Abstract
Deletion of the yeast homologue of frataxin, YFH1, results in mitochondrial iron accumulation and respiratory deficiency (petite formation). We used a genetic screen to identify mutants that modify iron-associated defects in respiratory activity in Δyfh1 cells. A deletion in the peroxisomal citrate synthase CIT2 in Δyfh1 cells decreased the rate of petite formation. Conversely, overexpression of CIT2 in Δyfh1 cells increased the rate of respiratory loss. Citrate toxicity in Δyfh1 cells was dependent on iron but was independent of mitochondrial respiration. Citrate toxicity was not restricted to iron-laden mitochondria but also occurred when iron accumulated in cytosol because of impaired vacuolar iron storage. These results suggest that high levels of citrate may promote iron-mediated tissue damage.
The yeast YFH1 gene and its mammalian orthologue, frataxin, are nuclear genes that encode mitochondrial proteins (1). In humans, mutations in frataxin are responsible for the neurologic and cardiac disease Friedreich's ataxia (2). The most common mutation of frataxin is an expansion of a GAA triplet within the first intron of the frataxin gene (3). The effect of the triplet expansion is to reduce frataxin transcription and protein concentration. Although the frataxin protein is highly conserved in eukaryotes, there is no consensus regarding the function of Yfh1p/frataxin. Suggested roles for the protein include mitochondrial iron storage (4, 5), iron–sulfur biosynthesis (2, 6), regulation of respiration (7), and control of antioxidant defenses (8). Although the function of Yfh1p/frataxin is unknown, it is thought that the pathophysiology of the human disorder results from a mitochondrial defect (9). In yeast, a deficit of Yfh1p leads to accumulation of mitochondrial iron, which reacts with oxygen metabolites to generate oxygen radicals, resulting in a respiratory deficit (1, 10). We used a genetic screen to identify genes that would preserve the respiratory activity of Δyfh1 cells. We report that deletion of CIT2, a gene that encodes an extramitochondrial citrate synthase, can markedly attenuate the iron toxicity in Δyfh1 cells and preserve respiratory activity. Alternatively, overexpression of CIT2 can induce toxicity when iron accumulates in either the mitochondria or in the cytosol, the latter as a result of impaired iron storage. Although extracellular citrate is an iron chelator, these results suggest that intracellular citrate iron complexes are toxic.
Materials and Methods
Strains, Growth Media, and Plasmids.
DY150 [MATa, ura3-52, leu2-3,112, trp1-1, his3-11, ade2-1, can1-100 (oc)] and DY1457 [MATα, ura3-52, leu2-3,112, trp1-1, his3-11, ade6, can1-100 (oc)] were derived from the W303 strain of Saccharomyces cerevisiae. The METYFH1 strain [Δyfh1, pMET3YFH1[URA3] (a yfh1 strain that has a plasmid containing the YFH1 gene controlled by the MET3 promoter in a vector that has a URA3 gene)] was generated by crossing the Δyfh1 [MATa, ura3-52, leu2-3,112, trp1-1, ade2-1, can1-100 (oc), yfh1:HIS3] strain with DY1457 as described (10). The expression of YFH1 was inhibited by growing cells in complete synthetic media (CM) supplemented with methionine (330 μg/ml).
Gene deletion of CIT2 and RTG2 was created by an insertion of the kanamycin resistance gene (11). Deletion candidates were selected on yeast extract/peptone/dextrose (YPD) supplemented with G418 (200 μg/ml) and verified by Southern blot. CIT2 plasmid was generated in which the CIT2 gene was placed under the control of the ADH1 promoter. CIT2 DNA was synthesized by PCR with primers annealing to the 5′ sequence adjacent to the ATG (076: TCC CCC GGG ATG ACA GTT CCT TAT CTA AAT TCA AAC A) and 3′ sequence adjacent to the stop codon of the ORF (077: CCC AAG CTT CTA TAG TTT GCT TTC AAT GTT TTT GAC CAA). The PCR product was cloned into a 2-μ vector to create a high copy CIT2 plasmid, termed pRS425ADHCIT2, and into a centromeric vector to create a low copy CIT2 plasmid, termed pRS415ADHCIT2.
Cells were routinely grown in CM with 2% glucose. Appropriate media lacking specific nutrients were used to maintain plasmids within cells. To make iron-free CM, an iron chelator, bathophenanthroline sulfonate (BPS), was added at 40 μM; FeSO4 was added at 2.5 or 5 μM to make low-iron media of BPS (2.5) and BPS (5), respectively.
Suppressor Screen.
