two age-related genes, SIM1 and HST3. HST3 is a homolog of the gene SIR2, which encodes NAD-dependent histone deacetylase and regulates the replicative life span of yeast (20). Interestingly, ARG4 was found in one LAP of SIR2. In addition, from the LAPs of the last urea cycle gene, CAR1, we found GLN3, CPA1, and CPA2 , which took us back to what was discussed earlier. On the other hand, we notice that glutamate, a product of most amino acid deamination, is at the head of the ornithine/arginine biosynthesis/urea cycle pathway (Fig. 4). From the LAPs of CYR1, we find five genes involved in glutamate metabolism and nitrogen utilization: PUT2, LYS9, ARO9, GAP1, and MEP2. Standard profile-similarity analysis also helps; SCH9 is positively correlated with CAR2. Thus, lowering the SCH9 expression is concomitant with the down-regulation of CAR2, thereby preventing the leakage of ornithine from the urea cycle. Before leaving this topic, we note that what is described here is not about the physiology for deletion mutants in the nondividing state. For our data, the yeast cells harvested for mRNA samples have just re-entered the cell cycle from the arrested states; they are growing and dividing.
Electron Transport.
Mitochondria generate energy via the process of oxidative phosphorylation (21). During the process, electrons are passed along a series of respiratory enzyme complexes located in the inner mitochondrial membrane by using the released energy to pump protons across the membrane. The proton gradient then enables the making of ATP by ATP synthase (F1F0 ATPase, or complex V). Many of the encoding genes in the pathway indeed are coexpressed. One interesting example is cytochrome c1, which acts as the last leg for complex III (cytochrome bc1) in relaying electrons to cytochrome c. For S. cerevisiae, cytochrome c1 is encoded by a single gene, CYT1. From the profile-similarity analysis, we find one gene, QCR8 (ubiquinol cytochrome reductase subunit 8), from complex III and two genes, COX5a and COX8, from complex IV (cytochrome oxidase) to be among the top 20 genes with expression profiles most similar to CYT1. Complementing this finding, the LA method links CYT1 to genes further down the electron-transport pathway. From the 40 most significant LAPs of CYT1 (Table 2, which is published as supporting information on the PNAS web site), we find that two of them, ATP1 and ATP5, encode subunits from the mitochondrial ATP synthase. In fact, ATP1 (F1 α subunit) appears 11 times.
Calmodulin.
Calmodulin is a ubiquitous Ca+-binding protein that regulates a wide range of proteins and processes in all eukaryotes. It is encoded by CMD1 in S. cerevisiae (22). The best known binding target of calmodulin in mitosis is Nuf1p. Nuf1p is a component of spindle pole body, the yeast microtubule-organizing center wherein calmodulin is localized. Despite the functional association between their gene products, CMD1 and NUF1 do not have similar expression profiles. NUF1 does not appear in the 40 LAPs of CMD1, either (Table 3, which is published as supporting information on the PNAS web site). Instead, we find it paired with an unknown gene, YGL149W, as a positive LAP of CMK1, one of the two genes encoding the calmodulin-regulated protein kinase. More interestingly, YGL149W also appears in one of the 20 positive LAPs of CMD1. Indeed, although the profiles of CMD1 and CMK1 are not similar, they do share nine genes in their LAPs. Furthermore, YGL149W has the protein–protein interaction with CRM1p (β-karyopherin, involved in exporting certain proteins from nucleus) that in turn interacts with NUF1p. Consistent with this connection, we find KAP120 (a member of karyopherin family) in the LAPs of CMD1 and SXM1 (putative β-karyopherin) in the LAPs of CMK1. From the joint LAP lists for CMD1 and CMK1, we further notice the convergence of a number of genes involved in the signal transduction pathways for various types of growth morphogenesis such as sporulation, pheromone response, pseudohyphal growth, RAS protein signal transduction, mitogen-activated protein kinase, cAMP-protein kinase A, and high-osmolarity glycerol-response pathways: IME1, RIM13, SSP1, SOK2, MUC1, TEC1, PTP3, PTC1, SIP4, CDA2, IME4, KAR4, KAR2, DOC1, AXL1, OPY1, and ERF2.
Glycolysis.
