Abstract
In vitro data show that the adenomatous polyposis coli (APC) protein associates with the mitotic spindle and that mouse embryonic stem cells with biallelic Apc mutations are karyotypically unstable. These findings led to suggestions that APC acts in chromosomal segregation and that APC inactivation leads to chromosomal instability (CIN). An alternative hypothesis based on allelic loss studies in colorectal adenomas proposes that CIN precedes and contributes to genetic changes at APC. We determined whether colorectal adenomas with two mutations at APC show features consistent with these models by studying 55 lesions (average size 5 mm; range 1–13 mm) from patients with familial adenomatous polyposis. A variety of methods was used depending on available material, including flow cytometry, comparative genomic hybridization, and loss of heterozygosity (LOH) analysis. Selected adenomas were assessed for proliferative activity by Ki-67 immunocytochemistry. Seventeen of 20 (85%) tumors were diploid, two were near-diploid, and one was hypotetraploid. Just one (near-diploid) tumor showed increased proliferative activity. LOH was found occasionally on chromosome 15q (2 of 49 tumors), but not on chromosome 18q (0 of 48). In 20 adenomas, LOH at APC was associated with loss at 5q but not 5p markers, with the former encompassing a minimum of 20 Mb. However, three of these lesions analyzed by comparative genomic hybridization displayed normal profiles, suggesting, together with other data, that the mechanism of LOH at APC is probably somatic recombination. Our results therefore do not support the hypothesis that CIN precedes APC mutations in tumorigenesis. Regarding the model in which APC mutations lead directly to CIN, if APC mutations do have this effect in vivo, it must be subtle. Alternatively, CIN associated with APC mutations might be essentially an in vitro phenomenon.
Most colorectal cancers, many sporadic adenomas, and all polyps in familial adenomatous polyposis (FAP) carry mutations in the adenomatous polyposis coli (APC) gene. APC encodes a 2,843-aa protein that is involved in several cellular processes, including the regulation of β-catenin, and that includes domains for binding to microtubules (reviewed in refs. 1 and 2). Disease-associated APC mutations fully or partially inactivate APC function, almost always by producing a truncated or absent protein (3–5). Truncated proteins are usually disrupted within their β-catenin binding/degradation domains and almost invariably lack the microtubule-binding sites, which are located at the C-terminal region of the molecule (6–10).
It is generally believed that APC mutations are selected, at least in part, for their effects on β-catenin levels (11–15): mutant APC cannot degrade β-catenin, leading to constitutive activation of the Wnt signaling pathway and to direct expansion of the mutant clone. The evidence that this effect of APC is critical has been derived from several sources, one being that colorectal tumors without APC mutations sometimes harbor mutations of β-catenin that prevent protein destruction and thus have effects similar to mutation of APC (16–18). It is unlikely, however, that inactivating APC mutations and activating β-catenin mutations are functionally identical, given, for example, that the former seem to be associated with a higher probability of progression from colorectal adenoma to carcinoma (19). Several groups have suggested, therefore, that loss of C-terminal APC functions provides a selective advantage additional to that which arises from constitutive Wnt signaling (20–24).
Fodde et al. (23) and Kaplan et al. (24) independently reported that APC may have a role in chromosomal segregation. Both groups studied mouse embryonic stem (ES) cells homozygous for a truncating Apc mutation (Min ES cells) and detected a marked increase in numerical and structural chromosome aberrations, as well as disorganized spindle microtubules. During mitosis, wild-type APC was found to be localized along kinetochore microtubules and at their ends adjacent to kinetochores. Together, these observations led the authors to suggest a new role for APC in kinetochore microtubule-chromosome attachment and therefore chromosome segregation, with mutations in Apc disrupting this function and resulting in chromosomal (or karyotypic) instability (CIN). In this way, APC mutations may be selected not only directly—through their effects on the Wnt pathway—but also indirectly, through the hypermutation which they engender in the form of CIN.
Shih et al. (25) analyzed 32 sporadic colorectal adenomas for loss of heterozygosity (LOH) by using digital single nucleotide polymorphism PCR and found relatively high frequencies of LOH on chromosomes 5q (55%), 1p (10%), 8p (19%), 15q (28%), and 18q (28%). Although digital SNP-PCR may provide some increase in sensitivity over microsatellite-based methods, owing to its use of confidence interval thresholds for scoring LOH, in reality most tumors with LOH in Shih et al.'s study (25) showed one of the SNP alleles to be at a frequency of 66% or more. This finding is equivalent to an allelic ratio of 2:1, the usual threshold for scoring LOH by using microsatellites. Most cases of LOH detectable by using digital SNP-PCR should, therefore, also be detectable by using microsatellite-based LOH. Although several explanations for their findings were discussed, Shih et al.'s (25) preferred interpretation of their data was that karyotypic instability occurred early in colorectal tumorigenesis, preceding and leading to APC mutations.
