Abstract
The Forkhead Box (Fox) proteins are an extensive family of transcription factors that shares homology in the winged helix DNA-binding domain and whose members play essential roles in cellular proliferation, differentiation, transformation, longevity, and metabolic homeostasis. Liver regeneration studies with transgenic mice demonstrated that FoxM1B regulates the onset of hepatocyte DNA replication and mitosis by stimulating expression of cell cycle genes. Here, we demonstrate that albumin-promoter-driven Cre recombinase-mediated hepatocyte-specific deletion of the Foxm1b Floxed (fl) targeted allele resulted in significant reduction in hepatocyte DNA replication and inhibition of mitosis after partial hepatectomy. Reduced DNA replication in regenerating Foxm1b−/− hepatocytes was associated with sustained increase in nuclear staining of the cyclin-dependent kinase (Cdk) inhibitor p21Cip1 (p21) protein between 24 and 40 h after partial hepatectomy. Furthermore, increased nuclear p21 levels and reduced expression of Cdc25A phosphatase coincided with decreases in Cdk2 activation and hepatocyte progression into S-phase. Moreover, the significant reduction in hepatocyte mitosis was associated with diminished mRNA levels and nuclear expression of Cdc25B phosphatase and delayed accumulation of cyclin B1 protein, which is required for Cdk1 activation and entry into mitosis. Cotransfection studies demonstrate that FoxM1B protein directly activated transcription of the Cdc25B promoter region. Our present study shows that the mammalian Foxm1b transcription factor regulates expression of cell cycle proteins essential for hepatocyte entry into DNA replication and mitosis.
Keywords: knock-out mouse, Cdc25A, Cdc25B, cyclin-dependent kinase inhibitor p21Cip1
Cell division is a tightly regulated process, especially at the initiation of DNA replication (S-phase) and at the entry into mitosis (M-phase). Progression through the cell cycle is regulated by temporal activation of multiple cyclin-dependent kinases (Cdk). In addition to assembly with a cyclin-regulatory subunit, Cdk activity requires dephosphorylation of the Cdk catalytic subunit by the Cdc25A, Cdc25B, or Cdc25C phosphatase protein (1–3) and is negatively regulated by Cdk inhibitor p21Cip1 (p21), p27Kip1, and p16INK4A (4). Cdk2 activity in complex with cyclin E and cyclin A is critical for S-phase progression because cyclin E/A-Cdk2 cooperates with cyclin D-Cdk4/6 to phosphorylate the retinoblastoma (RB) protein, which releases bound E2F transcription factor and allows it to stimulate expression of proliferation-specific target genes (5, 6). Likewise, phosphorylation of critical target proteins by the active cyclin B-Cdk1 complex mediates progression into mitosis (7).
The Forkhead Box (Fox) transcription factors are an extensive family of transcription factors, consisting of more than 50 mammalian proteins (8) that share homology in the winged helix DNA-binding domain (9). Its members play important roles in regulating expression of genes involved in cellular proliferation (10), differentiation (11–13), apoptosis (14), transformation (15), longevity (16), and metabolic homeostasis (17). The mammalian liver is one of the few adult organs capable of completely regenerating itself in response to injury, and which is mediated by the release of growth factors and cytokines that stimulate reentry of terminally differentiated hepatocytes into the cell cycle (18–21). In regenerating liver, increased hepatic expression of FoxM1B levels occurs at the G1/S transition of the cell cycle; its levels remain elevated throughout the period of proliferation, suggesting that the delayed early FoxM1B transcription factor plays a role in cell cycle progression (22). Premature expression of FoxM1B (HFH-11B) levels in regenerating liver of transgenic (TG) mice accelerated the onset of hepatocyte DNA replication and mitosis through stimulating earlier expression of cell cycle genes (10, 23). Analysis of cDNA microarrays shows that diminished proliferation exhibited by human fibroblasts from either the elderly or genetically aged patients with Hutchinson-Gilford progeria compared with proliferating young human fibroblasts is associated with reduced expression of FoxM1B and its cell cycle target genes (24). Recent liver regeneration studies indicate that maintaining hepatocyte expression of FoxM1B in 12-month old (old-aged) TG mice is sufficient to increase hepatocyte DNA replication and mitosis and reestablish expression of cell cycle regulatory genes to levels found in young regenerating mouse liver (25, 26). These results suggest the hypothesis that Foxm1b controls the transcriptional network of genes essential for cell cycle progression.
In this study, we performed partial hepatectomy (PHx) operations to induce hepatic regeneration, and we demonstrated that albumin-promoter-driven Cre recombinase (Alb-Cre)-mediated hepatocyte-specific deletion of the Foxm1b Floxed (fl/fl) allele resulted in significant reduction in hepatocyte DNA replication and inhibition of mitosis. Diminished hepatocyte proliferation in regenerating Alb-Cre Foxm1b−/− liver was associated with altered expression of proteins that limit Cdk1 and Cdk2 activity required for normal cell cycle progression into DNA replication and mitosis.
Materials and Methods
Generation of Mice with fl Foxm1b-Targeted Allele and Hepatocyte-Specific Deletion.
