Abstract
Protecting the fetus and placenta from the maternal immune system has long been considered a function of placental trophoblasts. Here, we present two related lines of evidence that contradict this assumption. First, we show that transformed mouse trophoblast cell lines akin to human choriocarcinomas form tumors in syngeneic and immunodeficient mice, yet are rejected in immunocompetent allogeneic mice. Second, we show that wild-type trophoblasts are rapidly killed after i.v. injection into allogeneic mice. In both cases, the pattern of trophoblast killing in different strains of immunodeficient mice indicated that rejection involved host natural killer cells, and this was corroborated by in vitro killing assays. The apparent intrinsic susceptibility of mouse trophoblasts to immune attack strongly suggests that it is instead some property of the pregnant uterus that is of primary importance in preventing rejection of the fetus.
How the fetus and placenta escape immune rejection during pregnancy remains an outstanding issue in transplantation immunology (1, 2). Trophoblasts, the primary cell type of the placenta, are in direct contact with maternal tissue, yet do not invoke the cellular and humoral immune responses associated with other allografts. Because there has been little evidence to suggest that either the uterus is an immune-privileged site (3, 4) or that allograft rejection is systemically suppressed during pregnancy (5), reproductive immunologists have largely favored the idea that the placenta itself forms an immunological protective barrier around the fetus. Indeed, the available evidence has suggested that trophoblasts themselves might “intrinsically” resist immune-mediated attack (1, 6), which would render the uterine microenvironment and systemic pregnancy hormones unnecessary for the immunological protection of the fetus.
Perhaps the most compelling argument in favor of the idea of an intrinsic resistance mechanism comes from the existence of gestational choriocarcinomas in humans (7). These trophoblastic cancers originate during pregnancy and are semiallogeneic to the host as expected, yet clearly do not require the hormonal milieu of pregnancy or the uterine environment for continued growth, because they frequently become evident months after the antecedent pregnancy at metastatic sites outside the uterus. A similar argument comes from early studies showing that mouse ectoplacental cones (the part of the early extraembryonic tissue that gives rise to the placenta) transplanted to extrauterine sites in nonpregnant allogeneic mice form masses of differentiated trophoblast giant cells with apparently little inflammatory reaction, yet their counterpart embryos induce strong inflammatory responses in the same host (8). Lastly, the relative inability of cytotoxic T lymphocytes and natural killer (NK) cells to kill either human choriocarcinomas or isolated human and mouse placental cells in vitro has suggested that trophoblasts are resistant to killing by effector immune cells (9–12). This idea has gained support from the discovery of HLA-G, a nonclassical MHC class I molecule expressed by human trophoblasts that is thought to potentially protect these cells from NK cell-mediated attack (13).
The recent advent of mouse trophoblast stem cell lines (14) has afforded us the opportunity to test directly whether trophoblasts are intrinsically resistant to immune attack in vivo. Trophoblast stem cells can be grown indefinitely in culture in the presence of fibroblast growth factor 4 (FGF4) and an unidentified component of embryonic fibroblast conditioned media (EFCM). Removal of either factor causes rapid differentiation into trophoblast giant cells and spongiotrophoblasts, two postmitotic trophoblast subtypes. Surprisingly, we find that both transformed and nontransformed trophoblasts are rejected in nonpregnant allogeneic mice after extrauterine injection. Together, our data indicate that mouse trophoblasts are not intrinsically resistant to immune attack and point to a key role for NK cells in their rejection at ectopic sites.
Methods
Mice.
C57BL/6 (B6) (H-2b), BALB/c (H-2d), FVB/N (H-2q), Rag2−/−γc−/− (on a mixed B6 × C57BL/10 background), and Rag2−/− mice (B6 background) were obtained from Taconic Farms. Pfp−/− mice (B6 background) were from The Jackson Laboratory. NKD mice (B6 background; ref. 15) were the gift of W. Yokoyama (Washington University School of Medicine, St. Louis). Noon of the day of the copulation plug was counted as E0.5.
