Abstract
We performed a systematic screen of the set of ≈5,000 viable Saccharomyces cerevisiae haploid gene deletion mutants and have identified 103 genes whose deletion causes sensitivity to the DNA-damaging agent methyl methanesulfonate (MMS). In total, 40 previously uncharacterized alkylation damage response genes were identified. Comparison with the set of genes known to be transcriptionally induced in response to MMS revealed surprisingly little overlap with those required for MMS resistance, indicating that transcriptional regulation plays little, if any, role in the response to MMS damage. Clustering of the MMS response genes on the basis of their cross-sensitivities to hydroxyurea, UV radiation, and ionizing radiation revealed a DNA damage core of genes required for responses to a broad range of DNA-damaging agents. Of particular significance, we identified a subset of genes that show a specific MMS response, displaying defects in S phase progression only in the presence of MMS. These genes may promote replication fork stability or processivity during encounters between replication forks and DNA damage.
The budding yeast Saccharomyces cerevisiae has been an invaluable tool for studying DNA damage-response pathways. Many S. cerevisiae DNA damage-response genes have human homologues, and mutations in a number of these genes have been implicated in human diseases. Although several screens for S. cerevisiae DNA damage-response genes have been conducted over the past 30–40 years, additional genes are still being identified. The set of viable S. cerevisiae deletion mutants (1) has allowed for genome-wide studies to identify genes required for resistance to various cellular insults (2–6). Here we report a systematic analysis of the complete set of ≈5,000 viable gene deletion mutants to identify genes that are required for resistance to the DNA-damaging agent methyl methanesulfonate (MMS).
MMS is a monofunctional DNA alkylating agent and a known carcinogen (7, 8) and primarily methylates DNA on N7-deoxyguanine and N3-deoxyadenine (9). Although the N7-methylguanine adduct may be nontoxic and nonmutagenic, N3-methyladenine is a lethal lesion that inhibits DNA synthesis and needs to be actively repaired (8, 10). The three pathways responsible for the removal of most N3-methyladenine lesions are bypass repair (or postreplication repair), recombination repair, and base excision repair (11). All three pathways are required for wild-type resistance to MMS-induced DNA damage (11). In addition, checkpoint proteins are required to maintain cell viability in the presence of MMS (12, 13).
Several studies have found that cells are most sensitive to MMS during progression through S phase (13–15). Exposure to MMS causes a checkpoint-independent reduction in the rate of replication fork progression, likely due to a physical impediment of fork progression caused by alkylated DNA or some intermediate in lesion processing (13). rad53 and mec1 checkpoint mutants have high rates of replication fork termination, suggesting that damage-induced fork catastrophe is the cause of MMS sensitivity in checkpoint mutants (13). Thus, in addition to identifying proteins involved in repair of MMS lesions and in regulating cell cycle progression, a screen for MMS-sensitive mutants may reveal novel proteins required for DNA replication fork stability and processivity after alkylation damage.
Materials and Methods
Yeast Strains and Media.
Standard yeast media and growth conditions were used (16). Nonessential haploid deletion strains were made by the Saccharomyces Gene Deletion Project (1) and can be obtained from Research Genetics (Huntsville, AL) or EUROSCARF (Frankfurt, Germany). MMS (Aldrich) plates contained 0.035% (vol/vol) MMS in yeast extract/peptone/dextrose medium (YPD) and were used within 24 h of preparation. Hydroxyurea (HU) plates contained 200 mM HU in YPD. The genotype of the rad53 (mec2) mutant used in this study was MATα mec2–1-URA3 can1Δ:MFA1pr-HIS3 his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0. To construct this strain mec2–1 (12) was amplified by PCR and cointegrated with URA3 into Y3068 (17). The wild-type control strain used in MMS and HU sensitivity assays was BY4741 (18).
High-Throughput MMS Screen.
An ordered array of 4,644 MATa viable haploid yeast gene deletion mutants, in duplicate, at a density of 768 colonies per plate, was replica pinned onto YPD and YPD + 0.035% MMS. The screen was performed three times by using an automated system, as described (17). Plates were incubated at 30°C for 2 days before scoring.
Confirmation of Drug Sensitivity.
Putative MMS-sensitive strains were grown in YPD overnight at 30°C. Strains were diluted to a concentration of 1 × 107 cells/ml, and four additional 10-fold serial dilutions were made. Eight microliters of each serial dilution was spotted onto the indicated media and incubated at 30°C for 3 days.
α Factor Block and Release Experiment.
Yeast strains were grown at 30°C in YPD to an optical density at 600 nm of 0.3–0.4. Three hundred-microliter samples were removed from each culture for analysis by flow cytometry. The remainder of each culture was arrested in G1 phase by the addition of α mating factor to a final concentration of 5 μM. After 2.5 h of incubation, a sample was removed for analysis by flow cytometry (t = 0). The cultures were then split in half, harvested, washed once with YPD, and released from the cell cycle arrest by resuspension in YPD or YPD + 0.035% MMS. Samples were removed for analysis by flow cytometry at the indicated times after release. α factor block and release experiments were performed at least twice.
Flow Cytometry.
Cells were harvested and fixed in 70% ethanol. Samples were then resuspended in 0.5 ml of 0.1 mg/ml RNase A in 50 mM sodium citrate. After overnight incubation at 37°C, 0.5 ml of 2 μM SYTOX green (Molecular Probes) in 50 mM sodium citrate was added. The samples were sonicated briefly and analyzed using a Becton Dickinson FACSCalibur.