Mouse Foxm1b genomic DNA was isolated from mouse 129SvJ genomic library (Stratagene) and characterization of the intron and exon boundaries was determined as described (22). We constructed a triple-LoxP Foxm1b-targeting vector to generate an “fl” mouse Foxm1b-targeted locus consisting of a LoxP site inserted into the third Foxm1b intron before the winged helix DNA-binding domain and LoxP PGK-1 neomycin (neo) LoxP selection cassette placed 3′ to the final Foxm1b exon 7 (Fig. 1A). The PGK-1 promoter-driven herpes simplex virus-thymidine kinase (HSV-TK) gene was placed outside of the Foxm1b gene homology region for negative selection of nonhomologous recombination during targeted embryonic stem (ES) cell selection by using ganciclovir. We used this Foxm1b fl-targeting vector for electroporation of mouse ES cells (129SvJ; Genome Systems, St. Louis), which were propagated on mouse embryo fibroblasts feeder cells (Genome Systems) and selected for neo (G418) and HSV-TK (ganciclovir) resistance, as described (27). ES cells with the appropriate Foxm1b fl-targeted locus were identified by Southern blot analysis and were used to generate chimeric mice by injecting them into mouse blastocysts. Mice containing the Foxm1b fl targeted allele were determined by Southern blot analysis with 5′ and 3′ probes (Fig. 1 A–C) and PCR amplification with primers flanking the LoxP sequence located in the third exon (Fig. 1E). These Foxm1b primers included the sense 5′-taggagatacactgttatat-3′ and the antisense 5′-tgtgggaaaatgcttacaaaag-3′. The chimeric mice were bred with C57/B6 WT mice to produce Foxm1b fl/+ mice, which were backcrossed to generate viable Foxm1b fl/fl mice. Liver regeneration studies used Foxm1b fl/fl mice that were bred into the C57/B6 background for four generations. Hepatocyte-specific deletion of the Foxm1b fl/fl allele was accomplished through breeding with the Alb-Cre recombinase C57B6 transgenic mice (The Jackson Laboratory; ref. 28). Hepatocyte-specific deletion of the Foxm1b fl/fl allele was verified by using liver genomic DNA for Southern blot analysis (Fig. 1D).
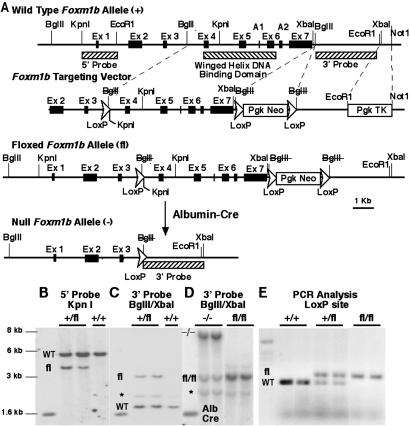
Fig 1.
fl Foxm1b-targeting vector, targeted Foxm1b allele, and deleted Foxm1b allele. (A) Diagrammatical representation of the mouse Foxm1b locus with seven exons (exons A1 and A2 are found in the Foxm1a splicing isoform, and exon A1 is found in the Foxm1c isoform). Also shown is the Foxm1b fl-targeting vector, the Foxm1b fl-targeted allele, and the deleted Foxm1b allele after introduction of the albumin promoter-driven Cre recombinase transgene (Alb-Cre). Hatched boxes indicate the position of the 5′ and 3′ Southern blot hybridization probes and the exons encoding the winged helix DNA-binding domain. Dotted lines indicate restriction enzymes used to introduce the KpnI-containing LoxP sequence (used for 5′ end screening), the LoxP PGK promoter-driven neomycin (neo) gene LoxP positive-selection cassette, and the PGK HSV-TK gene (used for negative selection for nonhomologous recombination). Insertion of the LoxP sequences disrupted the BglII restriction sites (BglII), and deletion by the Alb-Cre recombinase protein removes Foxm1b sequences flanking the two farthest LoxP sites, leaving one LoxP sequence. (B and C) Southern blot of genomic DNA from Foxm1b fl-targeted ES cells showing bands specific to WT and Foxm1b fl loci (pseudogene band indicated by *). (D) Southern blot of liver genomic DNA from Foxm1b fl/fl mice and the Alb-Cre recombinase transgene-mediated hepatocyte deletion (−/−) of the Foxm1b fl/fl allele creating the larger molecular weight DNA fragment. (E) PCR analysis of mouse tail genomic DNA with primers flanking the third intron LoxP sequence resulting in a larger PCR DNA product with the Foxm1b fl-targeted locus.
Partial Hepatectomy Surgery, Immunohistochemical Staining, Western Blot Analysis, and RNase Protection Assays (RPA).
Eight-week old Alb-Cre Foxm1b−/− mice or Foxm1b fl/fl littermates were subjected to PHx operations to induce liver regeneration (10, 25). Three mice at each time point were killed by using CO2 asphyxiation at the following intervals after PHx: 24, 28, 32, 36, 40, 44, 48, and 52 h and 7 days. The regenerating livers were harvested and divided into three portions (i) to isolate total RNA (10), (ii) to isolate total protein extract, and (iii) for paraffin embedding (29). DNA replication was monitored by immunohistochemical detection of 5-bromo-2′-deoxyuridine (BrdUrd, Sigma) incorporation as described (10, 25). By using three regenerating livers per time point, we counted the number of BrdUrd-positive nuclei per 1,000 hepatocytes to calculate the mean number of BrdUrd-positive cells (± SE), as described (10, 25). Three regenerating liver sections at 32, 36, 40, 44, 48, and 52 h after PHx were stained with hematoxylin and eosin and examined for hepatocyte mitotic figures (mitosis). Hepatocyte mitosis is expressed as the mean of the number of mitotic figures per 1,000 hepatocytes ± SD, as described (10, 25).
Antibodies specific to FoxM1B (10, 22), p21 (Calbiochem), or Cdc25B (Santa Cruz Biotechnology) proteins were used for immunohistochemical detection of paraffin-embedded 5-μm sections of regenerating liver by using methods described (10, 22, 26). For Western blot analysis, 50 μg of total liver protein (29) was separated on SDS/PAGE and transferred to Protran membrane (Schleicher & Schuell). The signals from primary p21 (Calbiochem; 1:750), Cdc25A (Santa Cruz Biotechnology, 1:200), cyclin B1 (Santa Cruz Biotechnology, 1:200) and Cdk1 phosphotyrosine-15-specific (Cell Signaling, Berkeley, CA; 1:1,000) antibodies were amplified by biotin-conjugated anti-rabbit IgG (Bio-Rad) and detected with Enhanced Chemiluminescence Plus (Amersham Pharmacia). Total RNA was prepared from mouse liver at indicated hours after PHx by using RNA-STAT-60 (Tel-Test, Friendswood, TX) and was used for RPA with antisense [α-32P]UTP-labeled probes specific to either cell cycle regulatory genes or the p21 gene, and expression levels were calculated as described (23, 25, 26).