Trophoblast Stem Cell Culture and Derivation of Transformed Trophoblast Cell Lines.
Trophoblast stem cells were maintained in trophoblast stem cell media as described (14), using a DMEM-based TS medium (14) supplemented with 25 ng/ml human FGF4 (R & D Systems), 1 μg/ml heparin (Sigma), and 80% (vol/vol) EFCM. EFCM was prepared by culturing mouse embryo fibroblasts in DMEM containing 10% FCS for 3 days (Specialty Media, Lavallette, NJ). The final 20% of the media was made up with an adjustment media that raised the final percentage of FCS (HyClone) to 20%. Differentiated trophoblasts were generated by culturing trophoblast stem cells in TS media without any supplements (differentiation media).
TSInk4a trophoblast stem cell lines were established as described (14) from culturing homozygous Ink4a/ArfΔ2,3 blastocysts (the gift of R. DePinho and N. Sharpless, Harvard Medical School). TSras1 was derived by transfecting the TSCMI cell line (n = 22) with the T24 bladder carcinoma Ha-rasV12 oncogene (16) by using the Effectene reagent (Qiagen, Valencia, CA) and selecting for continued growth upon discontinuation of FGF4 on day 2 after transfection. Once the TSras1 line was established, we also discontinued heparin treatment because it was not required for growth.
Real-Time RT-PCR Analysis.
Total RNA was isolated with Trizol (Invitrogen), and cDNA was prepared from 5 μg of RNA by using a first-strand synthesis kit (SuperScript, Invitrogen) and 10–100 ng of cDNA amplified by real-time PCR using an ABI Prism 7700 Sequence Detector (Applied Biosystems). The sequences of all primer/probe sets, reaction conditions, and data analysis are described in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Flow Cytometry and Cell-Surface Marker Expression.
The following antibodies were purchased from PharMingen: anti-H-2Kq-FITC (clone KH114), mouse IgG2a-FITC (G155–178), anti-NK-1.1-FITC (PK136), and anti-TCR-β-PE (H57–597). NK cell numbers in lung cell suspensions or spleen NK cell preparations were counted as the NK1.1+ TCR-β− population. We used a Becton Dickinson FACSCalibur and CELLQUEST software.
Tumorigenesis and Lung Clearance Assays.
Cells were removed from tissue culture plates with trypsin/EDTA, washed once in TS media and twice in Dulbecco's PBS (DPBS), and injected into the tail vein of 6- to 10-week-old virgin females. For tumorigenesis assays, we injected 0.2 × 106 TSras2 cells in 0.1 ml of DPBS; for lung clearance assays, we injected 0.1–0.5 × 106 TSGFP (see below) cells (n = 23–40) in 0.1 ml of DPBS containing 0.5 mM EDTA to prevent cell clumping. The preparation of lung cell suspensions, adapted from a protocol originally designed for the isolation of dendritic cells (17), and their analysis by flow cytometry are described in detail in Supporting Text.
In Vitro Cytotoxicity Assays.
Standard 51Cr release assays were performed by using B6-derived IL-2-activated lymphokine-activated killers (LAKs) (18) as effector cells. Trophoblast stem cell and differentiated giant cell targets were generated by preplating TSGFP in stem cell media for 16 h or in differentiation media for 60 h before the addition of effector cells, respectively.
Statistical Analysis.
Student's t test was used for all comparisons, with a P value of <0.05 taken as statistically significant.
Results
Derivation and Characterization of Transformed Mouse Trophoblast Cell Lines.