Results and Discussion
Screening for MMS-Sensitive Deletion Mutants.
We performed a high-throughput MMS sensitivity screen by robotically pinning an ordered array of ≈4,700 haploid yeast deletion mutants onto YPD or YPD plus 0.035% MMS (Fig. 1). From the three screens, 244 mutants were scored as MMS sensitive at least once, and 92 were scored at least twice. Strains scored as sensitive at least two of three times were verified by spotting serial dilutions of the cells onto media containing 0.035% MMS (Fig. 2). An additional 38 mutants (19 that were scored as sensitive in one of three screens, and 19 that were not scored in any of the screens) were also chosen for verification, because mutations in the deleted genes were previously reported to be MMS sensitive. Thus, a total of 130 deletion mutants were tested by spotting serial dilutions, of which 103 were confirmed to be MMS sensitive (Table 1). In total, 40 genes with no previously known role in MMS response were identified (Table 1, indicated in bold), including 15 uncharacterized genes. We also identified 48 genes that were not identified in genome-wide screens for sensitivity to UV or ionizing radiation (IR) (3, 4), illustrating the utility of performing screens with different DNA-damaging agents. Fifty-five of the MMS-resistance genes have readily identifiable human homologues (Table 1, indicated in italics), including four previously uncharacterized genes. Twelve of the MMS-resistance genes have an established link to a human disease (19).
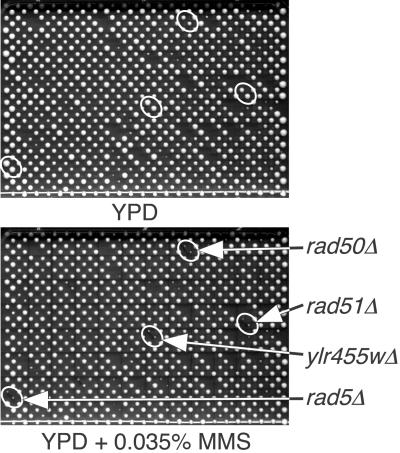
Fig 1.
High-throughput MMS screen. The complete set of haploid yeast deletion mutants was arrayed in duplicate onto 16 plates and pinned onto YPD media or YPD + 0.035% MMS (array plate 11 of 16 is shown). Putative MMS-sensitive mutants lead to the formation of smaller colonies when grown on MMS-containing media.
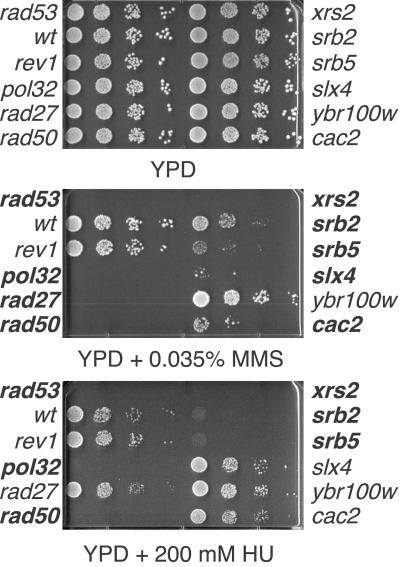
Fig 2.
Confirmation of MMS sensitivity. Putative MMS-sensitive strains were grown in YPD overnight at 30°C. Serial 10-fold dilutions were spotted onto YPD, YPD + 0.035% MMS, or YPD + 200 mM HU and incubated at 30°C for 3 days. Strains in bold were scored as sensitive. A rad53 mutant was used as a positive control.
Table 1.
MMS-sensitive deletion strains
| Gene | Hits | MMSS | Cellular role | Gene | Hits | MMSS | Cellular role |
|---|---|---|---|---|---|---|---|
| AAT2 | 3 | +++ | Amino acid metabolism | RAD1 | 0 | + | DNA repair |
| ANC1 | 0 | +++ | Pol II transcription | RAD5/REV2 | 2 | +++ | DNA repair |
| AOR1 | 1 | +++ | Unknown | RAD6 | 0 | +++ | DNA repair |
| APN1 | 0 | +++ | DNA repair | RAD9 | 3 | ++ | Cell cycle control |
| ARO1 | 2 | +++ | Amino acid metabolism | RAD17 | 1 | +++ | Cell cycle control |
| ARO7 | 2 | +++ | Amino acid metabolism | RAD18 | 0 | + | DNA repair |
| ASF1 | 3 | +++ | DNA synthesis | RAD24 | 2 | +++ | Cell cycle control |
| BDF1 | 1 | +++ | Meiosis | RAD27 | 3 | +++ | DNA synthesis |
| BUD25 | 3 | +++ | Cell polarity | RAD50 | 3 | +++ | DNA repair |
| BUR2 | 0 | +++ | Pol II transcription | RAD51 | 3 | +++ | DNA repair |
| CAC2 | 1 | +++ | DNA repair | RAD52 | 3 | +++ | DNA repair |
| CDC40 | 0 | +++ | Meiosis | RAD54 | 3 | +++ | DNA repair |
| CDC50 | 2 | +++ | Cell cycle control | RAD55 | 3 | +++ | DNA repair |
| CHL1 | 0 | ++ | Mitosis | RAD57 | 3 | +++ | DNA repair |
| CIK1 | 2 | +++ | Meiosis | RAD59 | 1 | +++ | DNA repair |
| CSE2 | 3 | + | Mitosis | REM50 | 3 | +++ | DNA repair |
| CTF18 | 0 | +++ | Cell cycle control | REV3 | 0 | +++ | DNA repair |
| CTF4/POB1 | 2 | +++ | DNA synthesis | RPB9 | 2 | ++ | Pol II transcription |
| CTF8 | 1 | +++ | Chromatin/chromosome structure | RRN10 | 3 | + | Pol I transcription |
| DCC1 | 1 | +++ | Chromatin/chromosome structure | RTT101 | 2 | +++ | Protein modification |
| DDC1 | 1 | +++ | Cell cycle control | SAE2 | 2 | +++ | Meiosis |
| DEG1 | 2 | ++ | Protein synthesis | SEC66 | 2 | +++ | Cell cycle control |
| DOA1 | 2 | +++ | Protein degradation | SGS1 | 3 | +++ | DNA repair |
| DUN1 | 1 | +++ | DNA repair | SIT4 | 2 | + | Cell cycle control |