Evidently, the findings of Fodde et al. (23) and Kaplan et al. (24) using in vitro methods may not apply in vivo, and Shih et al. (25) made no direct assessment of chromosomal-scale changes in their tumors. We have, therefore, studied a set of 55 colorectal adenomas (average size = 5 mm; range = 1–13 mm) from 18 FAP patients with a variety of germ-line mutations and characterized second hits at APC. Using a variety of analytical methods [flow cytometry (FCM), LOH analysis, and comparative genomic hybridization (CGH)], we searched for evidence of aneuploidy and polyploidy in these tumors. In addition, Ki-67 immunocytochemistry was used to assess the proliferative activity in a subset of these lesions. To elucidate the molecular mechanism underlying allelic loss at APC, the extent of LOH on chromosome 5 was determined in 20 adenomas.
Patients and Methods
Study Population.
This study examined 55 colorectal adenomas and four normal biopsies from 18 patients diagnosed with FAP and with a known germ-line APC mutation. All tumors were tubular adenomas with mild dysplasia (average size = 5 mm; range = 1–13 mm) that had either been fresh-frozen at colectomy (n = 47) or fixed in formalin and embedded in paraffin (n = 8). A minimum of 60% neoplastic material was present in each biopsy as assessed by the analysis of hematoxylin and eosin-stained sections. The second hit at APC had been determined in all lesions by using standard mutation detection techniques [single-strand conformation polymorphism (SSCP) analysis, DNA sequencing] and LOH analysis at microsatellite markers close to the APC locus (D5S346, D5S656, and D5S421). Details of the colorectal adenomas are summarized in Table 1.
Table 1.
Patient ID, APC mutation status, and size of the colorectal adenomas analyzed in this study, as well as analytical methods applied
| Patient ID | Adenoma ID | Germ line APC mutation (nucleotide; codon) | Somatic APC mutation (nucleotide; codon) | Adenoma size, mm | Analytical method |
|---|---|---|---|---|---|
| N-1144 | 249a | 502 A>T; R168X | 4192 del 2bp; 1398 FS | 4 | L15/18 |
| 249b | 502 A>T; R168X | 4316 del 1bp; 1439 FS | 3.5 | L15/18 | |
| 315 | 502 A>T; R168X | 4132 C>T; Q1371X | 5.5 | L15/18, FCM, Ki-67 | |
| N-1154 | 203 | 1495 C>T; R499X | 4466 ins 2bp; 1489 FS | 4.5 | L15/18, FCM, Ki-67 |
| 298 | 1495 C>T; R499X | 4393 del 2bp; 1465 FS | 8 | L15/18, FCM, Ki-67 | |
| 300 | 1495 C>T; R499X | 4012 C>T; Q1338X | 6 | L15/18 | |
| 301 | 1495 C>T; R499X | 4317 del 1bp; 1439 FS | 5 | L15/18, FCM, Ki-67 | |
| 312 | 1495 C>T; R499X | 3927 del 5bp; 1309 FS | 7.5 | L15/18, FCM, Ki-67, CGH | |
| 340 | 1495 C>T; R499X | 3916 G>T, E1306X | 5 | L15/18, FCM, Ki-67 | |
| 352 | 1495 C>T; R499X | 4348 del 4bp; 1450 FS | 5 | L15/18, FCM, Ki-67, CGH | |
| 240 | 1495 C>T; R499X | 3927 del 5bp; 1309 FS | 6.5 | L15/18, FCM, Ki-67 | |
| 243 | 1495 C>T; R499X | 3927 del 5bp; 1309 FS | 5.5 | L15/18 | |
| 259 | 1495 C>T; R499X | 4216 C>T; Q1406X | 6 | L15/18, FCM, Ki-67 | |
| 264 | 1495 C>T; R499X | 3927 del 5bp; 1309 FS | 5 | L15/18 | |
| 350 | 1495 C>T; R499X | 4277 del 1bp; 1426 FS | 6 | L15/18, FCM, Ki-67 | |
| 374 | 1495 C>T; R499X | 4466 del 1bp; 1489 FS | 5 | L15/18, FCM, Ki-67 | |
| 194b | 1495 C>T; R499X | 4316 del 1bp; 1439 FS | 6 | L15/18 | |
| N-117 | 155a | 1842 ins 1 bp; 614 FS | 4484 ins 1bp; 1495 FS | 2.5 | L15/18 |
| 155b | 1842 ins 1 bp; 614 FS | 4306 del 13bp; 1436 FS | 2.5 | L15/18 | |
| 155c | 1842 ins 1 bp; 614 FS | 4446 del 10bp; 1482 FS | 1 | L15/18 | |
| N-1263 | 17 | 3183 del 5 bp; 1061 FS | 4312 del 1bp; 1438 FS | 2 | L15/18 |
| 135b | 3183 del 5 bp; 1061 FS | 3905 del 1bp; 1302 FS | 1.5 | L15/18 | |
| N-1016 | 308 | 3863 del 1bp; 1287 FS | LOH | 5.5 | FCM, Ki-67 |
| N-609 | 1 | 3887 ins 13 bp; 1296 FS | LOH | 3 | L15/18 |
| 2 | 3887 ins 13 bp; 1296 FS | LOH | 3 | L15/18 | |
| 5 | 3887 ins 13 bp; 1296 FS | LOH | 3 | L15/18 | |
| 6 | 3887 ins 13 bp; 1296 FS | LOH | 3 | L15/18 | |
| 8 | 3887 ins 13 bp; 1296 FS | LOH | 3 | L15/18 | |
| 10 | 3887 ins 13 bp; 1296 FS | LOH | 3 | L15/18 | |