From the outset, we reasoned that if trophoblasts were intrinsically resistant to rejection, their growth and survival at ectopic locations in nonpregnant allogeneic mice would not depend on the immune status of the host. We initially tried to test this hypothesis in a straightforward fashion by histologically comparing the s.c. growth of wild-type trophoblast stem cells injected into either immunodeficient or immunocompetent strain-matched allogeneic mice. These experiments, however, were difficult to interpret because the trophoblasts always differentiated into postmitotic giant cells over a 5- to 10-day period and rapidly degenerated thereafter (presumably because of FGF4 or EFCM withdrawal), forming structures variable in size (0.5–5.0 mm in diameter) with variable degrees of necrosis even in the same host strain. To circumvent these difficulties, we set out to derive a transformed mouse trophoblast cell line that no longer required exogenous FGF4 and EFCM for continuous growth. Such a line would allow us to establish a system in which we could instead score for endpoint tumor growth as an indication of trophoblast acceptance or rejection and test directly whether a gestational choriocarcinoma was intrinsically resistant to immune rejection outside the influence of an antecedent pregnancy.
To create a transformed mouse trophoblast cell line, we first derived a new trophoblast stem cell line (TSInk4a) from Ink4a/ArfΔ2,3 mice (19) harboring a targeted deletion of common exons used by the p16Ink4a and p19ARF tumor suppressor genes. By disrupting both p16INK4a and p53 tumor suppressor axes, this mutation allows dominant oncogenes to transform primary cells directly rather than induce their premature senescence (20). Like wild-type trophoblast stem cells (14), low passage TSInk4a trophoblasts, which were on a FVB/N background, grew as tight epithelial colonies with a low level of spontaneous differentiation into trophoblast giant cells (Fig. 4, which is published as supporting information on the PNAS web site). As expected, the cells ceased proliferating and fully differentiated into giant cells upon removal of either FGF4 or EFCM (not shown). At higher passage (n = 18), we noted that a very small percentage (≪1%) of cells continued to divide after removal of the EFCM, and we were able to derive a conditioned media-independent subline (TSCMI) from these cells that no longer required EFCM for continuous growth. These cells, however, still fully differentiated into giant cells upon removal of FGF4 and did not form tumors in syngeneic mice.
Next, we stably transfected TSCMI with the Ha-rasV12 oncogene and selected for continued cell proliferation upon FGF4 withdrawal, reasoning that constitutive Ras signaling would compensate for loss of upstream signaling by the FGF receptor. One colony grew out, named TSras1, which expressed Ha-rasV12 by RT-PCR analysis (data not shown). This cell line was able to form tumors in syngeneic mice. A final cell line, named TSras2, was derived from the reculturing of one such tumor. TSras1 and TSras2 continued to show a low level of spontaneous differentiation into giant cells (Fig. 4), and this level could be greatly increased if the cells were either maintained at confluence for a few days or treated with retinoic acid, an agent previously shown to induce wild-type trophoblast stem cell differentiation even in the presence of FGF4 and EFCM (21). Thus, both TSras1 and TSras2 retained the capacity for terminal trophoblast differentiation despite their transformed phenotypes.
Trophoblast Marker and MHC Expression by Transformed Trophoblasts.