The genome-wide LA-scouting ability of a gene can be assessed according to the number of its LAPs with scores exceeding a threshold. This ability varies substantially from gene to gene (Table 4, which is published as supporting information on the PNAS web site). Reasoning that each surviving species must evolve a delicate expression system to manage the intricate interplay in the gene products, we speculate that the LA-scouting ability of a gene should reflect the importance of its encoded protein. To support this statement, we compare the genes involved in the most well studied metabolic pathway, glycolysis. The principal rate-limiting enzyme in glycolysis is 6-phosphofructokinase (PFK), a heterooctamer enzyme of four α and four β chains. It turns out that PFK1 and PFK2, encoding the α and β subunits of PFK, do detect far more LAPs than any other genes listed by the Munich Information Center for Protein Sequences (MIPS) under the energy category of glycolysis and gluconeogenesis (see Table 5, which is published as supporting information on the PNAS web site).
LA-Scouting Leaders.
Using some highly stringent criteria, we have selected a set of 66 genes with the highest LA-scouting ability (Table 6, which is published as supporting information on the PNAS web site). Some of these LA-scouting leaders are well studied genes [for instance, CYR1, PFK1, TPS1 (trehalose-6-phosphate synthase), TPS2 (trehalose-6-phosphate phosphatase), GSY1 (glycogen synthase isoform 1), GLC3 (α-1,4-glucan branching enzyme), ATP1, PPA1 (subunit of vacuolar ATPase), QCR9 (ubiquinol cytochrome c reductase subunit 9), CYC7 (cytochrome c, isoform 2), ERG13 (3-hydroxy-3methylglutaryl CoA synthase), AAT2 (aspartate aminotransferase), APC1 (largest subunit of anaphase-promoting complex), and IME1 (positive regulator of sporulation genes)]. The selection of LA-scouting leaders is by no means definitive. GSY2 (dominant isoform of glycogen synthase) just misses the cut. PFK2 and YAP6 would have been included if not for much missing data in one of the four cell-cycle experiments. YAP6 encodes a transcription factor homologous to YAP1p (basic leucine zipper, yeast homolog of c-Jun). Overexpression of YAP6 increases resistance to cisplatin (23), one of the most widely used drugs for cancer chemotherapy. The LA-scouting leaders are often found to scout each other. For example, among the top 10 LA-negative and 10 LA-positive scouting genes for (CYR1,PFK1), 7 are LA-scouting leaders (see also Fig. 9, which is published as supporting information on the PNAS web site).
Prediction.
The LA system provides a portrait of the cellular context in which a gene is likely to be involved, which is useful for predicting the functions of little known genes. For example, consider one of the LA-scouting leaders, ECM1. The literature on ECM1 is quite thin, and its functional annotation is still vaguely put as “involved in cell wall biosynthesis” by MIPS and SGD (Saccharomyces Genome Database) (wording slightly different). A year ago when the LA idea was first conceived, we were wondering why, among the scouting targets of ECM1, several are involved in translation; for example, NIP7 (required for efficient 60S ribosome subunits biogenesis, delete-lethal), appears six times (Table 7, which is published as supporting information on the PNAS web site). Now we find this to be consistent with the recent finding that ECM1p tagged with GFP is localized in the nucleus with a mild enrichment in the nucleolus, and that ECM1p genetically interacts with MTR2p in the 60S ribosomal protein subunit export (24). Coincidentally, we find that NIP7p appears as one of the 23 proteins copurified with NUG1p, the main gene characterized in ref. 24. The chance for this coincidence to occur by a fluke is <0.5%. This reassures the biological relevance of the LA approach in protein-function prediction.
YNL101W is another LA-scouting leader that is still considered as an unknown gene in SGD and MIPS at this writing. From the list of LAPs for YNL101W (Table 8, which is published as supporting information on the PNAS web site), we find several genes involved in autophagy, protein degradation, and transport: AUT7 (essential for autophagy, appearing six times), PRE6 and PUP1 (20S proteasome subunits), RPT6 (26S proteasome-regulatory subunit), CLC1 (clathrin light chain), YPT52 (GTP-binding protein of the RAB family), and UBP2 (ubiquitin-specific proteinase). This seems consistent with the newly identified role of YNL101W (newly named AVT4) as a membrane transporter responsible for the efflux of tyrosine and other large neutral amino acids from the vacuole (25).