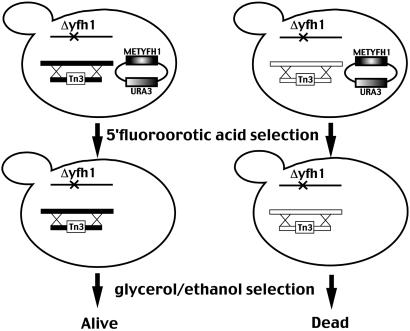
The METYFH1 strain (Δyfh1, pMET3YFH1[URA3]) was mutagenized by transformation with a minitransposon-inserted yeast genomic library (generously provided by Michael Snyder, Yale University, New Haven, CT), and transformants were selected on media lacking leucine (12). Cells were replica-plated to CM lacking leucine but containing 5-fluoroorotic acid to ensure loss of the pMET3YFH1 plasmid. Cells then were replica-plated to glycerol–ethanol medium containing 500 μM FeSO4 (13). Colonies able to grow then were streaked to BPS media, which contains low iron. Only cells able to grow on BPS media were selected for further study. The inserted transposon was rescued by using the procedure of Burns et al. (12). The rescued plasmid was sequenced to determine the location of transposon insertion into the yeast genome.
Citrate Analysis.
The citrate content of cells was determined as described (14). Briefly, yeast was grown in CM and harvested at mid-log phase (5–8 × 106 cells per ml). Citrate was extracted in perchloric acid, neutralized, and analyzed with a citric acid analysis kit (Roche Molecular Biochemicals). Between 0.5 and 1 ml of citrate extract was incubated with reaction buffer containing NADH, lactate dehydrogenase (LDH), and malate dehydrogenase (MDH). Absorbance at 340 nm was measured to determine the background level of NADH clearance. Citrate lysate then was added to the reaction to determine citrate-dependent clearance of NADH.
Iron-Dependent Petite Formation.
Cultures of METYFH1 cells were grown in CM in the presence or absence of methionine (330 μg/ml) for 15–18 h. The media also were supplemented with iron (FeSO4) at the specified amounts. At the end of the incubation, cells were plated on CM plates lacking methionine. Cells were allowed to grow for 2 days before being replica-plated to yeast extract/peptone/glycerol/ethanol (YPGE) (2% glycerol and 2% ethanol) to determine the number of petite colonies. All experiments were repeated at least three times. Although there is variation in the absolute degree of petite formation, the relative degree of petite formation in cells with either CIT2 deletions or CIT2-containing plasmids compared with control cells or Δyfh1 cells was consistent.
Quantification of mRNA Analysis by S1 Nuclease Assay.
Yeast cells were harvested at mid-log phase, and total RNA was extracted by vigorous vortex mixing with glass beads in phenol/chloroform/isoamyl alcohol (25:24:1) solvent. DNA oligonucleotides, with complementary sequence to the transcripts of CIT2 and CMD1 (calmodulin, an internal control), were end labeled with 32P by using T4 polynucleotide kinase. The CIT2 oligo (088) sequence is: GCC AAC CCG TTC AAA CCT GAT GCA AGG GAC AGA TAA GGTGAT GAT AGT GCT GAC CCC CAC CTC. The CMD1 oligo (CMD1OLIGO) sequence is: GGG CAA AGG CTT CTT TGA ATT CAG CAA TTT GTT CGG TGG AGC C. The 32P-labeled oligos then were hybridized with 25 μg of total RNA in Hepes buffer (38 mM Hepes, pH 7.6/0.3 M NaCl/1 mM EDTA/0.1% Triton X-100) at 55°C for at least 12 h. The reaction mixtures were digested with S1 nuclease, and the undigested double-strand oligos with specific mRNA were ethanol-precipitated and denatured by heat in formamide buffer. The oligo DNA then was separated on an 8 M urea/polyacrylamide gel in modified TBE buffer (134 mM Tris/44 mM boric acid/3.5 mM EDTA, pH 8.0), followed by autoradiography.
Iron Uptake Assay and 59Fe Pulse–Chase and Subcellular Fractionations.
METYFH1 cells were grown in either the presence or absence of methionine for 15 to 18 h. The cells were harvested and then washed three times with cold assay buffer [low-iron medium (LIM)-EDTA] (15). Cells (5 × 106) were incubated with 59Fe (0.5 μM in the presence of 1 mM ascorbate in LIM-EDTA buffer) at 30°C for 10 min, and iron transport activity was assayed as described (15).
Accumulation of 59Fe in mitochondria was assayed as described (13). Briefly, cells (1 × 109) were incubated with 59Fe (0.5 μM in the presence of 1 mM ascorbate in 100 ml of LIM-EDTA) at 30°C for 10 min. The cells were washed three times with cold EDTA-containing buffer and then incubated for 2 h in fresh media in the absence of radioactive iron. At the end of the incubation, cells were harvested, spheroplasts were prepared, and membranes were recovered by centrifugation of a postnuclear supernatant at 15,000 × g for 30 min. The membranes were layered onto a 0–25% iodixanol gradient, which was centrifuged at 12,000 × g for 2 h. The gradient then was fractionated into 16 fractions. The amount of 59Fe in each fraction was determined. The radioactivity in each fraction was expressed as a percentage of the total radioactivity in the postnuclear supernatant. As shown previously (10), mitochondria are found in fractions 3–9 of an iodixanol gradient and are well separated from other organelles.
Results
Identification of CIT2 as a Modifier of Respiratory Function in Δyfh1 Cells.