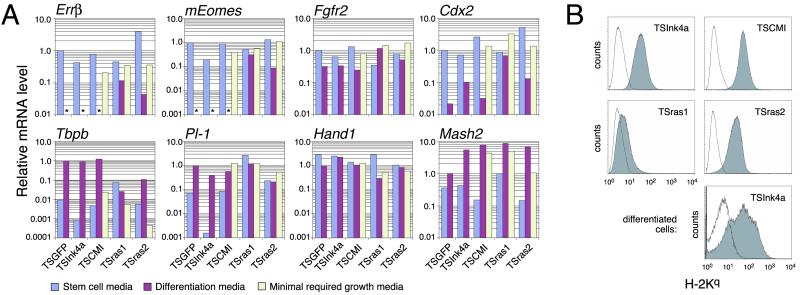
The expression pattern of several trophoblast-specific markers (14), determined by quantitative real-time RT-PCR, further established the trophoblastic identity of TSInk4a, TSCMI, TSras1, and TSras2. As shown in Fig. 1A, all four cell lines grown in the presence of FGF4 and EFCM (stem cell media) expressed a number of trophoblast stem cell markers, namely Errβ, mEomes, Fgfr2, and Cdx2, at levels very comparable to the levels expressed by outbred wild-type GFP-labeled trophoblast stem cells [EGFP-TS3.5, a gift of J. Rossant (University of Toronto, Toronto), which we refer to here for convenience as TSGFP; ref. 14]. Upon removal of FGF4 and EFCM (shift to differentiation media), wild-type stem cells, TSInk4a, and TSCMI all dramatically down-regulated the stem cell markers ERRβ, mEomes, and Cdx2, and conversely up-regulated the spongiotrophoblast and giant cell differentiation markers Tbpb and placental lactogen 1 (Pl-1), respectively, consistent with the previous description of trophoblast stem cell differentiation in culture (14). A similar shift in culture conditions induced a more blunted response in TSras1 and TSras2, presumably because constitutive Ras signaling in these cells attenuated the effect of FGF4 removal. Importantly, stem cell marker expression was maintained in TSCMI, TSras1, and TSras2 when these lines were cultured in the minimal conditions that respectively allowed for their continuous growth. As previously shown for wild-type trophoblasts (14), the giant cell marker Hand1 was similarly expressed in all culture conditions, and the spongiotrophoblast developmental regulator Mash2 was up-regulated in all cell lines upon their differentiation. This higher level of expression was maintained even in minimal growth media. Taken together, these expression studies strongly support the trophoblastic identity of TSras1 and TSras2, making them the first transformed mouse trophoblast cell lines that retain both stem cell characteristics as well as a capacity to terminally differentiate (for comparison, see refs. 22 and 23). Thus, these cells represent the first mouse correlates of gestational choriocarcinoma to our knowledge.
Fig 1.
Trophoblast marker and MHC class I expression by TSInk4a and its transformed derivatives. (A) Quantitative RT-PCR performed on total RNA from cells growing in stem cell media (TS media plus FGF4 and EFCM), after a 4-day shift to differentiation media (TS media alone), or in the minimal media requirements for continuous growth (TS media plus FGF4 for TSCMI; TS media alone for TSras1 and TSras2). mRNA levels relative to β-actin (ΔCT values) were normalized to TSGFP reference ΔCTs, which were 5.54 (Errβ), 3.95 (mEomes), 6.95 (Fgfr2), and 8.75 (Cdx2), determined during stem cell growth, and 8.13 (Tbpb), 10.20 (Pl-1), 6.53 (Hand1), and 7.53 (Mash2), determined after differentiation. *, not detectable. (B) H-2Kq expression was determined by flow cytometry on cells growing in their minimal required growth media as above (Top and Middle, four panels) or after their differentiation for 4 days. Shaded and open histograms show H-2Kq and isotype control staining, respectively.
We used flow cytometry to determine the expression level of MHC molecules by TSInk4a and its transformed derivatives. As shown in Fig. 1B, TSInk4a stem cells expressed moderate levels of the MHC class I molecule H-2Kq, similar to the level of H-2K expression by a number of other inbred and F1 hybrid trophoblast stem cell lines we have derived (not shown). This level of expression was maintained in TSCMI but was substantially down-regulated in TSras1, which is consistent with the known effects of ras transformation (24). Interestingly, TSras2 (as well as other TSras1 sublines similarly passaged in vivo) again expressed levels of H-2Kq comparable to its nontransformed ancestors. Lastly, TSInk4a differentiated in vitro for 4 days also showed significant H-2Kq expression, with the high variance in expression level attributable to the large and heterogeneous size of differentiated trophoblasts and their high degree of autofluorescence. By using another trophoblast cell line derived from B6 mice, we also found that trophoblast stem cells and differentiated trophoblasts expressed insignificant levels of MHC class II and Qa-1b, the mouse homologue of HLA-E (data not shown).
NK Cell-Dependent Rejection of Transformed Mouse Trophoblasts in Allogeneic Mice.