Discussion
From the modeling point of view, how to infer the underlying cellular program from the mRNA data is extremely challenging because (i) microarray measurements are surely noisy, (ii) protein abundance may not be reflected well enough by the mRNA level, because other factors such as tRNA charging, protein stability, and so on are not considered, and (iii) intrinsic variables such as the protein phosphorylation or other modification status, localization of transcription factors, and quantities of important molecules such as ATP, NADPH, cAMP, etc. are difficult to predict. Because mathematical models seem intractable, statistical approaches become popular. Many similarity-based methods have proved very useful in elucidating the expression data.
Taking one step further, the LA method conducts a genome-wide search and identifies the most critical cellular players that may affect the coexpression pattern for any two genes. This method can be applied in any large microarray study. The aim of LA is to explore and exploit the dynamic, as opposed to the static, aspect of gene expression in cells. Our method eliminates the need to specify the cellular state before applying it. Instead, the method provides results for portraying the intrinsic state that facilitates the coexpression changes. This in turn can be used to guide the specification of experimental conditions for conducting more microarray profiling.
We are able to demonstrate some applications of LA using the yeast data. We show how the proper flow of ornithine in the urea cycle is mediated through a delicate switch between coexpression and contraexpression of ARG2 and CAR2. The switch depends on the expression level of CPA2, and it reflects well the cellular need for arginine. We also find that the regulation of GLN3 on CAR1 is linked to ARG4. Moreover, the expression of SCH9 is associated with the change in the coregulation pattern of ARG1 and ARG2. A possible connection of efficient expression of the urea cycle to yeast longevity is inferred.
For mammals, the urea cycle is activated in the liver to excrete excessive nitrogen resulting from the metabolic breakdown of amino acids. Our body cannot use the arginine synthesized in the liver because it is immediately cleaved to form urea, which then is sequestered by the kidney for secretion in the urine. In contrast, arginine biosynthesis is very important for single-celled organisms such as yeast. Thus it would be interesting to find out how different the coexpression pattern is in the liver if such expression data are available in the future.
This work points to a new source of information hidden in the microarray data. Methodologically, it can be viewed as one that offers a strategy of data refinery. The original expression data are processed, and the information about the LA activities is distilled. For N genes, the algorithm returns a huge amount of message, in the order of N3, that can be stored and used in a variety of ways to meet different researchers' needs. In our illustrations, we only use a small portion of high-scoring LAPs. In general, the better the LA score is, the more likely we can detect the LA pattern when visually examining the profile plots. It is in this sense that leading LA-scouting genes are better surrogates of the relevant intrinsic cellular-state variable. But how we use these surrogates to infer the cellular state depends on the available biological knowledge. The state variable can serve as a conceptual device for bringing out plausible biological hypotheses that can be cross-examined by using other bioinformatic resources such as the transcription factor database TRANSFAC (26) or the protein–protein interaction/complex database of MIPS.
The postrefinery statistical analysis can be extended in several directions. In addition to P values and visual inspection, one may bring in methods from multiple comparison/false discovery, an area with renewed interest fueled by microarray analysis. Similarity-based methods such as principal component analysis, also known as singular value decomposition, can be applied to high LA-scoring genes. In addition to the yeast data, our method can be applied to other large microarray studies on cancers, cell lines, and drug sensitivity (Supporting Text).
Supplementary Material
Acknowledgments
I am grateful to David Eisenberg, Samuel Karlin, and Michael Waterman for inspiring discussion; Chun-Houh Chen and Robert Yuan for computing assistance; Don Ylvisaker and Jack and Jennie Li for editing; and two referees for useful suggestions on presentation. This work is supported in part by National Science Foundation Grants DMS-9803459, 0104038, and 0201005.