Immunoprecipitation Kinase Assays.
Either the Cdk1 (2 μg, Labvision) or Cdk2 antibody (1 μg, Santa Cruz Biotechnology) and protein A Sepharose beads (Amersham Pharmacia) were used to immunoprecipitate these kinases from 200 μg of total protein extracts from regenerating liver. The immunoprecipitated Cdk1 and Cdk2 proteins were used for kinase assays with [32P]γ-ATP and either the Cdk2 substrate Rb protein (Santa Cruz Biotechnology) or the Cdk1 substrate histone H1 protein (Santa Cruz Biotechnology), as described (25, 27). One half of the kinase reaction was separated by SDS/PAGE and exposed to a phosphorimager screen, scanned with the Storm 860 phosphorimager, and phosphorylated bands were quantitated with the IMAGEQUANT program (Amersham Pharmacia).
U2OS Cell Cotransfection Assays.
U2OS cells were transfected with 50 ng of either CMV (cytomegalovirus)-FoxM1B (1–748) cDNA, CMV-FoxM1B (1–688) cDNA, or CMV-empty expression vectors and 1500 ng of the −200-bp mouse Cdc25B-promoter luciferase plasmid using Fugene6 (Roche Molecular Biochemicals). Protein extracts were prepared from transfected U2OS cells at 24 h after DNA transfection, and the Dual-Luciferase Assay System (Promega) was used to measure luciferase enzyme activity, as described (25). Results are presented as mean fold induction of Cdc25B promoter activity ± SD from two separate experiments in duplicate, with the CMV-empty value set at 1.0.
Results
Alb-Cre Recombinase Mediates Hepatocyte-Specific Deletion of the Mouse Foxm1b fl/fl-Targeted Allele.
To generate mice with a hepatocyte-specific deletion of the mouse Foxm1b gene, we used a triple-LoxP Foxm1b fl-targeting vector (Fig. 1A) for electroporation of mouse ES cells. We selected G418 (neo) and ganciclovir (HSV-TK)-resistant ES cells containing a Foxm1b fl-targeted allele (27) and used them to generate Foxm1b fl/fl mice (see Materials and Methods for details). Albumin promoter-driven Alb-Cre mediates hepatocyte-specific deletion of the mouse Foxm1b fl/fl-targeted allele, removing the entire winged helix DNA-binding domain and the C-terminal transcriptional-activation domain (22), thereby preventing expression of functional Foxm1b protein (Fig. 1A). Mouse ES cells containing homologous recombination with the Foxm1b fl-targeting vector were identified by Southern blot analysis. By using the 5′ genomic probe and KpnI digestion (Fig. 1A), the Foxm1b fl allele is detected by a smaller molecular weight DNA fragment because of the additional KpnI site engineered at the end of the LoxP sequence (Fig. 1B). By using the 3′ genomic probe and BglII/XbaI digestion (Fig. 1A), the Foxm1b fl allele is detected by a larger band because of the additional 1.6-kb PGK-neo positive selection cassette (Fig. 1C). After mouse blastocyst injection of the Foxm1b fl/+ ES cells, we obtained chimeric mice with germ line transmission and bred them to generate viable Foxm1b fl/fl C57/B6 mice (Fig. 1D). PCR amplification of mouse tail genomic DNA with primers that flanked the third intron LoxP site detected the larger molecular weight product derived from the Foxm1b fl allele (Fig. 1E). Breeding the Alb-Cre recombinase transgene into the Foxm1b fl/fl mouse genetic background allowed hepatocyte-specific deletion of the Foxm1b locus within 6 weeks after birth (28), as detected by a larger molecular weight band in Southern blots of BglII/XbaI-digested liver genomic DNA (Fig. 1D, band indicated by −/−).
Foxm1b Is Required for Normal Hepatocyte DNA Replication and Mitosis in Regenerating Liver.
Adult Alb-Cre Foxm1b−/− livers were histologically normal before the PHx and exhibited no difference in liver weight/body weight ratio compared with Foxm1b fl/fl controls. Eight-week old Alb-Cre Foxm1b−/− mice or Foxm1b fl/fl littermates were subjected to PHx, and their regenerating livers were harvested at different intervals between 24 and 52 h and 7 days after surgery. Regenerating liver from Foxm1b fl/fl littermates displayed normal induction of Foxm1b mRNA and abundant hepatocyte nuclear staining of Foxm1b protein at 40 h after PHx (Fig. 2 A and B). Confirming hepatocyte-specific deletion of the Foxm1b fl/fl allele, we found that regenerating Alb-Cre Foxm1b−/− liver failed to induce high levels of Foxm1b mRNA and displayed no hepatocyte nuclear staining of Foxm1b protein in any of the micrographs examined (Fig. 2 B and C and data not shown). The Foxm1b fl/fl mice exhibited a bifunctional S-phase peak in hepatocyte DNA replication, as evidenced by BrdUrd incorporation between 32 and 44 h after PHx (Fig. 3A). In contrast, regenerating Alb-Cre Foxm1b−/− liver displayed a significant reduction in hepatocyte DNA replication, but BrdUrd incorporation was still detectable during these time points (Fig. 3A). A more significant reduction in hepatocyte mitosis was observed in regenerating Alb-Cre Foxm1b−/− liver, as evidenced by the paucity of mitotic figures between 36 to 52 h after PHx compared with regenerating Foxm1b fl/fl liver (Fig. 3B). This diminished hepatocyte mitosis in regenerating Alb-Cre Foxm1b−/− liver caused hepatocyte hypertrophy and compensatory increase in liver size at 7 days after PHx, but they did not exhibit any increase in apoptosis, as measured by terminal deoxynucleotidyltransferase-mediated dUTP assay (see Supporting Text and Fig. 7, which are published as supporting information on the PNAS web site, www.pnas.org). These studies suggest that Foxm1b is critical for mediating normal levels of hepatocyte DNA replication and is essential for progression into mitosis.