Surprisingly, despite consistent tumor growth of 1–2 × 106 TSras1 and TSras2 trophoblasts injected s.c. into nonpregnant syngeneic FVB/N mice, these cells failed to form tumors in B6 or BALB/c mice (0/25 TSras1 and 0/8 TSras2 tumors in B6 mice; 0/22 TSras1 tumors in BALB/c mice). Thus, TSras1 and TSras2 were immunologically rejected in vivo, and we conclude that these cells are not intrinsically resistant to rejection.
Interestingly, we noticed that the latency for TSras2 tumor growth in FVB/N mice was ≈25 days, yet it was ≈40 days for TSras1. This finding suggested that TSras1 growth in vivo entailed selection for more tumorigenic subpopulations, such as the one we grew as TSras2. Although the selection may have in part been for improved intrinsic growth characteristics, the reexpression of MHC class I by TSras2 vs. TSras1 suggested that an immunological selection also occurred, particularly for variants that could resist killing by syngeneic NK cells.
To directly characterize the immune cell types involved in rejecting TSras2, we monitored tumor formation in various mouse strains after i.v., rather than s.c., injection. This approach had the dual advantage of modeling both the hematogenous spread and pulmonary metastasis of gestational choriocarcinoma in humans (7), as well as the phenomenon of “trophoblast deportation” from the placenta during normal human pregnancy and the trapping of these embolic trophoblasts within the lung (25). As a positive control, we used immunodeficient Rag2−/−γc−/− mice, which completely and specifically lack all lymphoid cell populations (i.e., B, T, NKT, and NK cells) because of their combined defects in the Rag2 recombinase required for B cell receptor (BCR) and T cell receptor (TCR) gene rearrangements (26) and the common γ-chain required for IL-2, -4, -7, -9, and -15 receptor signaling (27). These mice, which were on a mixed B6 × C57BL/10 allogeneic background, uniformly formed tumors and died ≈30–40 days after the i.v. injection of 0.2 × 106 TSras2 cells (Fig. 2). Tumors were found mainly in the lung, mediastinum, liver, and occasionally in the peritoneal cavity, ovary, and s.c. tissue, and are histologically shown in Fig. 5, which is published as supporting information on the PNAS web site.
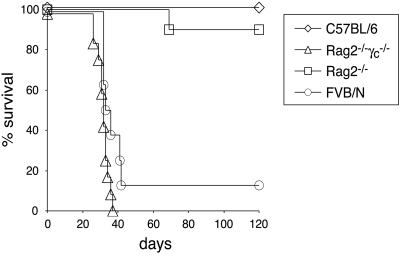
Fig 2.
TSras2 tumor formation in Rag2−/−γc−/− mice but not Rag2−/− mice. (A) Allogeneic Rag2−/−γc−/− (12/12) and syngeneic FVB/N (7/8) mice developed tumors within 30–40 days; however, allogeneic Rag2−/− (1/10) and B6 (0/12) mice rarely developed tumors. Data represent the compilation of three independent experiments, each with two to four mice per genotype.
The rapid kinetics of tumor formation in Rag2−/−γc−/− mice were very similar to those seen with syngeneic FVB/N mice (Fig. 2), suggesting that Rag2−/−γc−/− mice had no other immune system defects relevant to our system besides the ones involved in allorecognition described above. In contrast, both wild-type B6 control mice and, importantly, Rag2−/− mice on a B6 background were resistant to tumor formation, with only 1 of 10 injected Rag2−/− mice developing tumors after 60 days. Because Rag2−/− lack B, T, and NKT cells yet retain NK cells, the differential ability of these mice vs. Rag2−/−γc−/− mice to resist TSras2 tumor formation strongly suggested that NK cells alone were able to reject TSras2. Of note, the growth of TSras1 and TSras2 in syngeneic mice demonstrates that the transformation of these cells did not generate neoantigens strong enough to artifactually induce their rejection.
NK Cell-Dependent Rejection of Wild-Type Trophoblasts in Nonpregnant Allogeneic Mice.