Abbreviations
MIPS, Munich Information Center for Protein Sequences
TR, thyroid hormone receptor
LA, liquid association
LAP, LA pair
PFK, 6-phosphofructokinase
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Spellman P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D. & Futcher, B. (1998) Mol. Biol. Cell 9, 3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alon U., Barkai, N., Notterman, D. A., Gish, K., Ybarra, S., Mack, D. & Levine, A. J. (1999) Proc. Natl. Acad. Sci. USA 96, 6745-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scherf U., Ross, D. T., Waltham, M., Smith, L. H., Lee, J. K., Tanabe, L., Kohn, K. W., Reinhold, W. C., Myers, T. G., Andrews, D. T., et al. (2000) Nat. Genet. 24, 236-244. [DOI] [PubMed] [Google Scholar]
- 4.Bhattaxharjee A., Richards, W. G., Staunton, J., Li, C., Monti, S., Vasa, P., Ladd, C., Beheshti, J., Bueno, R., Gillette, M., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giordano T. J., Shedden, K. A., Schwartz, D. R., Kuick, R., Taylor, J. M. G., Lee, N., Misek, D. E., Greenson, J. K., Kardia, S. L. R., Beer, D. G., et al. (2001) Am. J. Pathol. 159, 1231-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisen M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamayo P., Slonim, D., Mesirov, J., Zhu, Q., Kitareewan, S., Dmitrovsky, E., Lander, E. S. & Golub, T. R. (1999) Proc. Natl. Acad. Sci. USA 96, 2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alter O., Brown, P. O. & Botstein, D. (2000) Proc. Natl. Acad. Sci. USA 97, 10101-10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown M. P. S., Grundy, W. N., Lin, D., Cristianini, N., Sugnet, C. W., Furey, T. S., Ares, M., Jr. & Haussler, D. (2000) Proc. Natl. Acad. Sci. USA 97, 262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcotte E. M., Pellegrini, M., Thompson, M. J., Yeates, T. O. & Eisenberg, D. (1999) Nature 402, 83-86. [DOI] [PubMed] [Google Scholar]
- 11.Weaver R., (2002) Molecular Biology (McGraw–Hill, NY), pp. 406–407.
- 12.Stein C. (1981) Ann. Stat. 9, 1135-1151. [Google Scholar]
- 13.Dubois E. & Messenguy, F. (1997) Mol. Gen. Genet. 253, 568-580. [DOI] [PubMed] [Google Scholar]
- 14.Smart W. C., Coffman, J. A. & Cooper, T. G. (1996) Mol. Cell. Biol. 16, 5876-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulkarni A. A., Abul-Hamd, A. T., Rai, R., El Berry, H. & Cooper, T. G. (2001) J. Biol. Chem. 276, 32136-32144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abadjieva A., Pauwels, K., Hilven, P. & Crabeel, M. (2001) J. Biol. Chem. 276, 42869-42880. [DOI] [PubMed] [Google Scholar]
- 17.Fabrizio P., Pozza, F., Pletcher, S. D., Gendron, C. M. & Longo, V. D. (2001) Science 292, 288-290. [DOI] [PubMed] [Google Scholar]
- 18.Crauwels, M., Donaton, M. C., Pernambuco, M. B., Winderickx, J., de Winde, J. H. & Thevelein, J. M. (1997) Microbiology 2627–2637. [DOI] [PubMed]
- 19.Finch C. E. & Ruvkun, G. (2001) Annu. Rev. Genomics Hum. Genet. 2, 435-462. [DOI] [PubMed] [Google Scholar]
- 20.Lin S., Defossez, P. & Guarente, L. (2000) Science 289, 2126-2128. [DOI] [PubMed] [Google Scholar]
- 21.Saraste M. (1999) Science 283, 1488-1493. [DOI] [PubMed] [Google Scholar]
- 22.Cyert M. S. (2001) Annu. Rev. Genet. 35, 647-672. [DOI] [PubMed] [Google Scholar]
- 23.Furuchi T., Ishikawa, H., Miura, N., Ishizuka, M., Kajiya, K., Kuge, S. & Naganuma, A. (2001) Mol. Pharmacol. 59, 470-474. [DOI] [PubMed] [Google Scholar]
- 24.Bassler J., Grandi, P., Gadal, O., Lessmann, T., Petfalski, E., Tollervey, D., Lechner, J. & Hurt, E. (2001) Mol. Cell 8, 517-529. [DOI] [PubMed] [Google Scholar]
- 25.Russnak R., Konczal, D. & McIntire, S. L. (2001) J. Biol. Chem. 276, 23849-23857. [DOI] [PubMed] [Google Scholar]
- 26.Wingender E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Prüβ, M., Reuter, I. & Schacherer, F. (2000) Nucleic Acids Res. 28, 316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.