A strain of yeast, METYFH1, which has a deletion in the YFH1 gene transformed with a plasmid containing a MET3 promoter-regulated YFH1 gene, retains normal mitochondrial function when grown in the absence of methionine (10, 13). This strain was “mutagenized” by transformation with a transposon-inserted genomic library (12). Cells that could grow on respiratory substrates in the absence of the pMET3YFH1 plasmid were selected (Fig. 1). Because deletions in genes required for high-affinity iron transport also preserve respiratory activity in a Δyfh1 strain, only cells capable of growth on low-iron media were selected for further study. Thirty thousand colonies were screened, and 17 colonies were able to grow on respiratory substrate. Of the 17 candidates, 3 were able to grow on low-iron plates. One colony, termed sup6, was studied in detail. Southern blot analysis revealed that only one copy of the transposon had integrated into the genome. The integrated genomic construct was rescued and sequenced, revealing that the transposon had integrated into the RTG2 gene and was expected to lead to a gene disruption.
Fig 1.
Scheme of Δyfh1 suppressor screen. See text for details. Tn3, minitransposon.
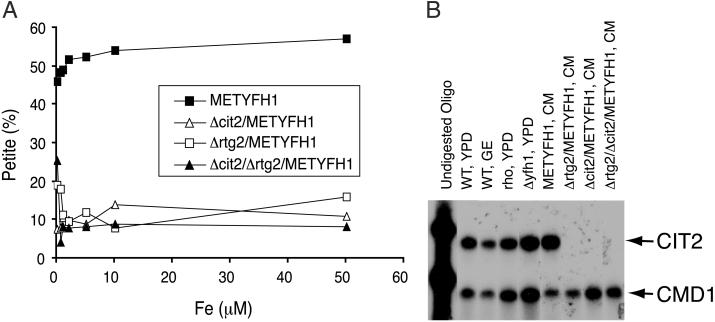
To confirm that disruption of RTG2 was responsible for the ability of Δyfh1 cells to grow on glycerol–ethanol, we generated a targeted deletion of RTG2 in the METYFH1 strain. When Yfh1p is expressed (absence of methionine), both METYFH1 and Δrtg2/METYFH1 strains were respiratory-competent even in the presence of high levels of iron. When Yfh1p is absent (presence of methionine), there is a marked increase in the number of petite cells in METYFH1 cells (10, 13). Deletion of RTG2 in the absence of Yfh1p expression resulted in preservation of respiratory activity, as shown by the reduced number of petite cells (Fig. 2A). With iron concentrations below 50–100 μM, Δrtg2/METYFH1 cells retained respiratory activity. With iron concentrations above 100 μM, both METYFH1 and the Δrtg2/METYFH1 cells had decreased viability in both glucose and glycerol–ethanol media, which precludes a quantitative assay of petite formation (data not shown).
Fig 2.
Disruption of RTG2 or CIT2 suppresses loss of respiratory activity in Δyfh1 cells. (A) Cells (METYFH1, Δrtg2/METYFH1, Δcit2/METYFH1, and Δcit2/Δrtg2/METYFH1) were grown to mid-log phase and then were transferred to media supplemented with methionine and iron for 18 h. The cells then were plated on glucose-containing plates lacking methionine. The number of cells that lost respiratory activity was determined by replica plating to YPGE. (B) CIT2 mRNA level was determined by S1 nuclease assay. Total RNA was extracted from a wild-type strain, DY150, grown in glucose (YPD)- or glycerol–ethanol (GE)-based media; a rho0 isogenic strain derived from the wild type, grown in YPD; Δyfh1 grown in YPD; and four other strains: METYFH1, Δrtg2/METYFH1, Δcit2/METYFH1, and Δrtg2/Δcit2/METYFH1 grown in CM lacking both methionine and uracil. S1 nuclease analysis was performed with oligomers specific to CIT2 and to CMD1 (calmodulin) as described in Materials and Methods.
The product of the RTG2 gene is a component of a signaling pathway that regulates the transcription of a specific set of nuclear genes in response to mitochondrial dysfunction (16). This interorganelle signaling pathway is called retrograde regulation. It is thought that the RTG-dependent pathway encodes proteins that provide essential metabolites, which compensate for those lost when mitochondrial function is compromised. Cit2p was identified as a peroxisomal citrate synthase, although the amino-terminal sequence of the protein also may permit a mitochondrial localization (17). Transcription of CIT2 is regulated by the RTG-dependent pathway (16). Deletion of CIT1, which encodes the mitochondrial citrate synthase, results in reduced respiratory activity, whereas deletion of CIT2 has no effect on respiratory activity. Because citrate is a known iron chelator, we determined whether the ability of the RTG2 deletion to preserve the respiratory activity of Δyfh1 occurred through its effect on CIT2 transcription. In cells not expressing Yfh1p, deletion of CIT2 reduced iron-dependent petite formation to the same extent as deletion of RTG2. Deletion of both CIT2 and RTG2 in a METYFH1 strain did not lead to increased protection (Fig. 2A). In some yeast strains the RTG-dependent pathway is induced by conditions that lead to a respiratory deficit, and there is little transcription of CIT2 in cells with respiratory-competent mitochondria (18). In yeast strains derived from W303 (including the strain used in these studies), the RTG2-dependent signaling pathway seems to be constitutively active, and CIT2 transcription is unaffected by respiratory activity or carbon source (19) (Fig. 2B). Deletion of RTG2, however, results in the absence of CIT2 transcription, indicating that in this yeast strain, CIT2 is regulated by RTG2, independent of mitochondrial respiration.