Although the above results showed that TSras2 could be rejected in vivo via a mechanism that very likely required NK cells, we were concerned that the transformed nature of these trophoblasts might have altered their immunological properties. Therefore, we sought to substantiate our findings with wild-type, nontransformed trophoblasts, employing a variation of the standard lung clearance assay historically used to detect NK activity in vivo toward syngeneic and allogeneic tumor cells (28–30). In our modified assay, we monitored the clearance of GFP-labeled target cells (rather than radiolabeled target cells) by comparing the number of GFP-labeled trophoblasts present in the lungs of allogeneic mice <1 h after their i.v. injection to the number present 24 h after injection. As shown in Fig. 3A, wild-type (outbred) TSGFP trophoblast stem cells were easily visualized <1 h after injection by flow cytometry of cell suspensions prepared from B6 mouse lungs, where they were presumably trapped in the pulmonary vasculature. After 24 h, however, only 1% of these GFP+ cells were still present. This rapid clearance had an immunological component, because TSGFP retention in the lungs of Rag2−/−γc−/− mice 24 h after injection increased 10-fold compared with B6 mice (Fig. 3B). On the other hand, the high degree of TSGFP clearance in wild-type mice was not the artifactual result of the removal of a key surface molecule during cell preparation with trypsin plus EDTA, because it was similarly seen with cells removed from tissue culture plastic with EDTA alone (five experiments; data not shown). In the experiment shown in Fig. 3B, an equivalent number of TSGFP cells was injected into both mouse strains at the same time, leading to similar numbers of pulmonary GFP+ cells <1 h after injection but to disparate numbers 24 h after injection. However, we found that we could clearly compare between different sets of experiments if we plotted the number of GFP+ cells retained after 24 h (per lung parenchymal cell) vs. the number found <1 h after injection (Fig. 3C), or lastly, if we simply plotted the retention of GFP+ cells detected 24 h after injection as a percentage of cells detected <1 h after injection (Fig. 3D). These analyses showed that the increased retention of TSGFP in Rag2−/−γc−/− vs. wild-type mice occurred over a wide range of cells detected at the initial time point and revealed a remarkable consistency in the percentage retention in B6 and Rag2−/−γc−/− mice between experiments. Lastly, we found that trophoblasts differentiated predominantly into giant cells 2.5–3 days in vitro were also differentially cleared in B6 vs. Rag2−/−γc−/− mice (three experiments; data not shown). Taken together, the differential clearance of both wild-type trophoblast stem cells as well as differentiated trophoblast giant cells in B6 vs. Rag2−/−γc−/− mice strongly suggested that these cells, like their transformed counterparts, were rejected in vivo.
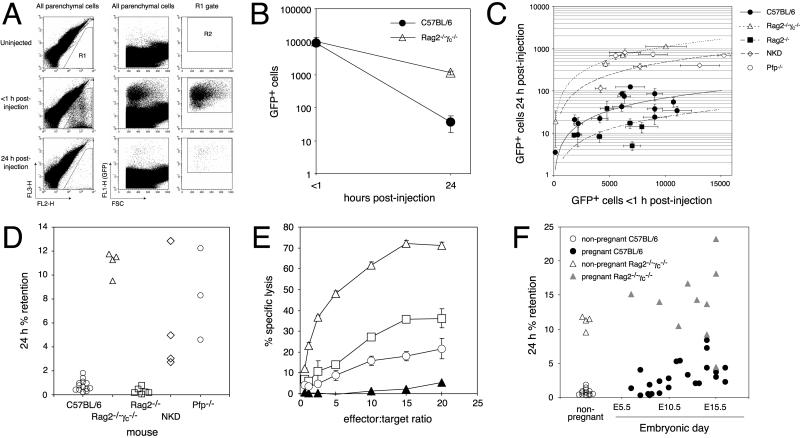
Fig 3.