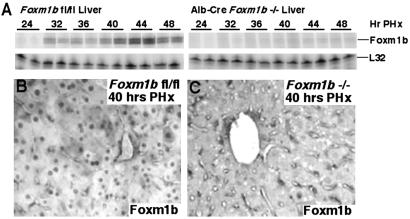
Fig 2.
Liver regeneration studies demonstrate that Foxm1b-deficient (−/−) hepatocytes lack induction of the Foxm1b mRNA and protein. (A) RPA demonstrate significant reduction in expression of Foxm1b mRNA. Total RNA was prepared from regenerating livers from either fl/fl mice or Alb-Cre Foxm1b−/− mice at indicated hours after PHx (numbers above panels) and was used for RPA to demonstrate that regenerating Foxm1b−/− hepatocytes failed to induce high levels of Foxm1b mRNA. Immunohistochemical staining of regenerating liver sections (40 h after PHx) with Foxm1b antibody demonstrates that regenerating Alb-Cre Foxm1b−/− hepatocytes (C) display undetectable nuclear staining of Foxm1b protein compared with regenerating Foxm1b fl/fl hepatocytes (B).
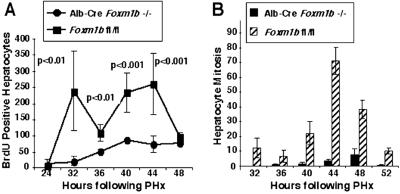
Fig 3.
Regenerating Alb-Cre Foxm1b−/− mouse liver displays significant decreases in hepatocyte DNA replication and mitosis. (A) Graphic representation of diminished BrdUrd incorporation during mouse liver regeneration in Foxm1b-deficient hepatocytes. Graphically presented is the BrdUrd incorporation (DNA replication) detected by immunohistochemical staining (10, 25) at the indicated hours after PHx with either 8-week-old Alb-Cre Foxm1b−/− mice or Foxm1b fl/fl littermates. The mean of the number of BrdUrd-positive hepatocyte nuclei per 1,000 hepatocytes ± SE was calculated for each time point (three mice per time point). (B) Graphic representation of diminished hepatocyte mitosis in regenerating livers of Foxm1b-deficient vs. fl Foxm1b mouse liver between 32 and 52 h after PHx. Hepatocyte mitosis is expressed as the mean of the number of mitotic figures found per 1,000 hepatocytes ± SD using three mice per time point (10, 25).
Regenerating Alb-Cre Foxm1b−/− Liver Displays Increased Levels of p21 and Diminished Cdc25A Protein Expression Leading to Decreased Cdk2 Activity.
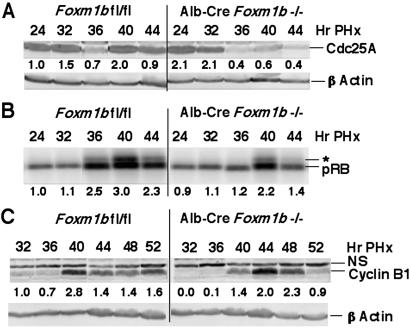
Previous CCl4 liver injury studies demonstrated that increased hepatocyte levels of FoxM1B mediated reduced expression of Cdk inhibitor p21 protein before S-phase (23). Because Alb-Cre Foxm1b−/− liver displayed minimal changes in either cyclin D or cyclin E mRNA levels after PHx (data not shown), we next examined whether regenerating Alb-Cre Foxm1b−/− liver exhibited altered expression of the p21 protein. Western blot analysis of protein extracts from regenerating Alb-Cre Foxm1b−/− livers showed that they displayed increased p21 protein levels compared with the Foxm1b fl/fl control liver (Fig. 4A). Furthermore, whereas regenerating Foxm1b fl/fl hepatocytes exhibited only a transient increase in nuclear p21 staining at 32 h after PHx (Fig. 4 B–E), Foxm1b−/− hepatocytes displayed a sustained increase in nuclear p21 protein levels between 24 to 40 h after PHx (Fig. 4 F–I). Moreover, the hepatocyte levels of nuclear p21 protein were not uniform, a finding consistent with detectable levels of hepatocyte DNA replication in regenerating Alb-Cre Foxm1b−/− liver. Because regenerating p21−/− liver displayed increased Cdc25A expression and earlier nuclear localization of Cdc25A (30), we examined whether increased p21 levels resulted in diminished protein expression of Cdc25A phosphatase required for Cdk2 activity (5). Western blot analysis demonstrated that regenerating Alb-Cre Foxm1b−/− livers displayed reduced Cdc25A protein levels before S-phase that initiated at 36 h after PHx (Fig. 5A). Thus, both decreased Cdc25A protein expression and increased nuclear expression of p21 protein are consistent with reduced Cdk2 kinase activity necessary for S-phase progression (5). Regenerating liver protein extracts prepared from either Foxm1b fl/fl or Alb-Cre Foxm1b−/− mice were, therefore, immunoprecipitated with Cdk2 antibody and then used to phosphorylate recombinant RB protein substrate. These experiments demonstrated that regenerating Alb-Cre Foxm1b−/− liver displayed little hyperphosphorylation of the RB protein (Fig. 5B, indicated by *), thereby demonstrating reduced Cdk2 kinase activity compared with that of regenerating Foxm1b fl/fl liver. Furthermore, active cyclin A2-Cdk2 kinase complex is required to phosphorylate the Cdh1 subunit of the ubiquitin–ligase anaphase-promoting complex (APC), which prevents APC-mediated degradation of cyclin B at the end of S-phase and thus allows cyclin B accumulation to promote entry into mitosis (5). Consistent with this concept, Western blot analysis with the cyclin B1 antibody showed delayed S-phase accumulation of cyclin B1 protein between 32 and 40 h after PHx in liver extracts prepared from Alb-Cre Foxm1b−/− mice compared with Foxm1b fl/fl littermates (Fig. 5C). Collectively, diminished DNA replication in Foxm1b−/− liver is associated with increased Cdk inhibitor p21 protein and decreased Cdc25A phosphatase levels, resulting in a reduction of cyclin A/E-Cdk2 kinase activity required for S-phase progression.