| N-1026 | 258 | 3907 C>T; Q1303X | LOH | 4.5 | L15/18, FCM, Ki-67 |
| 335 | 3907 C>T; Q1303X | LOH | 6 | L15/18, FCM, Ki-67 | |
| N-283 | 347 | 3927 del 5bp; 1309 FS | LOH | 4.5 | L15, FCM, Ki-67 |
| N-1066 | 187 | 3927 del 5bp; 1309 FS | LOH | 7 | L15/18 |
| N-1633 | 292 | 3927 del 5bp; 1309 FS | LOH | 6 | L15/18 |
| N-220 | 399 | 3927 del 5bp; 1309 FS | LOH | 7 | FCM, Ki-67 |
| N-127 | 128a | 3927 del 5bp; 1309 FS | LOH | 5 | L15/18 |
| 128b | 3927 del 5bp; 1309 FS | LOH | 5 | L15/18 | |
| 128c | 3927 del 5bp; 1309 FS | LOH | 5 | L15/18, CGH | |
| 129 | 3927 del 5bp; 1309 FS | LOH | 1 | L15/18, CGH | |
| 130 | 3927 del 5bp; 1309 FS | LOH | 5 | L15/18 | |
| 131b | 3927 del 5bp; 1309 FS | LOH | 3 | L15/18 | |
| 131c | 3927 del 5bp; 1309 FS | LOH | 3 | L15/18, CGH | |
| 131d | 3927 del 5bp; 1309 FS | LOH | 3 | L15/18 | |
| N-907 | 3 | 3927 del 5bp; 1309 FS | LOH | 3 | L15/18 |
| 5 | 3927 del 5bp; 1309 FS | LOH | 3 | L15/18 | |
| 8 | 3927 del 5bp; 1309 FS | LOH | 3 | L15/18 | |
| N-205 | 206 | 3927 del 5bp; 1309 FS | LOH | 1 | L15/18 |
| 207 | 3927 del 5bp; 1309 FS | LOH | 1 | L15/18 | |
| 208 | 3927 del 5bp; 1309 FS | LOH | 1 | L15/18 | |
| 209 | 3927 del 5bp; 1309 FS | LOH | 1 | L15/18 | |
| 1974/92/B | 3927 del 5bp; 1309 FS | LOH | 11 | FCM | |
| N-458 | 1 | 3927 del 5bp; 1309 FS | LOH | 12 | FCM |
| 52701 | 2929-M | 3927 del 5bp; 1309 FS | LOH | 6 | FCM |
| N-610 | 5 | 3927 del 5bp; 1309 FS | LOH | 12.5 | FCM |
| N-351 | 2 | 4392 del 2bp; 1464 FS | LOH | 3 | L15/18 |
L15/18, LOH analysis at chromosomes 15q/18q; FCM, flow cytometry; Ki-67, Ki-67 immunocytochemistry; CGH, comparative genomic hybridization. FS denotes frameshift mutations.
FCM and Ki-67 Immunocytochemistry.
Multiparameter FCM was performed on 4 paraffin-embedded and 16 fresh-frozen tumors, as well as 2 respective normal biopsies. All fresh-frozen tissue was simultaneously assayed for expression of the Ki-67 antigen by using FITC-labeled monoclonal mouse antibody (Dako). The appropriate FITC-labeled mouse IgG1 antibody (Dako) was used as isotype control. In brief, a small piece (about 4 mm3) of fresh-frozen tissue was disaggregated into a cellular suspension by using the Dako Medimachine System. Cells were fixed in 70% ethanol for 40 min at 4°C, washed twice in PBS, 0.5% Tween 20 (pH 7.2), and resuspended in 80 μl of PBS, 0.5% Tween 20, 0.5% BSA (pH 7.2). Incubation with 20 μl of antibody was performed for 30 min at 4°C. Cells were washed twice in PBS, 0.5% Tween 20, 0.5% BSA (pH 7.2), treated with 100 μg/ml RNase, and stained with 50 μg/ml propidium iodide (PI). The suspension was filtered through a 70-μm nylon filter and immediately analyzed on a FACSCalibur (Becton Dickinson). Cells were excited by the argon laser emitting at 488 nm. Fluorescence from FITC-labeled antibodies was detected by using a 530/30-nm band pass filter, and PI fluorescence was detected by using a 670-nm long pass filter. Forward and right angle light scatter were used to set a gate including all cells, but excluding debris. A second gate set on area and width of PI fluorescence was used to further define the single cell population. Both FITC and PI fluorescence were collected in linear mode, and acquisition was stopped after 8,000 gated events had been acquired. Data were analyzed for proliferative activity (percentage of Ki-67-positive cells normalized against the isotype control) and aneuploidy [DNA index (DI)] by using dedicated modfit software (Verity Software House, Topsham, ME). The DNA diploid peak was set by using the two normal samples. Paraffin-embedded tissue was prepared for analysis by cutting a 50-μm section from each block, placing it into a histopathology cassette between two sheets of Whatman 3MM filter paper, and dewaxing it in xylene overnight. The section was rehydrated in an ethanol series and rinsed twice in water. The tissue was digested in 0.4% pepsin for 30 min at 37°C, and the digestion was stopped in 0.2% glycine, 2× PBS (pH 7.2). Cells were washed twice in PBS (pH 7.2) before FCM analysis.