| ERG3 | 3 | ++ | Lipid, fatty acid, sterol metabolism | SLX4 | 2 | +++ | DNA repair |
| ESC4 | 3 | +++ | Chromatin/chromosome structure | SOD1 | 3 | +++ | Amino acid metabolism |
| GRR1 | 3 | +++ | Amino acid metabolism | SPT4 | 1 | + | Recombination |
| HOF1 | 3 | ++ | Cytokinesis | SRB2 | 2 | + | Pol II transcription |
| HPR1 | 0 | +++ | Recombination | SRB5 | 2 | ++ | Pol II transcription |
| HPR5/SRS2 | 2 | +++ | DNA repair | SW16 | 2 | +++ | Cell cycle control |
| HTL1 | 0 | +++ | Unknown | TOM37 | 2 | ++ | Protein translocation |
| ISC1 | 3 | + | Lipid, fatty acid, sterol metabolism | TOP3 | 3 | +++ | DNA repair |
| KIM3/MMS1 | 2 | +++ | Cell stress | UBC13 | 3 | +++ | DNA repair |
| KRE22 | 1 | +++ | Unknown | UBP6 | 2 | +++ | Protein modification |
| LSM1 | 2 | +++ | RNA turnover | UME6 | 3 | + | Meiosis |
| LSM6 | 2 | ++ | RNA splicing | VID21 | 1 | +++ | Unknown |
| LYS7 | 2 | +++ | Amino acid metabolism | VID31 | 1 | +++ | Unknown |
| MAG1 | 2 | +++ | DNA repair | VMA21 | 2 | +++ | Small molecule transport |
| MEC3 | 1 | +++ | Cell cycle control | VPS36 | 2 | ++ | Amino acid metabolism |
| MED1 | 2 | ++ | Unknown | XRS2 | 3 | +++ | DNA repair |
| MET18/MMS19 | 0 | +++ | Amino acid metabolism | YBL006C | 1 | ++ | Unknown |
| MMS2 | 3 | +++ | DNA repair | YBR099C | 2 | +++ | Unknown |
| MMS22 | 3 | +++ | DNA repair | YCK3 | 2 | +++ | Unknown |
| MMS4/SLX2 | 2 | +++ | DNA repair | YEL045C | 2 | +++ | Unknown |
| MRE11 | 3 | +++ | DNA repair | YJL161W | 1 | + | Unknown |
| MUS81 | 3 | +++ | DNA repair | YLR218C | 3 | + | Unknown |
| NAT3 | 3 | +++ | Protein modification | YLR235C | 3 | +++ | Unknown |
| NCE4 | 2 | +++ | Cell wall maintenance | YLR376C | 2 | +++ | Unknown |
| NPL6 | 2 | + | Nuclear–cytoplasmic transport | YMC2 | 2 | +++ | Small molecule transport |
| NUP133 | 0 | + | Nuclear–cytoplasmic transport | YMR031W-A | 2 | +++ | Unknown |
| NUP84 | 1 | +++ | Nuclear–cytoplasmic transport | YOR275C | 3 | +++ | Cell stress |
| POL32 | 2 | +++ | DNA synthesis |
Previously uncharacterized MMS resistance genes are indicated in bold.
Genes with human homologues.
Genes involved in human disease.
Cellular role as indicated in YPD (www.proteome.com/).
We estimate that 63 MMS-resistance genes have been reported in the literature (4, 6, 20–50), not including genes whose deletion mutants were reported as weakly sensitive to MMS (4), or genes for which the haploid deletion mutant was not sensitive in our study. We successfully identified 78% of the known MMS-resistance genes in at least one of three screens. If we focus on those genes that were scored as sensitive in at least two of three screens, we identified 72 mutants, 56% of known MMS-resistance genes, and had a false-positive rate of 21.7%. We estimate that ≈26 MMS-resistance genes were not identified in our screen [(72/0.56) − 103 = 26]. Some of these 26 mutants may already be included in the mutants that were scored as sensitive in only one of three screens.
In the course of our studies, we noted that 10 deletion mutants that were reported to confer at least moderate MMS sensitivity in homozygous diploids (4) were not sensitive when tested as haploids. To ensure that this was not due to a general difference in sensitivity between haploids and diploids, we tested the MMS sensitivity of wild-type diploids (see Fig. 4A, which is published as supporting information on the PNAS web site, www.pnas.org). There was no difference in MMS sensitivity between the wild-type haploid and the corresponding diploid. We then tested the relevant homozygous diploid deletion mutants for MMS sensitivity (Fig. 4B). The cnm67Δ/cnm67Δ, dhh1Δ/dhh1Δ, yif2Δ/yif2Δ strains were found to be weakly sensitive to 0.035% MMS. The bem1Δ/bem1Δ, nup12Δ/nup12Δ, hfi1/Δhfi1Δ, gos1Δ/gos1Δ, rvs161Δ/rvs161Δ, rvs167Δ/rvs167Δ diploids were not sensitive to 0.035% MMS in our hands. We were unable to isolate a clc1Δ/clc1Δ strain. Thus only three deletions that confer MMS sensitivity in a diploid did not confer sensitivity in a haploid.