Citrate Levels Modify Iron Toxicity.
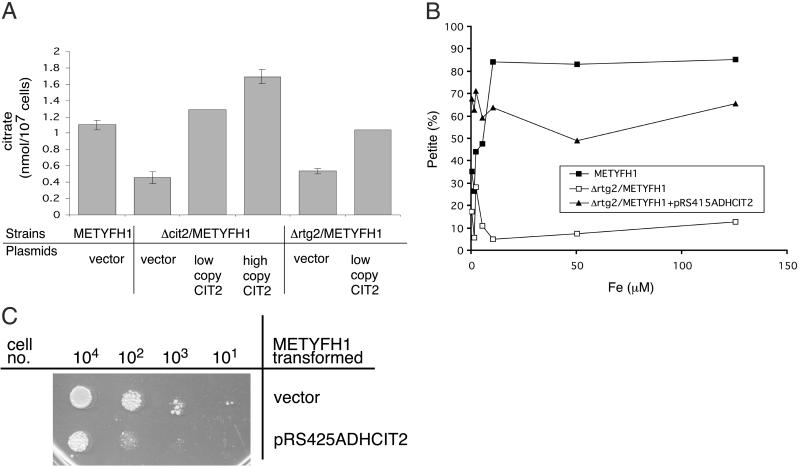
Measurement of citrate levels showed that deletion of CIT2 resulted in a 50% decrease in citrate concentration in cells expressing Yfh1p (Fig. 3A). Transformation of Δcit2/METYFH1 cells with a low copy plasmid containing an ADH1 promoter-regulated CIT2 restored citrate to near normal levels. In the absence of Yfh1p, these transformed cells showed a rate of iron-dependent petite formation identical to that of methionine-grown METYFH1 cells (data not shown). Transformation of Δrtg2/METYFH1 cells with a low copy CIT2 plasmid also resulted in a near normal level of citrate (Fig. 3A) and a restoration in iron-dependent petite formation (Fig. 3B). This result confirms that the preservation of respiratory activity caused by a deletion in the RTG pathway is due to the loss of CIT2 expression. Overexpression of CIT2 (high copy vector) elevated citrate concentration by 30–50% above wild-type levels. No phenotype was observed in wild-type cells (or METYFH1 cells grown in the absence of methionine) overexpressing CIT2. In the absence of Yfh1p, however, cells that overexpress CIT2 were unable to grow on glycerol–ethanol media (data not shown) and showed a severe growth defect even on glucose media (Fig. 3C).
Fig 3.
Expression of CIT2 restores iron-dependent toxicity. (A) Citrate levels were determined in cells. METYFH1, Δcit2/METYFH1, and Δrtg2/METYFH1 were transformed with various plasmids: control vector (pRS425ADH), high copy CIT2 plasmid (pRS425ADHCIT2), and low copy CIT2 plasmid (pRS415ADHCIT2). Total cellular citrate was acid-extracted from overnight cultures under the condition that permits Yfh1p expression. The amount of extracted citrate was determined by using a citric acid kit (Roche Molecular Biochemicals). (B) The rate of iron-dependent petite formation was measured in METYFH1, Δrtg2/METYFH1, and Δrtg2/METYFH1 transformed with a low copy CIT2 plasmid as described in the legend of Fig. 2. (C) Serial dilution of cells (METYFH1 and METYFH1 transformed with a high copy CIT2 plasmid, pRS425ADHCIT2) was spotted on CM plates supplemented with methionine (330 μg/ml) and FeSO4 (5 μM).
Citrate-Mediated Toxicity Is Iron-Dependent.
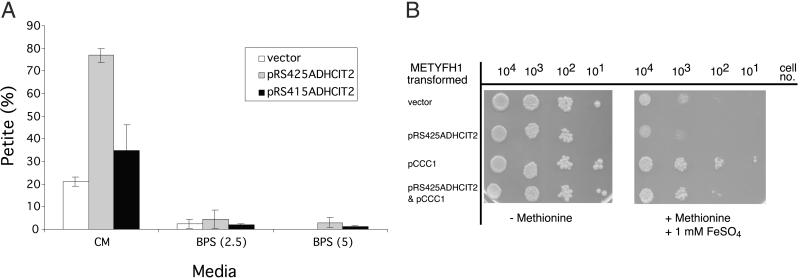
Two different experiments showed that the toxic effect of citrate was iron-dependent. First, as shown (10, 20), reduction in media iron prevents petite formation in Δyfh1 cells. Expression of either low copy or high copy CIT2 plasmids in METYFH1 cells increases petite formation in the absence of Yfh1p. Reduction of available media iron through the addition of the iron chelator BPS preserves respiratory activity, as shown by a reduction in petite formation (Fig. 4A). Second, overexpression of the vacuolar iron transporter CCC1 increases vacuolar iron storage, which lowers cytosolic iron. Decreased cytosolic iron prevents excessive mitochondrial iron accumulation in cells that do not express Yfh1p (13, 21). Overexpression of CCC1 protected pCIT2-transformed METYFH1 cells from the toxic effect of increased citrate concentration (Fig. 4B). Together, these results demonstrate that citrate toxicity is iron-dependent.