LOH (Allelic Loss) Analysis.
LOH analysis was performed at microsatellite markers on chromosome 15q (D15S995 and D15S1007; close to the CRAC1 locus) and chromosome 18q (D18S46 and D18S470; close to the SMAD4/MADH4 locus). In 20 adenomas with LOH as the second hit at APC, the extent of allelic loss on chromosome 5 was determined by using six microsatellite markers (D5S2845, 5p14.3; D5S1470, 5p13.3; D5S82, 5q21.3; D5S489, 5q22.3; D5S2117, 5q31.1; and D5S1456, 5q35.1). Standard methods of fluorescence-based genotyping were used on the ABI377 (Applied Biosystems) semiautomated sequencer. Allelic loss was scored if the area under one allelic peak in the tumor was reduced by 50% or more relative to the other allele, after correcting for the relative peak areas by using normal DNA.
CGH.
CGH was performed on five fresh-frozen tumors containing at least 60% neoplastic material, as described (26). Briefly, 50–100 ng of tumor and reference DNA were amplified by degenerate oligonucleotide-primed PCR (DOP-PCR) and fluorescently labeled by nick translation (27). Labeled tumor and normal DNA were precipitated in the presence of 50 μg Cot1 DNA (Life Technologies, Grand Island, NY) and dissolved in hybridization buffer (50% formamide/10% dextran sulfate/2× SSC). The mixture was denatured at 75°C for 5 min, left to preanneal for 30 min, and applied to denatured metaphase spreads (Vysis, Downers Grove, IL) prepared from normal male peripheral blood lymphocytes. The metaphase spreads were denatured in 70% formamide, 2× SSC at 73°C and dehydrated in an ethanol series. Slides were left to hybridize for 2–3 days at 37°C, then washed in 50% formamide, 2× SSC, followed by a wash in 2× SSC. After air drying, the slides were counterstained with 4,6-diamino-2-phenylindole (DAPI). Images were captured with a charge-coupled device (CCD) camera attached to a Zeiss axioskop microscope and analyzed by using quips (Vysis, Downers Grove, IL) software. Between 5 to 10 metaphases were analyzed for each tumor. Negative control hybridizations were included in each batch of experiments. A chromosomal region was considered to be lost or gained if the mean hybridization ratio between tumor and normal DNA was <0.85:1 or >1.15:1, respectively (26).
Results
A total of 55 colorectal adenomas with two characterized mutational hits at APC was analyzed for CIN. A variety of techniques was used, depending on the amount and type of material available. Where possible, the proliferative activity of these lesions was also assessed.
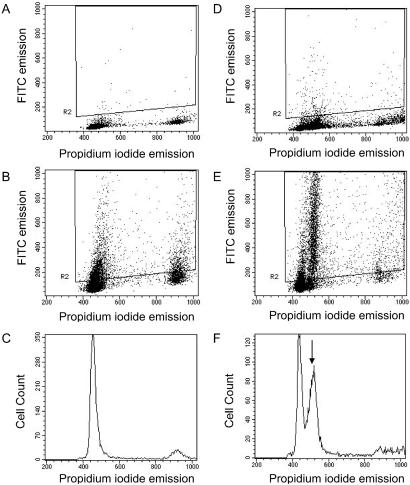
Four paraffin-embedded and 16 fresh-frozen tumors were analyzed for aneuploidy/polyploidy by multiparameter FCM (Table 2). Three of 20 (15%) adenomas contained subpopulations of cells displaying changes in ploidy, with two being near-diploid (DI = 1.2 and DI = 0.8) and one being hypotetraploid (DI = 1.8) (Fig. 1). In addition, Ki-67 immunocytochemistry was performed on all fresh-frozen lesions, as well as two normal biopsies, to assess their proliferative activity (percentage of Ki-67-positive cells). No apparent difference in the proportion of Ki-67-expressing cells was observed between diploid tumors and normal colonic tissue. However, one of the two aneuploid polyps studied displayed an increase in the proportion of Ki-67-expressing cells (53.1%), as compared with the euploid biopsies (average = 16.8%; range = 5.0–35.7%). Results were of similar quality for paraffin-embedded and fresh-frozen material, with the mean coefficient of variation (CV) of the G1/G0 peak being 4.9 ± 1.2 and 4.1 ± 0.5, respectively.
Table 2.