Cross-Sensitivity of MMS-Sensitive Mutants to UV, IR, and HU.
We also tested the MMS-sensitive mutants for sensitivity to HU by spotting serial dilutions of cultures onto YPD plates containing 200 mM HU (Fig. 2). Two additional genome-wide screens have recently been reported, one for deletion mutants that confer sensitivity to UV radiation (3) and one for deletion mutants that confer sensitivity to IR (4). We combined our data for MMS and HU sensitivity with the reported data for UV and IR sensitivity to cluster the MMS-sensitive mutants on the basis of their cross-sensitivity to UV, IR, and HU (Table 2). Because both these genome-wide studies missed known UV- or IR-sensitive mutants, we supplemented the data with literature reports (4, 5, 49, 51–62). We noted several interesting properties of these mutants. First, although several lines of evidence indicate that both HU and MMS exert their effects largely during S phase and cause stalling of replication forks, 13 of the 103 MMS-sensitive mutants displayed no significant sensitivity to HU. This indicates that MMS and HU cause a different spectrum of DNA damage and suggests that resistance to alkylation damage requires some activities that are distinct from those involved in HU resistance. Despite this clear difference, we also noted that a large cluster (37 genes of 103) contained genes required for both MMS and HU resistance but not for UV or IR resistance, indicating that MMS and HU are more alike in their action than they are to that of UV or IR. This cluster also contains most of the genes with unknown function that were identified in the MMS screen. These genes may have S phase-specific roles in DNA-damage response. Finally, we found just four genes whose deletion conferred MMS sensitivity but not sensitivity to HU, UV, or IR. These genes may function specifically to preserve viability after alkylation damage, and all have known roles in DNA metabolism. MAG1 encodes a glycosylase that initiates base excision repair of N3-methyladenine (26, 63, 64). The CAC2 gene product is a member of the chromatin assembly factor I (CAF-I) (65, 66). REV3 encodes the catalytic subunit of DNA polymerase ζ and is required for DNA damage-induced mutagenesis (67, 68). SLX4 was identified in a screen for mutants that are synthetically lethal with deletion of the DNA helicase gene SGS1 (23).
Table 2.
Cross-sensitivity of MMS-sensitive deletion mutants to HU, IR, and UV
| HU | IR | UV | n | Gene/ORF |
|---|---|---|---|---|
| S | S | S | 41 | ASF1, BUR2, CDC40, CTF4, CTF8, DCC1, DUN1, GRR1, HOF1, HPR1, HTL1, KRE22, MEC3, MMS2, MMS4, MMS22, MRE11, MUS81, NPL6, NUP84, POL32, RAD5, RAD6, RAD17, RAD18, RAD24, RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, REM50, RPB9, SGS1, SRS2, VID31, XRS2, YBR099C, YLR235C |
| R | 7 | ANC1, ERG3, NAT3, SOD1, TOM37, UME6, YBL006C | ||
| R | S | 5 | DDC1, LSM1, MET18, MMS1, TOP3 | |
| R | 37 | AAT2, AOR1, ARO1, ARO7, BDF1, BUD25, CDC50, CIK1, CSE2, CTF18, DEG1, DOA1, ESC4, ISC1, LSM6, LYS7, MED1, NCE4, RRN10, RTT101, SEC66, SIT4, SPT4, SRB2, SRB5, SWI6, UBP6, VMA21, VPS36, YCK3, YMC2, YEL045C, YJL161W, YLR218C, YLR376C, YMR031W-A, YOR275C | ||
| R | S | S | 8 | NUP133, CHL1, RAD1, RAD9, RAD27, SAE1, UBC13, VID21 |
| R | 1 | APN1 | ||
| R | S | 0 | ||
| R | 4 | CAC2, MAG1, REV3, SLX4 |
As expected, we identified a large number of RAD genes in the MMS screen, and all of these are sensitive to at least one other DNA-damaging agent. Clustering of the cross-sensitivity data revealed a core subset of DNA damage-response genes that are required for resistance to HU, MMS, UV, and IR (Table 2, bold). This core includes genes involved in recombination repair (MRE11, RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, and XRS2) (69); bypass repair (RAD5, RAD6, RAD18, and MMS2) (70); DNA damage checkpoint activation (RAD17, MEC3) (71); and processing of repair or recombination intermediates (MMS4, MUS81, SGS1, and SRS2) (72–76). Other genes present in this core have less defined roles in the repair of DNA damage. ASF1, which is regulated by the checkpoint kinase Rad53, may function in chromatin assembly during DNA repair (77). CTF4, CTF8, and DCC1 encode proteins that are required for sister chromatid cohesion (78, 79), defects in which might affect recombination repair (80). Deletion of YBR099c and YLR235c removes the 3′ ends of the MMS4 and TOP3 ORFs, and so the sensitivity of these mutants likely reflects loss of Mms4 or Top3 function.
Comparison of the MMS Transcriptional Profile with the Set of Genes Required for MMS Resistance.