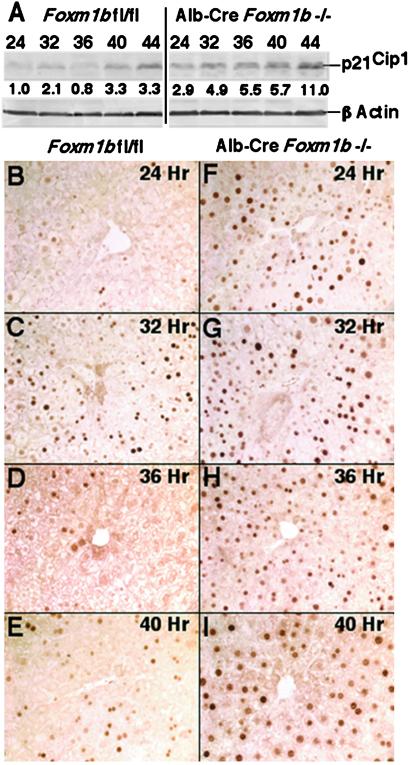
Fig 4.
Regenerating Alb-Cre Foxm1b−/− liver display increased p21 protein levels and nuclear staining. (A) Western blot analysis with p21 antibody reveals increased p21 protein levels in regenerating Alb-Cre Foxm1b−/− liver. Total protein extracts were isolated from regenerating liver of Foxm1b fl/fl and Alb-Cre Foxm1b−/− mice and analyzed for protein expression of p21 by Western blot analysis. Shown below the panels is the fold increase in expression levels with respect to regenerating Foxm1b fl/fl liver at the 24-h time point. Immunohistochemical staining of regenerating liver sections at the indicated time points after PHx with p21 antibody demonstrates increased hepatocyte nuclear staining of p21 protein in Alb-Cre Foxm1b−/− liver (F–I) compared with Foxm1b fl/fl littermates (B–E). (B–I, ×100 magnification.)
Fig 5.
Regenerating Alb-Cre Foxm1b−/− liver exhibit diminished Cdc25A protein levels and Cdk2 activity. (A) Western blot analysis with Cdc25A antibody reveals diminished Cdc25A phosphatase protein levels in regenerating Alb-Cre Foxm1b−/− liver. (B) Diminished Cdk2 kinase activity in regenerating Alb-Cre Foxm1b−/− liver. Total protein extracts were isolated from regenerating liver, immunoprecipitated with Cdk2 antibody, and used for Cdk2 kinase assays with RB protein substrate. Position of the phosphorylated and hyperphosphorylated (*) RB protein is indicated on panels. (C) Western blot analysis with cyclin B1 antibody reveals delayed cyclin B1 protein levels in regenerating Alb-Cre Foxm1b−/− liver. A nonspecific band reacting with the cyclin B1 antibody is labeled by NS. Shown below the panels is the fold increase in expression levels with respect to regenerating Foxm1b fl/fl liver at the 24- or 32-h time points.
Regenerating Alb-Cre Foxm1b−/− Liver Displays Reduced Cdc25B Phosphatase Levels Leading to Diminished Cyclin B-Cdk1 Activity.
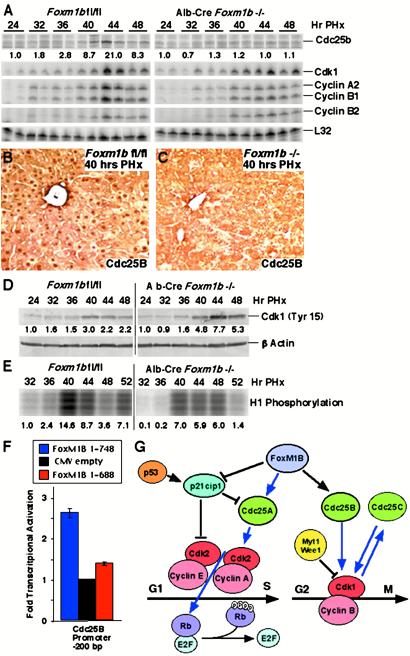
To identify mitotic regulatory genes whose expression is diminished in regenerating liver of Alb-Cre Foxm1b−/− mice, RPA were performed (Fig. 6A). Regenerating Alb-Cre Foxm1b−/− livers displayed significant reduction in induced levels of Cdc25B mRNA compared with regenerating Foxm1b fl/fl liver controls, whereas only slight decreases were found in regenerating hepatic levels of Cdk1, cyclin A2, and cyclin B1 mRNAs (Fig. 6A). Whereas Foxm1b fl/fl hepatocytes displayed abundant Cdc25B nuclear staining at 40 h after PHx (Fig. 6B), undetectable levels of nuclear Cdc25B protein was found in regenerating Foxm1b−/− hepatocytes (Fig. 6C). The Cdc25B phosphatase promotes M-phase progression by dephosphorylation of the Cdk1 tyrosine-15 and threonine-14 residues, in effect providing a mitotic checkpoint by regulating activity of the Cdk1/cyclin B kinase (1–3). Western blot analysis with Cdk-1-specific phosphotyrosine-15 antibody demonstrated increased Cdk-1 phosphorylation in regenerating liver extracts from Alb-Cre Foxm1b−/− mice (Fig. 6D), a finding consistent with diminished levels of the Cdc25B phosphatase protein (1–3). In support of diminished Cdk1 kinase activity, immunoprecipitation-kinase assays demonstrated that regenerating liver protein extracts from Alb-Cre Foxm1b−/− mice displayed reduced Cdk-1-dependent phosphorylation of the histone H1 substrate (Fig. 6E). These studies demonstrate that Foxm1b regulates nuclear expression of the Cdc25B phosphatase protein, an essential activator of M-phase promoting Cdk1-cyclin B kinase.