Results of flow cytometry on colorectal adenomas with two mutational hits at APC and two normal controls
| Adenoma ID | DI | CV | Percentage of Ki-67 positive cells |
|---|---|---|---|
| Normal (N-283) | 1.0 | 3.7 | 21.3 |
| Normal (N-220) | 1.0 | 4.39 | 11.6 |
| 315 | 1.0 | 4.07 | 15.2 |
| 203 | 1.0 | 3.81 | 21.6 |
| 298 | 0.8 | 3.54 | 29.2 |
| 301 | 1.0 | 5.32 | 26.1 |
| 312 | 1.0 | 3.68 | 20.8 |
| 340 | 1.0 | 4.26 | 11.0 |
| 352 | 1.0 | 4.29 | 14.5 |
| 240 | 1.0 | 3.8 | 7.0 |
| 259 | 1.0 | 4.14 | 14.2 |
| 350 | 1.0 | 3.45 | 5.0 |
| 374 | 1.0 | 4.56 | 6.3 |
| 308 | 1.0 | 4.29 | 17.2 |
| 258 | 1.0 | 4.25 | 18.3 |
| 335 | 1.2 | 3.19 | 53.1 |
| 347 | 1.0 | 3.57 | 35.7 |
| 399 | 1.0 | 4.42 | 22.2 |
| 1974/92/B | 1.0 | 4.9 | — |
| 1 | 1.0 | 5.35 | — |
| 2929-M | 1.8 | 2.96 | — |
| 5 | 1.0 | 6.52 | — |
DI, DNA index; CV, coefficient of variation of the G1/G0 peak; —, no data.
Fig 1.
Representative results of flow cytometry on one normal biopsy (N-283; A–C) and one near-diploid colorectal adenoma with two mutational hits at APC (335; D–F). (A and D) PI fluorescence against Ki-67-FITC fluorescence of the isotype control sample. The box represents Ki-67-positive events. (B and E) PI fluorescence against Ki-67-FITC fluorescence for the monoclonal antibody-stained samples. (C and F) The PI histograms. The near-diploid subpopulation is indicated by the arrow.
LOH analysis was performed on DNA from 49 adenomas. Importantly, all of these lesions contained <40% contaminating normal tissue, and 27 had shown unequivocal LOH as the second hit at APC. We did detect LOH on other chromosomes in these adenomas, but at a low frequency: although 2 of 49 (4%) informative tumors showed LOH at markers on chromosome 15q, none of 48 (0%) showed LOH at markers on chromosome 18q. Our previously published data had shown evidence of LOH on chromosome 1p in 1 of 21 (5%) of these polyps (28). No evidence of microsatellite instability (MSI+) was found at any marker in any polyp.
The extent of allelic loss on chromosome 5 was determined in 20 adenomas with LOH as the second hit at APC (Table 3). Nineteen (95%) tumors showed LOH at all informative markers spanning chromosome 5q, with the minimal detectable region of allelic loss encompassing ≈20 Mb. The remaining tumor displayed a normal dosage at the telomeric marker D5S2117. In contrast, none of the 20 adenomas showed allelic loss at markers on chromosome 5p.
Table 3.
Results of LOH analysis on colorectal adenomas with allelic loss as the second mutational hit at APC, using microsatellite markers spanning chromosome 5
| Patient ID
|
Adenoma ID
|
D5S2845 5p14.3
|
D5S1470 5p13.3
|
D5S82 5q21.3
|
APC 5q22.2 | D5S489 5q22.3
|
D5S2117 5q31.1
|
D5S1456 5q35.1
|
||
|---|---|---|---|---|---|---|---|---|---|---|
| D5S346 | D5S656 | D5S421 | ||||||||
| N-127 | 128a | NL | NL | NI | NI | LOH | LOH | LOH | LOH | NI |
| 128b | NL | NL | NI | NI | LOH | LOH | LOH | LOH | NI | |
| 128c | NL | NL | NI | NI | LOH | LOH | LOH | LOH | NI | |
| 129 | NL | NL | NI | NI | LOH | LOH | LOH | LOH | NI | |
| 130 | NL | NL | NI | NI | LOH | LOH | LOH | NL | NI | |
| 131b | NL | NL | NI | NI | LOH | LOH | LOH | LOH | NI | |
| 131c | NL | NL | NI | NI | LOH | LOH | LOH | LOH | NI | |
| 131d | NL | NL | NI | NI | LOH | LOH | LOH | LOH | NI | |
| N-907 | 3 | NL | NL | LOH | LOH | LOH | LOH | LOH | LOH | LOH |
| 5 | NL | NL | LOH | LOH | LOH | LOH | LOH | LOH | LOH | |
| 8 | NL | NL | LOH | LOH | LOH | LOH | LOH | LOH | LOH | |
| N-609 | 1 | NL | — | LOH | LOH | LOH | LOH | LOH | LOH | LOH |
| 2 | NL | NL | LOH | LOH | LOH | LOH | LOH | LOH | LOH | |
| 5 | NL | NL | — | LOH | LOH | LOH | LOH | LOH | LOH | |
| 6 | NL | NL | LOH | — | LOH | LOH | LOH | LOH | LOH | |
| 8 | NL | NL | LOH | LOH | LOH | LOH | LOH | LOH | LOH | |
| 10 | NL | NL | LOH | LOH | LOH | LOH | LOH | LOH | LOH | |
| N-205 | 207 | NL | NL | LOH | LOH | LOH | LOH | LOH | LOH | LOH |
| 208 | NL | NL | — | LOH | LOH | LOH | LOH | LOH | LOH | |
| 209 | NL | NL | — | LOH | LOH | LOH | LOH | LOH | LOH | |
NI, noninformative; NL, no loss; —, no data. LOH at APC has been scored based on three markers in close proximity to the locus, D5S346, D5S656, and D5S421.