Exposure to DNA-damaging agents often results in differential gene expression (81–84). Two groups have independently examined the genomic expression response to MMS by DNA microarray analysis (85, 86). To determine the significance of the transcriptional response to MMS, we compared the MMS transcriptional profiles with the set of genes required for MMS resistance (see Table 3, which is published as supporting information on the PNAS web site). We found very little correlation between genes that are transcriptionally induced by MMS and genes that are required for MMS resistance. Of the 616 genes whose transcription was induced >1.5-fold by 0.02% MMS (86), there are only eight whose deletion confers MMS sensitivity. Furthermore, none of these genes were included in the 50 most highly induced genes. Similar results were seen when our MMS-sensitive data set was compared with those genes induced by 0.1% MMS (85), with only 7 of the 320 genes transcriptionally induced >4-fold conferring MMS sensitivity when deleted. Additionally, few MMS-induced genes are essential (1, 85, 86), so the poor overlap between the data sets is not due to the absence of essential genes on the deletion strain array. We conclude that the transcriptional response plays little role in resistance to MMS-induced cellular damage. Many of the genes induced by MMS may not be specifically required for a cellular response to MMS but are coincidentally up-regulated because the cells have been stressed (86). The important responses to MMS-induced cellular damage are likely effected immediately and require proteins that are already present in the cell. Consistent with our results, recent data have indicated that transcriptional response plays little, if any, role in sensitivity to IR, UV, cisplatin, and hydrogen peroxide exposure (5). Thus it appears that the cellular response to a wide range of DNA-damaging agents is largely independent of transcriptional regulation.
Cell Cycle Progression Analysis of Selected MMS-Sensitive Mutants.
Several studies have suggested that passage through S phase in the presence of MMS damage kills MMS-sensitive mutants (13–15). Hence, we examined the S phase progression of a subset of the MMS-sensitive mutants (Fig. 3). We selected representative genes from pathways known to be involved in repair of MMS damage, as well as several genes with poorly defined roles in MMS resistance. Cells were arrested in G1 by α mating factor treatment and then released synchronously into the cell cycle in either the presence or absence of MMS. Wild-type cells complete DNA replication within 60 min after release from the block in the absence of MMS. Consistent with previous reports, wild-type cells released in the presence of MMS show a slower progression through S phase (22, 87) due to a slowing of replication fork progression and a checkpoint-dependent inhibition of late replication origin firing (13), completing S phase by 120 min (Fig. 3 Upper).
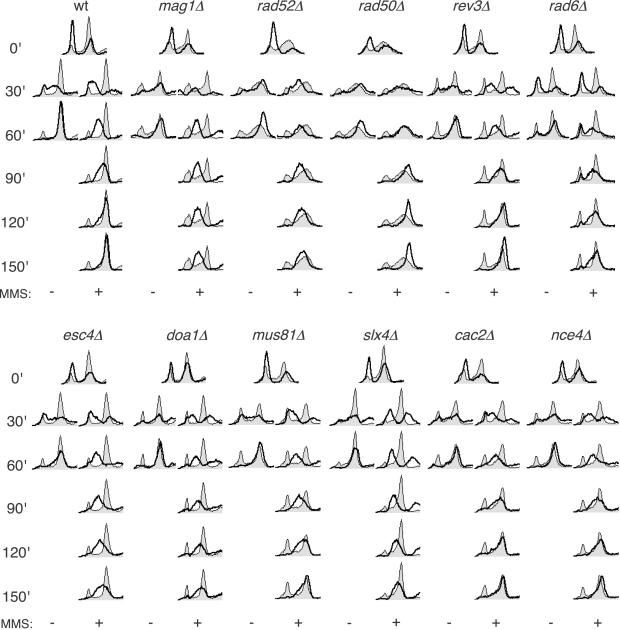
Fig 3.
S phase progression analysis of selected MMS mutants. Cells were arrested in G1 and released in either the presence or absence of 0.035% MMS. Shaded histograms represent the cell cycle distribution of asynchronous culture before cell cycle synchronization. Overlaid histograms represent the cell cycle distribution after release from G1 arrest ± 0.035% MMS for the indicated times.
The mag1Δ mutant (defective in base excision repair) progresses normally in the absence of MMS but arrests with an intermediate DNA content in the presence of MMS. This arrest was maintained for at least 20 h (data not shown), indicating that removal of N3-methyladenine is essential for the completion of S phase, consistent with polymerases being unable to synthesize through or bypass this lesion (10). The rad6Δ and rad52Δ mutant strains (defective in bypass repair and homologous recombination repair, respectively) displayed significantly slower progression through S phase in the presence of MMS compared with the wild-type strain, suggesting that both bypass repair and recombination repair are required for replication through MMS-induced lesions. The replication defects in rad6Δ and rad52Δ were not as severe as in mag1Δ, perhaps indicating that Rad6 and Rad52 play a secondary role in processing MMS-induced lesions, downstream of Mag1. The rad50Δ strain (defective in nonhomologous end-joining and homologous recombination repair) progressed through S phase more rapidly than the wild-type strain, consistent with a role for RAD50 in activating the intra-S phase checkpoint response to MMS. A similar result was obtained with mre11Δ (data not shown). RAD50 and MRE11 are required for Rad53 activation in response to double-strand DNA breaks (88, 89), and our data suggest a similar requirement after MMS damage. We found no significant role for the bypass polymerase gene REV3 in S phase progression in the presence of MMS despite a role for RAD6. The RAD6 epistasis group consists of two major pathways, one involved in postreplication repair and one in mutagenesis (90, 91). Rev3 is involved in mutagenesis, whereas Rad6 plays a critical role in both pathways (90, 91), which may explain the differences seen between rad6Δ and rev3Δ in the S phase progression assay.