Fig 6.
Regenerating Alb-Cre Foxm1b−/− liver exhibits diminished expression and nuclear staining of cdc25B phosphatase protein. (A) RPA demonstrate that regenerating Alb-Cre Foxm1b−/− liver exhibited reduced Cdc25B mRNA levels and slight decreases in expression of cyclin A2, cyclin B1, and Cdk1. (B and C) Undetectable hepatocyte nuclear Cdc25B protein staining in regenerating liver of Alb-Cre Foxm1b−/− mice. Immunohistochemical staining of regenerating liver sections (40 h after PHx) with Cdc25B antibody demonstrates that regenerating Alb-Cre Foxm1b−/− hepatocytes display undetectable Cdc25B nuclear staining (C) compared with regenerating Foxm1b fl/fl hepatocytes (B). (D) Increased phosphorylation of Cdk1 protein in regenerating Alb-Cre Foxm1b−/− liver. Western blot analysis with Cdk-1 phospho-tyrosine-15 antibody reveals increased Cdk-1 phosphorylation in regenerating Alb-Cre Foxm1b−/− liver resulting from diminished cdc25B phosphatase levels. (E) Diminished Cdk1-dependent phosphorylation of histone H1 protein in regenerating Alb-Cre Foxm1b−/− liver. The Cdk1 protein was immunoprecipitated from regenerating liver extracts and used for kinase assays with H1 protein phosphorylation substrate. Shown below the panels is the fold increase in expression levels with respect to regenerating liver at 24 h after PHx and (E) with respect to Foxm1b fl/fl 32-h time point. (F) Foxm1b directly activates transcription of the Cdc25B promoter in cotransfection assays. The osteosarcoma U2OS cell line was cotransfected with the mouse −200-bp Cdc25B promoter luciferase plasmid and CMV expression vector containing either human FoxM1B full-length cDNA (1–748) or transcriptionally inactive FoxM1B (1–688) deletion as described (25). Graphic presentation of normalized fold induction of Cdc25B promoter expression in response to FoxM1B cDNA cotransfection, with CMV-empty vector control set at 1.0. Two transfection experiments were performed in duplicate and used to determine mean fold induction ± SD. (G). Diagram depicting FoxM1B regulation of cell cycle genes. Blue arrows represent positive regulation and black lines represent negative regulation.
Cotransfection assays in osteosarcoma U2OS cells with the −200-bp mouse Cdc25B promoter luciferase plasmids and CMV human FoxM1B (1–748) cDNA expression vector demonstrated that FoxM1B protein stimulated expression of the Cdc25B promoter (Fig. 6F). In contrast, no transcriptional activation of the Cdc25B promoter was found with a C-terminal mutant FoxM1B (1–688) that deletes sequences critical for transcriptional activation (Fig. 6F). These results demonstrate that Foxm1b regulates transcription of Cdc25B phosphatase gene, whose expression is essential for activation of Cdk1-cyclin B kinase and M-phase progression.
Discussion
Liver regeneration studies with transgenic mice demonstrated that FoxM1B regulates the onset of hepatocyte DNA replication and mitosis by stimulating expression of cell cycle genes (10, 22, 23, 26). In this study, we used Alb-Cre recombinase to generate a hepatocyte-specific deletion of the Foxm1b gene and demonstrated that Foxm1b is required for normal levels of hepatocyte DNA replication and is essential for mitosis in regenerating liver. We found no significant increase in hepatocyte apoptosis in regenerating Alb-Cre Foxm1b−/− liver (data not shown), suggesting that Foxm1b is required for hepatocyte proliferation but not survival. Reduced DNA replication in regenerating Foxm1b−/− hepatocytes coincided with sustained increase in nuclear staining of the Cdk inhibitor p21 protein between 24 and 40 h after PHx. This increase in nuclear p21 levels and a reduction in Cdc25A phosphatase expression resulted in decreased activation of Cdk2 kinase (Fig. 6G). Cyclin E/A-Cdk2 complex cooperates with cyclin D-Cdk4/6 complex to phosphorylate the RB protein, which releases bound E2F transcription factor and allows it to stimulate expression of target genes essential for hepatocyte S-phase progression (5, 6). Furthermore, a significant reduction in hepatocyte mitosis was associated with reduced mRNA and nuclear protein levels of the Cdc25B phosphatase and delayed S-phase accumulation of cyclin B1 (Fig. 6G), both of which are necessary to stimulate Cdk1 activity that mediates phosphorylation of critical target proteins required for hepatocyte entry into mitosis (7). We demonstrated that the Cdc25B promoter region is a direct target for FoxM1B transcriptional activation in cotransfection assays. Our present study shows that the mammalian Foxm1b transcription factor is regulating expression of cell cycle proteins that stimulate Cdk2 and Cdk1 activity, which are essential for entry into DNA replication and mitosis, respectively. Consistent with these findings, elevated FoxM1B levels are found in numerous cell lines derived from tumors (22, 31, 32) and in human basal cell carcinomas (33), suggesting that FoxM1B is required for cellular proliferation in human cancers.