Fig 4.
Iron deprivation preserves respiratory activity in METYFH1 transformed with a high copy CIT2-containing vector. (A) METYFH1 transformed with either a high copy CIT2 plasmid (pRS425ADH1CIT2) or low copy CIT2 plasmid (pRS415ADH1CIT2) was grown in CM supplemented with methionine at 330 μg/ml (CM). An iron chelator, BPS (40 μM), and FeSO4 (2.5 or 5 μM) were added to the medium to make low-iron media of BPS (2.5) and BPS (5), respectively. Cells were harvested, diluted, and plated onto CM plates lacking methionine. Colonies then were replica-plated to YPGE to determine the number of petite cells. (B) METYFH1 cells, transformed with control vectors, high copy CIT2 plasmid (pRS425ADHCIT2), or a high copy CCC1 plasmid (pCCC1), were grown on CM plates as indicated.
The Protective Effect of a CIT2 Deletion Can Be Overcome by High Iron.
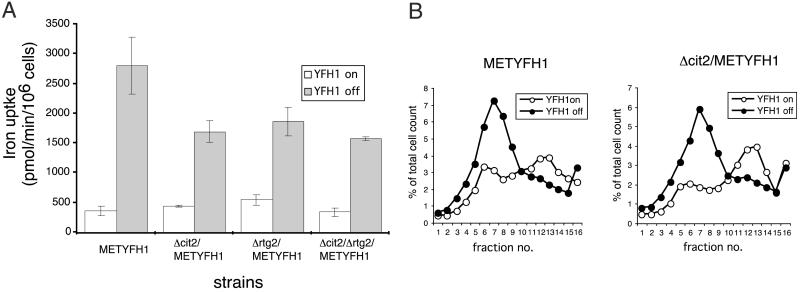
The loss of respiratory competence in Δyfh1 cells results from mitochondrial iron accumulation (10, 20). We tested the hypothesis that deletion of CIT2 preserves respiratory activity by lowering mitochondrial iron accumulation. We tested this hypothesis by measuring cellular iron transport, because mitochondrial iron accumulation lowers cytosolic iron, resulting in activation of the surface transport system (1, 10). Iron transport activity is low in CM-grown cells that express Yfh1p, whereas iron transport activity is increased dramatically in the absence of Yfh1p (Fig. 5A). If Yfh1p is expressed, deletion of CIT2 has little affect on iron transport activity, whereas in the absence of Yfh1p, Δcit2 cells show a reduced rate of iron transport. Deletion of RTG2 has the same effect on iron transport as deletion of CIT2. This result is consistent with preservation of mitochondrial respiratory activity caused by reduced levels of mitochondrial iron. High levels of media iron result in a high degree of petite formation in cells not expressing Yfh1p, even in the face of a deletion in CIT2. This high degree of petite formation correlates with mitochondrial iron accumulation, as shown by measuring either surface iron transport activity (data not shown) or, more directly, mitochondrial iron (Fig. 5B). In this experiment, cells were exposed to a high concentration of 59Fe in the presence of ascorbate for 10 min. Addition of ascorbate bypasses the need for a cell surface reductase and permits cells to accumulate a bolus of iron. After incubation with 59Fe, cells were washed and incubated in 59Fe-free medium for 2 h. The cells were homogenized, mitochondria were isolated with an iodixanol gradient, and the amount of 59Fe was determined. Under these conditions, the amount of mitochondrial iron accumulated was the same in Δcit2/METYFH1 and METYFH1 cells. Thus, when cells are exposed to high levels of iron, the protective effect of a CIT2 deletion is overcome.
Fig 5.
Effect of Δcit2 deletion on mitochondrial iron accumulation. (A) METYFH1, Δcit2/METYFH1, and Δrtg2/METYFH1 were grown in CM with and without methionine (YFH1 “off” and YFH1 “on,” respectively) for 18 h. Cells were washed, and iron transport activity was assayed by using 59Fe ascorbate. (B) METYFH1 and Δcit2/METYFH1 cells were grown in CM with or without methionine (YFH1 “off” and YFH1 “on,” respectively) for 18 h. The cells were incubated with 59Fe ascorbate for 10 min, washed, and then incubated for 2 h in 59Fe-free media. Cells were homogenized, and membranes were applied to an iodixanol gradient as described in Materials and Methods. The gradient was fractionated, and radioactivity in each fraction was determined relative to the total radioactivity.
The Toxic Effect of Iron Citrate Is Not Restricted to Mitochondria.