We next tested several mutants identified in the MMS screen with less defined roles in MMS response (Fig. 3 Lower). Deletions in CAC2 or NCE4 do not affect the rate of bulk DNA synthesis, suggesting that their roles in resistance to MMS-induced damage may be postreplicative. By contrast, both doa1Δ and esc4Δ are significantly defective in S phase progression in the presence of MMS, displaying cells with incompletely replicated DNA 120 and 150 min after release from the G1 block. DOA1 and ESC4 might be required for replication fork stability or processivity when forks are stalled by DNA damage. Consistent with this idea, DOA1 and ESC4 are both members of the cluster of genes required for resistance to MMS and HU, drugs specifically affecting S phase progression, but not UV or IR (Table 2). mus81Δ and slx4Δ are also defective in S phase progression, but to a lesser extent. MUS81 encodes a subunit of an endonuclease that is thought to act on stalled replication forks (72–74). Interestingly, the sensitivity of mec1 and rad53 mutants to MMS is a result of replication forks terminating irreversibly at a high rate (13). Furthermore, Mus81 physically interacts with Rad53 (92, 93). MUS81 may be acting in the same pathway as the checkpoint genes MEC1 and RAD53 in stabilizing or restarting stalled replication forks. Slx4, like Mus81, is required for viability in the absence of the DNA helicase Sgs1 (23). However, mus81Δ mutants are sensitive to HU, UV, and IR, whereas slx4Δ is not, suggesting that Slx4 may be required to process DNA structures that arise specifically in the presence of alkylated DNA. Thus, by using a high-throughput genome-wide screen for previously uncharacterized MMS-resistance genes, we have identified a class of genes that are required for S phase progression in the presence of DNA damage.
Supplementary Material
Acknowledgments
We thank George Brush and Susan Forsburg for comments on the manuscript; Amy Fung for performing flow cytometry; Hong Xu for help with the screens; and Tim Hughes, Mike Tyers, and Brenda Andrews for strains. This work was supported by grants from the Canadian Institutes of Health Research (to C.B.) and the National Cancer Institute of Canada (to G.W.B. and C.B.). G.W.B. is a Research Scientist of the National Cancer Institute of Canada.
Abbreviations
MMS, methyl methanesulfonate
IR, ionizing radiation
HU, hydroxyurea
YPD, yeast extract/peptone/dextrose
References
- 1.Winzeler E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 2.Chan T. F., Carvalho, J., Riles, L. & Zheng, X. F. (2000) Proc. Natl. Acad. Sci. USA 97, 13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birrell G. W., Giaever, G., Chu, A. M., Davis, R. W. & Brown, J. M. (2001) Proc. Natl. Acad. Sci. USA 98, 12608-12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett C. B., Lewis, L. K., Karthikeyan, G., Lobachev, K. S., Jin, Y. H., Sterling, J. F., Snipe, J. R. & Resnick, M. A. (2001) Nat. Genet. 29, 426-434. [DOI] [PubMed] [Google Scholar]
- 5.Birrell G. W., Brown, J. A., Wu, H. I., Giaever, G., Chu, A. M., Davis, R. W. & Brown, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 8778-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanway D., Chin, J. K., Xia, G., Oshiro, G., Winzeler, E. A. & Romesberg, F. E. (2002) Proc. Natl. Acad. Sci. USA 99, 10605-10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawley P. D. (1989) Mutat. Res. 213, 3-25. [DOI] [PubMed] [Google Scholar]
- 8.Beranek D. T. (1990) Mutat. Res. 231, 11-30. [DOI] [PubMed] [Google Scholar]
- 9.Pegg A. E. (1984) Cancer Invest. 2, 223-231. [DOI] [PubMed] [Google Scholar]
- 10.Boiteux S., Huisman, O. & Laval, J. (1984) EMBO J. 3, 2569-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao W., Chow, B. L. & Rathgeber, L. (1996) Curr. Genet. 30, 461-468. [DOI] [PubMed] [Google Scholar]
- 12.Weinert T. A., Kiser, G. L. & Hartwell, L. H. (1994) Genes Dev. 8, 652-665. [DOI] [PubMed] [Google Scholar]
- 13.Tercero J. A. & Diffley, J. F. (2001) Nature 412, 553-557. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz J. L. (1989) Mutat. Res. 216, 111-118. [DOI] [PubMed] [Google Scholar]
- 15.Fung A. D., Ou, J., Bueler, S. & Brown, G. W. (2002) Mol. Cell. Biol. 22, 4477-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherman F. (1991) Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- 17.Tong A. H., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W., Bussey, H., et al. (2001) Science 294, 2364-2368. [DOI] [PubMed] [Google Scholar]
- 18.Brachmann C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P. & Boeke, J. D. (1998) Yeast 14, 115-132. [DOI] [PubMed] [Google Scholar]
- 19.Rebhan M., Chalifa-Caspi, V., Prilusky, J. & Lancet, D. (1997) Trends Genet. 13, 163. [DOI] [PubMed] [Google Scholar]