Foxm1b deficiency caused inhibition of regenerating hepatocyte mitosis, which was associated with undetectable nuclear levels of Cdc25B phosphatase, but exhibited normal levels of Cdc25C (data not shown) and correlated with decreased M-phase-promoting Cdk1 activity. Activation of Cdk1 kinase is controlled by multiple mechanisms and constitutes a regulatory mitotic checkpoint (1, 34). Stimulation of Cdk1 activity requires complex formation with cyclin B proteins and dephosphorylation of the Y15 and T14 residues of the Cdk1 protein by the Cdc25B and Cdc25C phosphatases (1–3). To prevent premature activation of Cdk1 kinase activity, these Cdk1 residues are phosphorylated by the Myt1 and Wee1 kinases, whose activities are inhibited by phosphorylation via the mitogen-activated protein kinases (RSK) pathway (1, 34). Stimulation of the Cdc25C phosphatase activity is mediated by Cdk1/cyclin B phosphorylation at mitosis, which in turn, dephosphorylates and further activates Cdk1 kinase mediating entry into mitosis (35, 36). In contrast, Cdc25B phosphatase activity appears at late S-phase and peaks during G2 phase; it may be involved in the initiation of the G2/M transition by activating the Cdk1/cyclin B kinase (37). Consistent with this hypothesis, microinjection of Cdc25B-specific antibody inhibits progression of Hs68 cells into mitosis (37). Furthermore, microinjection of the Cdc25B protein, but not the Cdc25C protein, in tissue culture cells can drive replicating cells into mitosis (38). Our current liver regeneration studies with Alb-Cre Foxm1b−/− mice are consistent with the hypothesis that deficiency in nuclear expression of Cdc25B protein is sufficient to inhibit hepatocyte mitosis (Fig. 6G).
Maintaining hepatocyte expression of FoxM1B in regenerating liver of old-aged TG CD-1 mice is sufficient to increase hepatocyte proliferation and expression of cyclin F, cyclin B1, cyclin B2, Cdc25B, and p55Cdc, all of which are required for mitosis (25). Despite the ability of FoxM1B to restore expression of these mitotic regulatory genes in regenerating old-aged mouse liver, our current studies show that FoxM1B is required only for Cdc25B expression in 2-month-old regenerating liver, suggesting that in young mice, redundant regulatory pathways exist for the other mitotic regulatory genes. Taken together, our studies demonstrate that Foxm1b regulates transcription and nuclear expression levels of the Cdc25B phosphatase, which is essential for hepatocyte entry into mitosis.
Reduced hepatocyte DNA replication in regenerating Alb-Cre Foxm1b−/− liver was associated with a transcriptional-independent stimulation of p21 protein expression (data not shown) and increased p21 nuclear protein, indicating that FoxM1B regulates p21 protein levels rather than influencing its promoter activity. One possible explanation for increased nuclear levels of p21 protein is that one of the Foxm1b target genes is involved in mediating p21 protein degradation. Elevated transgenic hepatic levels of p21 protein caused significant decreases in regenerating hepatocyte DNA replication (39), a finding consistent with diminished regenerating hepatocyte DNA replication in Alb-Cre Foxm1b−/− mice. However, regenerating Alb-Cre Foxm1b−/− liver exhibited detectable hepatocyte DNA replication. One explanation for this phenotype is that hepatocyte nuclear levels of p21 protein was not uniformly increased in regenerating Alb-Cre Foxm1b−/− liver, suggesting that a subset of hepatocytes displayed p21 levels that are insufficient to inhibit DNA replication. Collectively, these results suggest Foxm1b stimulated hepatocyte DNA replication through limiting p21 protein levels before S-phase and that Foxm1b mediated transcription and nuclear expression of Cdc25B phosphatase, which is essential for hepatocyte progression into mitosis.
Supplementary Material
Acknowledgments
We thank H. Ye for isolation of the Foxm1b genomic clone and R. Franks for injection of Foxm1b-targeted ES cells into mouse blastocysts. We also thank P. Raychaudhuri, K. Krupczak-Hollis, V. Kalinichenko, Y. Zhou, M. Major, and F. Rausa for critically reviewing the manuscript. This work was supported by National Institutes of Health Grant DK 54687 from National Institute of Diabetes and Digestive and Kidney Diseases (to R.H.C.).