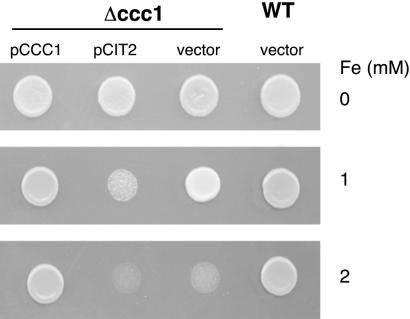
Citrate-mediated toxicity also is seen when iron inappropriately accumulates in the cytosol. Cells with a deletion in CCC1 are unable to transport iron into vacuoles, have higher cytosolic iron levels than wild-type cells, and are more sensitive to media iron (21). Overexpression of CIT2 in Δccc1 cells exacerbates iron toxicity (Fig. 6). Thus, citrate iron toxicity is not restricted to mitochondrial iron.
Fig 6.
Citrate-mediated iron toxicity is not restricted to mitochondrial iron. Wild-type cells and Δccc1 transformed with a control vector, a high copy CIT2 plasmid (pRS425ADHCIT2), or a high copy CCC1 plasmid (pCCC1) were spotted on iron-containing media. Cells were incubated at 30°C for 2 days before being photographed.
Citrate is an essential metabolite used by the tricarboxylic acid cycle. We considered that the increased citrate concentration might lead to an increase in respiration rate, resulting in the generation of H2O2, which might react with the mitochondrial iron resulting in increased amounts of oxidant damage. Two experiments, however, exclude this possibility. First, transformation of wild-type cells, grown in either glucose or glycerol–ethanol, with either a high or low copy CIT2 plasmid, did not affect the rate of oxygen consumption (data not shown). Second, the loss of cell viability attributable to overexpression of CIT2 is seen in cells lacking a mitochondrial genome (rho0), which are unable to respire. These results suggest that respiratory activity is not required for the toxic effect of citrate.
Discussion
Citrate has iron-chelating activity and can function physiologically as an iron carrier (22). Plants transport iron in xylem as iron citrate. When mammalian transferrin is highly saturated, plasma iron also can be found bound to citrate (23). Most eukaryotes can reduce extracellular iron citrate complexes, permitting the uptake of Fe2+ by specific transport systems. Although citrate is a physiologically relevant extracellular iron carrier, the role for citrate in intracellular iron metabolism remains unclear. A role for citrate in intracellular iron metabolism has been proposed because iron regulatory protein 1 (IRP1) in the presence of high iron is converted into a cytosolic aconitase (24, 25). Under conditions in which iron storage is promoted through increased ferritin synthesis, an active aconitase might lower cytosolic citrate levels. A reduction in citrate would lead to less iron-chelation capacity and might promote iron storage in ferritin (26). Despite these considerations, no phenotype is seen in the IRP1 knockout mouse (27).
Our previous studies have shown that iron causes mitochondrial damage in cells that do not express Yfh1p (10). When Δyfh1 cells are incubated in low to moderate amounts of iron (<100 μM), deletion of CIT2 leads to reduced toxicity, whereas overexpression of CIT2 leads to increased toxicity. Citrate-mediated iron toxicity is not restricted to Δyfh1 cells but also is seen in other conditions that result in deregulation of mitochondrial iron homeostasis. ISA1 is a nuclear gene that encodes a mitochondrial protein required for iron–sulfur cluster formation (28, 29). In the absence of ISA1, cells are respiratory-incompetent and accumulate mitochondrial iron. Overexpression of CIT2 in these cells leads to a loss of viability (data not shown). This result shows that the toxic effect of iron citrate is not specific to Yfh1p but is seen under other conditions associated with mitochondrial iron accumulation.
It is curious that overexpression of CIT2 has only a modest effect on elevating citrate levels yet leads to a severe effect on cell growth. Iron limitation, either by media deprivation or increased iron storage (overexpression of CCC1), eliminates citrate-dependent toxicity. We observed that mitochondrial iron accumulation correlated with the respiratory deficit. Deletion of CIT2 lowers mitochondrial iron accumulation and petite formation, whereas exposure of cells to high iron overcomes the protective effect of the CIT2 deletion, resulting in high levels of mitochondrial iron and a high degree of petite formation. We cannot determine whether citrate acts as an iron carrier increasing mitochondrial iron accumulation or whether citrate reacts with iron already present in mitochondria, increasing oxidant damage, which promotes further mitochondrial iron accumulation. Defects in iron–sulfur biosynthesis are known to lead to mitochondrial iron accumulation (for review see ref. 30). Loss of two proteins involved in the iron–sulfur pathway, Yfh1p and Nfs1p, leads to mitochondrial iron accumulation due to inhibition of mitochondrial iron export (31). One of the enzymes required for iron–sulfur biosynthesis, Yah1p, is an iron–sulfur-containing enzyme (32), and iron–sulfur proteins are highly susceptible to oxidant damage. Decreases in Yah1p lead to mitochondrial iron accumulation (33, 34). Thus, citrate iron could oxidize Yah1p, leading to a damaged protein unable to function in iron–sulfur cluster synthesis. The resulting decrease in iron–sulfur cluster synthesis then might lead to increased mitochondrial iron, promoting further oxidant damage.