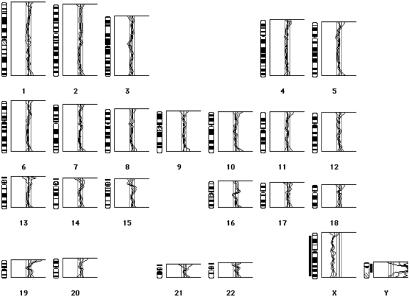
CGH analysis on five fresh-frozen tumors, two of which had been shown to be euploid by FCM, revealed normal CGH profiles (Fig. 2). Interestingly, three of the polyps had shown LOH at APC, with allelic loss involving at least 20 Mb of chromosome 5q (Table 3). One of these polyps had also shown LOH on chromosome 15q. Although the region of loss at 15q may have been below the resolution of CGH, the failure to detect a deletion of chromosome 5q is in accordance with our real-time quantitative multiplex PCR (RQM-PCR) results, showing that LOH at APC does not result from physical loss of material (unpublished data).
Fig 2.
A representative CGH result of one tumor, 131c, which showed loss of heterozygosity as the second mutational hit at APC by microsatellite analysis. Male tumor DNA was cohybridized with female reference DNA onto normal male metaphase spreads. The composite CGH profile shows 95% confidence intervals of the mean values from six metaphase spreads, with threshold values for chromosomal gain and loss of 1.15 and 0.85, respectively. Gains are indicated by bars to the right, losses by bars to the left, of the chromosomal ideograms. Tumor 131c shows no chromosomal imbalances, with the exception of the sex-mismatch control, a relative gain of the Y and loss of the X chromosome.
Discussion
Our results using a variety of experimental methods show that the majority of adenomas with two mutational hits at APC are diploid or near-diploid. Just 3 of 20 (15%) tumors displayed evidence of chromosomal (or karyotypic) instability as determined by FCM and CGH analysis, two being near-diploid and one being hypotetraploid. Three further samples showed normal DNA profiles by CGH analysis, indicating an absence of gross unbalanced karyotypic rearrangements. In our samples, CIN was not associated with the type of second hit (truncating mutation or LOH) at APC. One of the two near-diploid lesions showed an increase in the proportion of Ki-67-expressing cells, as compared with all euploid biopsies. Given that models of colorectal tumorigenesis predict increasingly aggressive features as tumors progress, it is likely that this tumor had become aneuploid as part of its progression rather than as a direct result of inactivation of APC.
Complementing these findings, we have previously identified two mutational hits at APC in near-diploid, microsatellite unstable (MSI+) colorectal cancer cell lines with only a few (<5) chromosomal rearrangements (2, 7, 29). Two of these cell lines (LoVo and VACO5) harbored biallelic truncating APC mutations, and two showed a truncating APC mutation and LOH (DLD1 and GP2d/GP5d).
Apart from at the APC locus, LOH was uncommon in our polyps, being found at a frequency of ≈5% at chromosomes 1p (28) and 15q, but being absent at chromosome 18q. In 20 analyzed adenomas, allelic loss at APC was associated with loss at markers on chromosome 5q but not 5p, with the minimal region of allelic loss encompassing ≈20 Mb. CGH analysis of three of these polyps, however, revealed no unbalanced chromosomal rearrangements. Together with our real-time quantitative multiplex (RQM)-PCR findings on colorectal adenomas showing that LOH at APC does not result from deletion of material (unpublished data), these data suggest that the molecular mechanism of allelic loss at APC is nearly always somatic recombination.
Our data are in agreement with a recent report by Haigis et al. (30), who analyzed 18 colorectal polyps from ApcMin mice and six human adenomas from patients without FAP that had uncertain APC mutation status. Haigis et al. (30) used interphase fluorescence in situ hybridization (FISH) analysis on selected mouse/human chromosomes and found no evidence of chromosomal gains or losses. In tumors from ApcMin mice, this result was confirmed by karyotypic analysis. Furthermore, allelic loss at Apc was shown to commonly occur by somatic recombination in Min adenomas. Other studies have found early colorectal adenomas to be near-diploid in most cases, although larger and/or more dysplastic lesions tend to become aneuploid/polyploid (31–37).
Like Shih et al. (25), we found LOH at sites on chromosomes 1p and 15q in a minority of colorectal adenomas, although LOH occurred at a lower frequency in our sample (10% vs. 5% at 1p; 28% vs. 4% at 15q) and was not found on chromosome 18q (28% vs. 0%). It is unlikely that this difference is due to an increased amount of contaminating normal tissue, because 27 of our 49 tumors had previously been shown to have LOH at the APC locus. The probable explanation is partly chance, but also that the two studies used different methods (with different specificity and sensitivity) and analyzed tumors of different origin (from sporadic cases and FAP patients, respectively). We disagree with the view of Shih et al. (25) that the LOH results indicate that karyotypic instability is common in early colorectal adenomas. First, we found three only cases of aneuploidy/polyploidy in 20 tumors analyzed by FCM and CGH analysis. Second, three polyps with detectable LOH were normal by CGH analysis. Third, we found that LOH at APC did not result from physical loss of material but probably from somatic recombination, inconsistent with Shih et al.'s (25) view that CIN precedes APC inactivation.