Abbreviations
Cdk, cyclin-dependent kinase
RB, retinoblastoma
Foxm1b, Forkhead Box m1b
PHx, partial hepatectomy
Alb-Cre, albumin-promoter-driven Cre recombinase
fl, Floxed
neo, neomycin
ES, embryonic stem
BrdUrd, 5 bromo-2′-deoxyuridine
p21, p21Cip1
RPA, RNase protection assays
CMV, cytomegalovirus
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nilsson I. & Hoffmann, I. (2000) Prog. Cell Cycle Res. 4, 107-114. [DOI] [PubMed] [Google Scholar]
- 2.Sebastian B., Kakizuka, A. & Hunter, T. (1993) Proc. Natl. Acad. Sci. USA 90, 3521-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trembley J. H., Ebbert, J. O., Kren, B. T. & Steer, C. J. (1996) Cell Growth Differ. 7, 903-916. [PubMed] [Google Scholar]
- 4.Sherr C. J. & Roberts, J. M. (1999) Genes Dev. 13, 1501-1512. [DOI] [PubMed] [Google Scholar]
- 5.Harbour J. W. & Dean, D. C. (2000) Genes Dev. 14, 2393-2409. [DOI] [PubMed] [Google Scholar]
- 6.Ishida S., Huang, E., Zuzan, H., Spang, R., Leone, G., West, M. & Nevins, J. R. (2001) Mol. Cell. Biol. 21, 4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zachariae W. & Nasmyth, K. (1999) Genes Dev. 13, 2039-2058. [DOI] [PubMed] [Google Scholar]
- 8.Kaestner K. H., Knochel, W. & Martinez, D. E. (2000) Genes Dev. 14, 142-146. [PubMed] [Google Scholar]
- 9.Clark K. L., Halay, E. D., Lai, E. & Burley, S. K. (1993) Nature 364, 412-420. [DOI] [PubMed] [Google Scholar]
- 10.Ye H., Holterman, A., Yoo, K. W., Franks, R. R. & Costa, R. H. (1999) Mol. Cell. Biol. 19, 8570-8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa R. H., Kalinichenko, V. V. & Lim, L. (2001) Am. J. Physiol. 280, L823-L838. [DOI] [PubMed] [Google Scholar]
- 12.Duncan S. A. (2000) Dev. Dyn. 219, 131-142. [DOI] [PubMed] [Google Scholar]
- 13.Zaret K. S. (2002) Nat. Rev. Genet. 3, 499-512. [DOI] [PubMed] [Google Scholar]
- 14.Burgering B. M. & Kops, G. J. (2002) Trends Biochem. Sci. 27, 352-360. [DOI] [PubMed] [Google Scholar]
- 15.Ma Y., Geerdes, D. W. & Vogt, P. K. (2000) Oncogene 19, 4815-4821. [DOI] [PubMed] [Google Scholar]
- 16.Lee R. Y., Hench, J. & Ruvkun, G. (2001) Curr. Biol. 11, 1950-1957. [DOI] [PubMed] [Google Scholar]
- 17.Kaestner K. H. (2000) Trends Endocrinol. Metab. 11, 281-285. [DOI] [PubMed] [Google Scholar]
- 18.Diehl A. M. (2000) Immunol. Rev. 174, 160-171. [DOI] [PubMed] [Google Scholar]
- 19.Fausto N. (2000) J. Hepatol. 32, 19-31. [DOI] [PubMed] [Google Scholar]
- 20.Michalopoulos G. K. & DeFrances, M. C. (1997) Science 276, 60-66. [DOI] [PubMed] [Google Scholar]
- 21.Taub R., Greenbaum, L. E. & Peng, Y. (1999) Semin. Liver Dis. 19, 117-127. [DOI] [PubMed] [Google Scholar]
- 22.Ye H., Kelly, T. F., Samadani, U., Lim, L., Rubio, S., Overdier, D. G., Roebuck, K. A. & Costa, R. H. (1997) Mol. Cell. Biol. 17, 1626-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Hung, N.-J. & Costa, R. H. (2001) Hepatology 33, 1404-1414. [DOI] [PubMed] [Google Scholar]
- 24.Ly D. H., Lockhart, D. J., Lerner, R. A. & Schultz, P. G. (2000) Science 287, 2486-2492. [DOI] [PubMed] [Google Scholar]
- 25.Wang X., Quail, E., Hung, N.-J., Tan, Y., Ye, H. & Costa, R. H. (2001) Proc. Natl. Acad. Sci. USA 98, 11468-11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., Krupczak-Hollis, K., Tan, Y., Dennewitz, M. B., Adami, G. R. & Costa, R. H. (2002) J. Biol. Chem. 277, 44310-44316. [DOI] [PubMed] [Google Scholar]
- 27.Kiyokawa H., Kineman, R. D., Manova-Todorova, K. O., Soares, V. C., Hoffman, E. S., Ono, M., Khanam, D., Hayday, A. C., Frohman, L. A. & Koff, A. (1996) Cell 85, 721-732. [DOI] [PubMed] [Google Scholar]
- 28.Postic C. & Magnuson, M. A. (2000) Genesis 26, 149-150. [DOI] [PubMed] [Google Scholar]
- 29.Rausa F. M., Tan, Y., Zhou, H., Yoo, K., Stolz, D. B., Watkins, S., Franks, R. R., Unterman, T. G. & Costa, R. H. (2000) Mol. Cell. Biol. 20, 8264-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaime M., Pujol, M. J., Serratosa, J., Pantoja, C., Canela, N., Casanovas, O., Serrano, M., Agell, N. & Bachs, O. (2002) Hepatology 35, 1063-1071. [DOI] [PubMed] [Google Scholar]
- 31.Korver W., Roose, J. & Clevers, H. (1997) Nucleic Acids Res. 25, 1715-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao K. M., Sha, M., Lu, Z. & Wong, G. G. (1997) J. Biol. Chem. 272, 19827-19836. [DOI] [PubMed] [Google Scholar]
- 33.Teh M. T., Wong, S. T., Neill, G. W., Ghali, L. R., Philpott, M. P. & Quinn, A. G. (2002) Cancer Res. 62, 4773-4780. [PubMed] [Google Scholar]
- 34.Ohi R. & Gould, K. L. (1999) Curr. Opin. Cell Biol. 11, 267-273. [DOI] [PubMed] [Google Scholar]
- 35.Izumi T. & Maller, J. L. (1993) Mol. Biol. Cell 4, 1337-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann I., Clarke, P. R., Marcote, M. J., Karsenti, E. & Draetta, G. (1993) EMBO J. 12, 53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lammer C., Wagerer, S., Saffrich, R., Mertens, D., Ansorge, W. & Hoffmann, I. (1998) J. Cell Sci. 111, 2445-2453. [DOI] [PubMed] [Google Scholar]
- 38.Karlsson C., Katich, S., Hagting, A., Hoffmann, I. & Pines, J. (1999) J. Cell Biol. 146, 573-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu H., Wade, M., Krall, L., Grisham, J., Xiong, Y. & Van Dyke, T. (1996) Genes Dev. 10, 245-260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.