We currently favor the hypothesis that citrate iron complexes lead to oxidant damage. Citrate-dependent iron toxicity is not restricted to mitochondrial iron accumulation. We observed that iron-dependent toxicity is increased by citrate when inappropriate iron accumulation occurs either in mitochondria or cytosol. In a variety of in vitro systems, citrate Fe3+ complexes lead to increased oxidant damage (35, 36). Most investigators have used these in vitro systems to model iron-dependent Fenton chemistry. Chelation of iron by citrate was found to promote the autooxidation of Fe2+ (Fe2+ + citrate → Fe3+ citrate + e−). Autooxidation of Fe2+ results in the peroxidation of lipids in test solutions and in isolated mitochondria. There is some debate as to which oxidant radical is involved, superoxide anion (36) and/or hydroxyl radical (37) production. Citrate iron toxicity is favored by intracellular conditions that provide a reducing environment, which, by providing a pool of Fe2+, would increase citrate-dependent iron oxidation.
The results of our studies on yeast may reflect mammalian iron pathophysiology. Our data offer a partial explanation for the tissue specificity seen in Friedreich's ataxia. Although all tissues show a deficit of frataxin, cardiac tissue particularly is affected. Nearly all patients with Friedreich's ataxia suffer from cardiac toxicity, and cardiac toxicity also is seen in the mouse model of the disease (2). Cardiac tissue has the highest reliance on respiration as an energy source and shows the highest concentration of citrate (38). Based on our results, we predict that increased mitochondrial iron due to frataxin deficiency would preferentially injure cardiac tissue. Increased iron-dependent toxicity due to citrate may explain the observation that biochemical parameters of damage in mouse cardiomyocytes occur before histochemically measurable iron accumulation (2). Much of the tissue damage in ischemia–reperfusion injury also is due to increased cytosolic iron, because iron chelation reduces tissue pathology (39–41), but during ischemia reperfusion there is also a 4- to 6-fold increase in citrate (42). Our results suggest that the combination of iron and citrate promotes tissue injury.
Acknowledgments
This work was supported by National Institutes of Health Grant NIDDK-52380. DNA sequencing and primer production were supported in part by National Institutes of Health Cancer Center Support Grant NCI-CCSGP30 CA 42014.
Abbreviations
CM, complete synthetic media
YPD, yeast extract/peptone/dextrose
BPS, bathophenanthroline sulfonate
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Babcock M., de Silva, D., Oaks, R., Davis-Kaplan, S., Jiralerspong, S., Montermini, L., Pandolfo, M. & Kaplan, J. (1997) Science 276, 1709-1712. [DOI] [PubMed] [Google Scholar]
- 2.Puccio H., Simon, D., Cossee, M., Criqui-Filipe, P., Tiziano, F., Melki, J., Hindelang, C., Matyas, R., Rustin, P. & Koenig, M. (2001) Nat. Genet. 27, 181-186. [DOI] [PubMed] [Google Scholar]
- 3.Pandolfo M. (1998) Neuromuscul. Disord. 8, 409-415. [DOI] [PubMed] [Google Scholar]
- 4.Adamec J., Rusnak, F., Owen, W. G., Naylor, S., Benson, L. M., Gacy, A. M. & Isaya, G. (2000) Am. J. Hum. Genet. 67, 549-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavadini P., O'Neill, H. A., Benada, O. & Isaya, G. (2002) Hum. Mol. Genet. 11, 217-227. [DOI] [PubMed] [Google Scholar]
- 6.Rotig A., de Lonlay, P., Chretien, D., Foury, F., Koenig, M., Sidi, D., Munnich, A. & Rustin, P. (1997) Nat. Genet. 17, 215-217. [DOI] [PubMed] [Google Scholar]
- 7.Ristow M., Pfister, M. F., Yee, A. J., Schubert, M., Michael, L., Zhang, C. Y., Ueki, K., Michael, M. D., II, Lowell, B. B. & Kahn, C. R. (2000) Proc. Natl. Acad. Sci. USA 97, 12239-12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chantrel-Groussard K., Geromel, V., Puccio, H., Koenig, M., Munnich, A., Rotig, A. & Rustin, P. (2001) Hum. Mol. Genet. 10, 2061-2067. [DOI] [PubMed] [Google Scholar]
- 9.Lodi R., Cooper, J. M., Bradley, J. L., Manners, D., Styles, P., Taylor, D. J. & Schapira, A. H. (1999) Proc. Natl. Acad. Sci. USA 96, 11492-11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radisky D. C., Babcock, M. C. & Kaplan, J. (1999) J. Biol. Chem. 274, 4497-4499. [DOI] [PubMed] [Google Scholar]
- 11.Longtine M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. & Pringle, J. R. (1998) Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- 12.Burns N., Grimwade, B., Ross-Macdonald, P. B., Choi, E. Y., Finberg, K., Roeder, G. S. & Snyder, M. (1994) Genes Dev. 8, 1087-1105. [DOI] [PubMed] [Google Scholar]
- 13.Chen O. S. & Kaplan, J. (2000) J. Biol. Chem. 275, 7626-7632. [DOI] [PubMed] [Google Scholar]
- 14.Bergmeyer H. U., (1965) Methods of Enzymatic Analysis (Academic, Orlando, FL), pp. 313–317.