Our results do not support the hypothesis that APC mutations are selected for effects on chromosomal mis-segregation manifesting as karyotypic instability in early stages of colorectal tumorigenesis, although we cannot exclude a minor tendency to CIN. It is evident, moreover, that at least some near-diploid colorectal carcinomas have two mutational hits at APC. Thus, whereas APC may well have a role in interacting with, or perhaps controlling, the mitotic spindle, loss of this C-terminal function does not inevitably lead to spindle dysfunction and genomic instability, even in late lesions in which cell cycle checkpoints are likely to be deranged.
In summary, our data and earlier results (6, 7) [together with the findings of Haigis et al. (30)] show that APC mutations are common in colorectal tumors because they provide cells with a direct selective advantage. The nature of that advantage remains largely unknown, but probably primarily involves changes in β-catenin levels. The APC protein may physically associate with components of the mitotic spindle, but its role, if any, in chromosomal segregation is not yet characterized. The model cell systems previously used to study the association of APC with chromosomal mis-segregation are themselves prone to spontaneous changes in chromosome number and structure, even in the presence of wild-type APC (38, 39). We cannot yet be certain whether or not APC mutations increase the tendency for chromosomal mis-segregation to occur in human tumors in vivo, but, if mutant APC does have this effect, its consequences do not generally manifest until later-stage tumorigenesis.
Acknowledgments
We thank all patients for their participation in this study, as well as their respective doctors and pathologists for contributing clinical information. We also thank the staff of the Cancer Research UK Equipment Park for their excellent support. This research was supported by Cancer Research UK, by a grant from the Boehringer Ingelheim Fonds (to O.M.S.), and by the Swiss National Science Foundation (to K.H.).
Abbreviations
APC, adenomatous polyposis coli
CIN, chromosomal instability
FAP, familial adenomatous polyposis
FCM, flow cytometry
CGH, comparative genomic hybridization
LOH, loss of heterozygosity
PI, propidium iodide
DI, DNA index
References
- 1.Sieber O. M., Tomlinson, I. P. & Lamlum, H. (2000) Mol. Med. Today 6, 462-469. [DOI] [PubMed] [Google Scholar]
- 2.Fearnhead N. S., Britton, M. P. & Bodmer, W. F. (2001) Hum. Mol. Genet. 10, 721-733. [DOI] [PubMed] [Google Scholar]
- 3.Miyoshi Y., Nagase, H., Ando, H., Horii, A., Ichii, S., Nakatsuru, S., Aoki, T., Miki, Y., Mori, T. & Nakamura, Y. (1992) Hum. Mol. Genet. 1, 229-233. [DOI] [PubMed] [Google Scholar]
- 4.Ichii S., Takeda, S., Horii, A., Nakatsuru, S., Miyoshi, Y., Emi, M., Fujiwara, Y., Koyama, K., Furuyama, J., Utsunomiya, J., et al. (1993) Oncogene 8, 2399-2405. [PubMed] [Google Scholar]
- 5.Cottrell S., Bicknell, D., Kaklamanis, L. & Bodmer, W. F. (1992) Lancet 340, 626-630. [DOI] [PubMed] [Google Scholar]
- 6.Lamlum H., Ilyas, M., Rowan, A., Clark, S., Johnson, V., Bell, J., Frayling, I., Efstathiou, J., Pack, K., Payne, S., et al. (1999) Nat. Med. 5, 1071-1075. [DOI] [PubMed] [Google Scholar]
- 7.Rowan A. J., Lamlum, H., Ilyas, M., Wheeler, J., Straub, J., Papadopoulou, A., Bicknell, D., Bodmer, W. F. & Tomlinson, I. P. (2000) Proc. Natl. Acad. Sci. USA 97, 3352-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith K. J., Levy, D. B., Maupin, P., Pollard, T. D., Vogelstein, B. & Kinzler, K. W. (1994) Cancer Res. 54, 3672-3675. [PubMed] [Google Scholar]
- 9.Munemitsu S., Souza, B., Muller, O., Albert, I., Rubinfeld, B. & Polakis, P. (1994) Cancer Res. 54, 3676-3681. [PubMed] [Google Scholar]
- 10.Deka J., Kuhlmann, J. & Muller, O. (1998) Eur. J. Biochem. 253, 591-597. [DOI] [PubMed] [Google Scholar]
- 11.Munemitsu S., Albert, I., Souza, B., Rubinfeld, B. & Polakis, P. (1995) Proc. Natl. Acad. Sci. USA 92, 3046-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubinfeld B., Albert, I., Porfiri, E., Fiol, C., Munemitsu, S. & Polakis, P. (1996) Science 272, 1023-1026. [DOI] [PubMed] [Google Scholar]