- 20.Hodges P. E., McKee, A. H., Davis, B. P., Payne, W. E. & Garrels, J. I. (1999) Nucleic Acids Res. 27, 69-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costanzo M. C., Crawford, M. E., Hirschman, J. E., Kranz, J.E., Olsen, P., Robertson, L. S., Skrzypek, M. S., Braun, B. R., Hopkins, K. L., Kondu, P., et al. (2001) Nucleic Acids Res. 29, 75-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulovich A. G., Margulies, R. U., Garvik, B. M. & Hartwell, L. H. (1997) Genetics 145, 45-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mullen J. R., Kaliraman, V., Ibrahim, S. S. & Brill, S. J. (2001) Genetics 157, 103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leem S. H., Ropp, P. A. & Sugino, A. (1994) Nucleic Acids Res. 22, 3011-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prakash L. & Prakash, S. (1977) Genetics 86, 33-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J., Derfler, B., Maskati, A. & Samson, L. (1989) Proc. Natl. Acad. Sci. USA 86, 7961-7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang M. E., de Calignon, A., Nicolas, A. & Galibert, F. (2000) Curr. Genet. 38, 178-187. [DOI] [PubMed] [Google Scholar]
- 28.Gellon L., Barbey, R., Auffret van der Kemp, P., Thomas, D. & Boiteux, S. (2001) Mol. Genet. Genom. 265, 1087-1096. [DOI] [PubMed] [Google Scholar]
- 29.Winston F., Chaleff, D. T., Valent, B. & Fink, G. R. (1984) Genetics 107, 179-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petukhova G., Van Komen, S., Vergano, S., Klein, H. & Sung, P. (1999) J. Biol. Chem. 274, 29453-29462. [DOI] [PubMed] [Google Scholar]
- 31.Ajimura M., Leem, S. H. & Ogawa, H. (1993) Genetics 133, 51-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rattray A. J., McGill, C. B., Shafer, B. K. & Strathern, J. N. (2001) Genetics 158, 109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nitiss J. L., Rose, A., Sykes, K. C., Harris, J. & Zhou, J. (1996) Ann. N.Y. Acad. Sci. 803, 32-43. [DOI] [PubMed] [Google Scholar]
- 34.Piruat J. I. & Aguilera, A. (1996) Genetics 143, 1533-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longhese M. P., Paciotti, V., Fraschini, R., Zaccarini, R., Plevani, P. & Lucchini, G. (1997) EMBO J. 16, 5216-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian Z., Huang, H., Hong, J. Y., Burck, C. L., Johnston, S. D., Berman, J., Carol, A. & Liebman, S. W. (1998) Mol. Cell. Biol. 18, 4783-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson C. W. (1994) Semin. Cell Biol. 5, 427-436. [DOI] [PubMed] [Google Scholar]
- 38.Xiao W. & Chow, B. L. (1998) Curr. Genet. 33, 92-99. [DOI] [PubMed] [Google Scholar]
- 39.Chua P. & Roeder, G. S. (1995) Mol. Cell. Biol. 15, 3685-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naiki T., Kondo, T., Nakada, D., Matsumoto, K. & Sugimoto, K. (2001) Mol. Cell. Biol. 21, 5838-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aguilera A. & Klein, H. L. (1990) Mol. Cell. Biol. 10, 1439-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X. & Wang, Z. (1999) Nucleic Acids Res. 27, 956-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyler J. K., Adams, C. R., Chen, S. R., Kobayashi, R., Kamakaka, R. T. & Kadonaga, J. T. (1999) Nature 402, 555-560. [DOI] [PubMed] [Google Scholar]
- 44.Sommers C. H., Miller, E. J., Dujon, B., Prakash, S. & Prakash, L. (1995) J. Biol. Chem. 270, 4193-4196. [DOI] [PubMed] [Google Scholar]
- 45.Aboussekhra A., Chanet, R., Zgaga, Z., Cassier-Chauvat, C., Heude, M. & Fabre, F. (1989) Nucleic Acids Res. 17, 7211-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nasim A. & Brychcy, T. (1979) Can. J. Genet. Cytol. 21, 129-137. [DOI] [PubMed] [Google Scholar]
- 47.Xiao W., Chow, B. L., Broomfield, S. & Hanna, M. (2000) Genetics 155, 1633-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chakraverty R. K., Kearsey, J. M., Oakley, T. J., Grenon, M., de La Torre Ruiz, M. A., Lowndes, N. F. & Hickson, I. D. (2001) Mol. Cell. Biol. 21, 7150-7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hryciw T., Tang, M., Fontanie, T. & Xiao, W. (2002) Mol. Genet. Genom. 266, 848-857. [DOI] [PubMed] [Google Scholar]
- 50.Fricke W. M., Kaliraman, V. & Brill, S. J. (2001) J. Biol. Chem. 276, 8848-8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fasullo M., Koudelik, J., AhChing, P., Giallanza, P. & Cera, C. (1999) Genetics 152, 909-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahne F., Jha, B. & Eckardt-Schupp, F. (1997) Nucleic Acids Res. 25, 743-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eckardt-Schupp F., Siede, W. & Game, J. C. (1987) Genetics 115, 83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore C. W. (1978) Mutat. Res. 58, 41-49. [DOI] [PubMed] [Google Scholar]