- 15.Davis-Kaplan S. R., Askwith, C. C., Bengtzen, A. C., Radisky, D. & Kaplan, J. (1998) Proc. Natl. Acad. Sci. USA 95, 13641-13645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chelstowska A. & Butow, R. A. (1995) J. Biol. Chem. 270, 18141-18146. [DOI] [PubMed] [Google Scholar]
- 17.Lee J. G., Cho, S. P., Lee, H. S., Lee, C. H., Bae, K. S. & Maeng, P. J. (2000) J. Biochem. (Tokyo) 128, 1059-1072. [DOI] [PubMed] [Google Scholar]
- 18.Epstein C. B., Waddle, J. A., Hale, W., IV, Davé, V., Thornton, J., Macatee, T. L., Garner, H. R. & Butow, R. A. (2001) Mol. Biol. Cell 12, 297-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foury F. & Talibi, D. (2001) J. Biol. Chem. 276, 7762-7768. [DOI] [PubMed] [Google Scholar]
- 20.Foury F. (1999) FEBS Lett. 456, 281-284. [DOI] [PubMed] [Google Scholar]
- 21.Li L., Chen, O. S., Ward, D. M. & Kaplan, J. (2001) J. Biol. Chem. 276, 29515-29519. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt W. (1999) New Phytol. 141, 1-26. [Google Scholar]
- 23.Grootveld M., Bell, J. D., Halliwell, B., Aruoma, O. I., Bomford, A. & Sadler, P. J. (1989) J. Biol. Chem. 264, 4417-4422. [PubMed] [Google Scholar]
- 24.Eisenstein R. S. & Blemings, K. P. (1998) J. Nutr. 128, 2295-2298. [DOI] [PubMed] [Google Scholar]
- 25.Rouault T. & Klausner, R. (1997) Curr. Top. Cell. Regul. 35, 1-19. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan J. & O'Halloran, T. V. (1996) Science 271, 1510-1512. [DOI] [PubMed] [Google Scholar]
- 27.LaVaute T., Smith, S., Cooperman, S., Iwai, K., Land, W., Meyron-Holtz, E., Drake, S. K., Miller, G., Abu-Asab, M., Tsokos, M., et al. (2001) Nat. Genet. 27, 209-214. [DOI] [PubMed] [Google Scholar]
- 28.Jensen L. T. & Culotta, V. C. (2000) Mol. Cell. Biol. 20, 3918-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaut A., Lange, H., Diekert, K., Kispal, G. & Lill, R. (2000) J. Biol. Chem. 275, 15955-15961. [DOI] [PubMed] [Google Scholar]
- 30.Lill R., Diekert, K., Kaut, A., Lange, H., Pelzer, W., Prohl, C. & Kispal, G. (1999) Biol. Chem. 380, 1157-1166. [DOI] [PubMed] [Google Scholar]
- 31.Chen O. S., Hemenway, S. & Kaplan, J. (2002) Proc. Natl. Acad. Sci. USA 99, 12321-12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutz T., Westermann, B., Neupert, W. & Herrmann, J. M. (2001) J. Mol. Biol. 307, 815-825. [DOI] [PubMed] [Google Scholar]
- 33.Barros M. H. & Nobrega, F. G. (1999) Gene 233, 197-203. [DOI] [PubMed] [Google Scholar]
- 34.Lange H., Kaut, A., Kispal, G. & Lill, R. (2000) Proc. Natl. Acad. Sci. USA 97, 1050-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prabhu H. R. & Krishnamurthy, S. (1993) Indian J. Biochem. Biophys. 30, 289-292. [PubMed] [Google Scholar]
- 36.Minotti G. & Aust, S. D. (1987) Free Radical Biol. Med. 3, 379-387. [DOI] [PubMed] [Google Scholar]
- 37.Gutteridge J. M. (1991) Free Radical Biol. Med. 11, 401-406. [DOI] [PubMed] [Google Scholar]
- 38.Williams D. H. & Bosnan, J. T. (1974) in Methods of Enzymatic Analysis, ed. Bergmeyer, H. U. (Academic, New York), Vol. 4, pp. 2266–2302. [Google Scholar]
- 39.Bel A., Martinod, E. & Menasche, P. (1996) Acta Haematol. 95, 63-65. [DOI] [PubMed] [Google Scholar]
- 40.Hershko C. (1994) Baillieres Clin. Haematol. 7, 965-1000. [DOI] [PubMed] [Google Scholar]
- 41.Hedlund B. E. & Hallaway, P. E. (1991) Klin. Wochenschr. 69, 1113-1117. [DOI] [PubMed] [Google Scholar]
- 42.Hassel B., Ilebekk, A. & Tonnessen, T. (1998) Acta Physiol. Scand. 164, 53-59. [DOI] [PubMed] [Google Scholar]