- 13.Rubinfeld B., Albert, I., Porfiri, E., Munemitsu, S. & Polakis, P. (1997) Cancer Res. 57, 4624-4630. [PubMed] [Google Scholar]
- 14.Korinek V., Barker, N., Morin, P. J., van Wichen, D., de Weger, R., Kinzler, K. W., Vogelstein, B. & Clevers, H. (1997) Science 275, 1784-1787. [DOI] [PubMed] [Google Scholar]
- 15.Kishida S., Yamamoto, H., Ikeda, S., Kishida, M., Sakamoto, I., Koyama, S. & Kikuchi, A. (1998) J. Biol. Chem. 273, 10823-10826. [DOI] [PubMed] [Google Scholar]
- 16.Ilyas M., Tomlinson, I. P., Rowan, A., Pignatelli, M. & Bodmer, W. F. (1997) Proc. Natl. Acad. Sci. USA 94, 10330-10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morin P. J., Sparks, A. B., Korinek, V., Barker, N., Clevers, H., Vogelstein, B. & Kinzler, K. W. (1997) Science 275, 1787-1790. [DOI] [PubMed] [Google Scholar]
- 18.Sparks A. B., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Cancer Res. 58, 1130-1134. [PubMed] [Google Scholar]
- 19.Samowitz W. S., Powers, M. D., Spirio, L. N., Nollet, F., van Roy, F. & Slattery, M. L. (1999) Cancer Res. 59, 1442-1444. [PubMed] [Google Scholar]
- 20.Nathke I. S., Adams, C. L., Polakis, P., Sellin, J. H. & Nelson, W. J. (1996) J. Cell Biol. 134, 165-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishidate T., Matsumine, A., Toyoshima, K. & Akiyama, T. (2000) Oncogene 19, 365-372. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki Y., Senda, T., Ishidate, T., Koyama, R., Morishita, T., Iwayama, Y., Higuchi, O. & Akiyama, T. (2000) Science 289, 1194-1197. [DOI] [PubMed] [Google Scholar]
- 23.Fodde R., Kuipers, J., Rosenberg, C., Smits, R., Kielman, M., Gaspar, C., van Es, J. H., Breukel, C., Wiegant, J., Giles, R. H. & Clevers, H. (2001) Nat. Cell Biol. 3, 433-438. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan K. B., Burds, A. A., Swedlow, J. R., Bekir, S. S., Sorger, P. K. & Nathke, I. S. (2001) Nat. Cell Biol. 3, 429-432. [DOI] [PubMed] [Google Scholar]
- 25.Shih I. M., Zhou, W., Goodman, S. N., Lengauer, C., Kinzler, K. W. & Vogelstein, B. (2001) Cancer Res. 61, 818-822. [PubMed] [Google Scholar]
- 26.Roylance R., Gorman, P., Harris, W., Liebmann, R., Barnes, D., Hanby, A. & Sheer, D. (1999) Cancer Res. 59, 1433-1436. [PubMed] [Google Scholar]
- 27.Telenius H., Carter, N. P., Bebb, C. E., Nordenskjold, M., Ponder, B. A. & Tunnacliffe, A. (1992) Genomics 13, 718-725. [DOI] [PubMed] [Google Scholar]
- 28.Lamlum H., Papadopoulou, A., Ilyas, M., Rowan, A., Gillet, C., Hanby, A., Talbot, I., Bodmer, W. & Tomlinson, I. (2000) Proc. Natl. Acad. Sci. USA 97, 2225-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdel-Rahman W. M., Katsura, K., Rens, W., Gorman, P. A., Sheer, D., Bicknell, D., Bodmer, W. F., Arends, M. J., Wyllie, A. H. & Edwards, P. A. (2001) Proc. Natl. Acad. Sci. USA 98, 2538-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haigis K. M., Caya, J. G., Reichelderfer, M. & Dove, W. F. (2002) Proc. Natl. Acad. Sci. USA 99, 8927-8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss H., Wildner, G. P., Jacobasch, K. H., Heinz, U. & Schaelicke, W. (1985) Oncology 42, 33-41. [DOI] [PubMed] [Google Scholar]
- 32.van den Ingh H. F., Griffioen, G. & Cornelisse, C. J. (1985) Cancer Res. 45, 3392-3397. [PubMed] [Google Scholar]
- 33.Quirke P., Fozard, J. B., Dixon, M. F., Dyson, J. E., Giles, G. R. & Bird, C. C. (1986) Br. J. Cancer 53, 477-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goh H. S. (1987) Ann. Acad. Med. Singapore 16, 535-538. [PubMed] [Google Scholar]
- 35.Hamada S., Namura, K., Itoh, R. & Fujita, S. (1987) Jpn. J. Cancer Res. 78, 826-832. [PubMed] [Google Scholar]
- 36.Giaretti W., Sciallero, S., Bruno, S., Geido, E., Aste, H. & Di Vinci, A. (1988) Cytometry 9, 238-244. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki S., Mizuno, M., Tomoda, J., Ohmori, M. & Tsuji, T. (1995) Gastroenterology 109, 1098-1104. [DOI] [PubMed] [Google Scholar]
- 38.Longo L., Bygrave, A., Grosveld, F. G. & Pandolfi, P. P. (1997) Transgenic Res. 6, 321-328. [DOI] [PubMed] [Google Scholar]
- 39.Liu X., Wu, H., Loring, J., Hormuzdi, S., Disteche, C. M., Bornstein, P. & Jaenisch, R. (1997) Dev. Dyn. 209, 85-91. [DOI] [PubMed] [Google Scholar]