- 55.Rattray A. J. & Symington, L. S. (1995) Genetics 139, 45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Interthal H. & Heyer, W. D. (2000) Mol. Gen. Genet. 263, 812-827. [DOI] [PubMed] [Google Scholar]
- 57.Bai Y. & Symington, L. S. (1996) Genes Dev. 10, 2025-2037. [DOI] [PubMed] [Google Scholar]
- 58.Wang H. & Elledge, S. J. (2002) Genetics 160, 1295-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lauder S., Bankmann, M., Guzder, S. N., Sung, P., Prakash, L. & Prakash, S. (1996) Mol. Cell. Biol. 16, 6783-6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bressan D. A., Baxter, B. K. & Petrini, J. H. (1999) Mol. Cell. Biol. 19, 7681-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kozhin S. A., Chepurnaya, O. V. & Korolev, V. G. (1995) Yeast 11, 1211-1213. [DOI] [PubMed] [Google Scholar]
- 62.Lee J. H., Choi, I. Y., Kil, I. S., Kim, S. Y., Yang, E. S. & Park, J. W. (2001) Biochim. Biophys. Acta 1526, 191-198. [DOI] [PubMed] [Google Scholar]
- 63.Chen J., Derfler, B. & Samson, L. (1990) EMBO J. 9, 4569-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berdal K. G., Bjoras, M., Bjelland, S. & Seeberg, E. (1990) EMBO J. 9, 4563-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaufman P. D., Kobayashi, R. & Stillman, B. (1997) Genes Dev. 11, 345-357. [DOI] [PubMed] [Google Scholar]
- 66.Game J. C. & Kaufman, P. D. (1999) Genetics 151, 485-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson J. R., Lawrence, C. W. & Hinkle, D. C. (1996) Science 272, 1646-1649. [DOI] [PubMed] [Google Scholar]
- 68.Lawrence C. W. & Maher, V. M. (2001) Biochem. Soc. Trans. 29, 187-191. [DOI] [PubMed] [Google Scholar]
- 69.Game J. C. (2000) Mutat. Res. 451, 277-293. [DOI] [PubMed] [Google Scholar]
- 70.Broomfield S., Hryciw, T. & Xiao, W. (2001) Mutat. Res. 486, 167-184. [DOI] [PubMed] [Google Scholar]
- 71.Lowndes N. F. & Murguia, J. R. (2000) Curr. Opin. Genet. Dev. 10, 17-25. [DOI] [PubMed] [Google Scholar]
- 72.Kaliraman V., Mullen, J. R., Fricke, W. M., Bastin-Shanower, S. A. & Brill, S. J. (2001) Genes Dev. 15, 2730-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X. B., Melchionna, R., Denis, C. M., Gaillard, P. H., Blasina, A., Van de Weyer, I., Boddy, M. N., Russell, P., Vialard, J. & McGowan, C. H. (2001) Mol. Cell 8, 1117-1127. [DOI] [PubMed] [Google Scholar]
- 74.Boddy M. N., Gaillard, P. H., McDonald, W. H., Shanahan, P., Yates, J. R., III & Russell, P. (2001) Cell 107, 537-548. [DOI] [PubMed] [Google Scholar]
- 75.Lee S. K., Johnson, R. E., Yu, S. L., Prakash, L. & Prakash, S. (1999) Science 286, 2339-2342. [DOI] [PubMed] [Google Scholar]
- 76.Klein H. L. (2001) Genetics 157, 557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Emili A., Schieltz, D. M., Yates, J. R., III & Hartwell, L. H. (2001) Mol. Cell 7, 13-20. [DOI] [PubMed] [Google Scholar]
- 78.Hanna J. S., Kroll, E. S., Lundblad, V. & Spencer, F. A. (2001) Mol. Cell. Biol. 21, 3144-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mayer M. L., Gygi, S. P., Aebersold, R. & Hieter, P. (2001) Mol. Cell 7, 959-970. [DOI] [PubMed] [Google Scholar]
- 80.Hartsuiker E., Vaessen, E., Carr, A. M. & Kohli, J. (2001) EMBO J. 20, 6660-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kiser G. L. & Weinert, T. A. (1996) Mol. Biol. Cell 7, 703-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herrlich P., Blattner, C., Knebel, A., Bender, K. & Rahmsdorf, H. J. (1997) Biol. Chem. 378, 1217-1229. [DOI] [PubMed] [Google Scholar]
- 83.Zhan Q., Carrier, F. & Fornace, A. J., Jr. (1993) Mol. Cell. Biol. 13, 4242-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fornace A. J., Alamo, I., Jr. & Hollander, M. C. (1988) Proc. Natl. Acad. Sci. USA 85, 8800-8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jelinsky S. A. & Samson, L. D. (1999) Proc. Natl. Acad. Sci. USA 96, 1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gasch A. P., Huang, M., Metzner, S., Botstein, D., Elledge, S. J. & Brown, P. O. (2001) Mol. Biol. Cell 12, 2987-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paulovich A. G. & Hartwell, L. H. (1995) Cell 82, 841-847. [DOI] [PubMed] [Google Scholar]
- 88.Grenon M., Gilbert, C. & Lowndes, N. F. (2001) Nat. Cell Biol. 3, 844-847. [DOI] [PubMed] [Google Scholar]
- 89.D'Amours D. & Jackson, S. P. (2001) Genes Dev. 15, 2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Prakash S., Sung, P. & Prakash, L. (1993) Annu. Rev. Genet. 27, 33-70. [DOI] [PubMed] [Google Scholar]
- 91.Friedberg E. C., Walker, G. C. & Siede, W., (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC).
- 92.Ho Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180-183. [DOI] [PubMed] [Google Scholar]
- 93.Boddy M. N., Lopez-Girona, A., Shanahan, P., Interthal, H., Heyer, W. D. & Russell, P. (2000) Mol. Cell. Biol. 20, 8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.