NK cell-dependent killing of wild-type trophoblasts in vivo. (A) Visualization and clearance of TSGFP after i.v. injection into B6 mice. Single-cell lung suspensions were previously gated by their forward- and side-scatter characteristics to include all viable cells except the lymphoid population (not shown), because this latter population would be expected to vary between host strains. The remaining nonlymphoid parenchymal cell population was then gated to exclude autofluorescent host cells (the R1 gate in the left set of FL3-H vs. FL2-H plots); the final tally of true GFP+ cells used the R2 gate in the right set of plots and gave a false-positive background rate of one cell per million parenchymal cells in uninjected mice (n = 4). (B) Immune-mediated clearance of TSGFP. B6 and Rag2−/−γc−/− mice were injected with TSGFP, and the number of GFP+ cells per million parenchymal cells was determined <1 h and 24 h after injection. Each point represents the mean ± SEM of three mice. (C and D) NK cell- and perforin-dependent clearance of TSGFP. (C) A compilation of all of our experiments with trypsinized TSGFP stem cells injected into virgin females. Each point represents the mean ± SEM GFP+ cell number per million parenchymal cells for a group of five to six mice, with half killed at each time point. Plotted lines were determined by linear regression. (D) Data in C now plotted for each genotype as the percentage retention of GFP+ cells over the 24-h time period after injection. Increased retention (i.e., decreased clearance) compared with wild-type B6 mice (n = 13 experiments) was seen in Rag2−/−γc−/− (n = 4 experiments, P < 0.0001), NKD (n = 4 experiments; P < 0.001), and Pfp−/− mice (n = 3 experiments; P < 0.0001); decreased retention was seen in Rag2−/− mice (n = 6 experiments; P < 0.05). (E) Susceptibility of mouse trophoblasts to NK cell-mediated killing in vitro. 51Cr release assays with LAK effector cells were performed on TSGFP stem cells (□) or TSGFP trophoblasts differentiated into giant cells (○). LAK-sensitive YAC-1 (▵) and LAK-resistant BB88 (▴; ref. 38) targets served as positive and negative controls, respectively. Each point is the mean ± SD of triplicate samples. The data shown are representative of three independent experiments. (E) Ectopic trophoblast clearance in pregnant mice. Data for nonpregnant mice are the same as shown in Fig. 3D. E11.5–E16.5 pregnant B6 mice (n = 12 mice) showed increased retention compared with nonpregnant B6 mice (n = 13 experiments; P < 0.0001), but still less retention than E11.5–E16.5 pregnant Rag2−/−γc−/− mice (n = 8 mice; P < 0.0001) or nonpregnant Rag2−/−γc−/− mice (n = 4 experiments; P < 0.0001).
To determine the immune basis of TSGFP clearance, we again turned to a number of other mouse strains with selective cellular immune defects, all on B6 backgrounds. As shown in Fig. 3 C and D, the impaired trophoblast clearance in Rag2−/−γc−/− mice could not be attributed to their lack of B, T, or NKT cells, because Rag2−/− mice did not show a similar impairment. Rather, we found that the Rag2−/−γc−/− phenotype was largely replicated in NKD mice, which have a selective NK deficiency caused by the presence of a Ly49A transgene under control of the mouse granzyme A promoter (15) and a 40–80% decrease in the number of pulmonary NK cells on our hands [NK cells represented 0.6 ± 0.3% (mean ± SD) of total viable lung cells in NKD mice (n = 6) vs. 1.8 ± 0.7% in B6 mice (n = 29)]. Indeed, Rag2−/− mice actually showed a 2-fold increase in trophoblast clearance (i.e., a decrease in retention) compared with B6 mice, consistent with their 3- to 5-fold increase in pulmonary NK cell number (7.4 ± 1.8% of total lung cells; n = 6). Collectively, these data strongly suggested that NK cells were largely, if not exclusively, responsible for the efficient clearance of ectopic trophoblasts. Lastly, trophoblast clearance occurred largely through a direct cytolytic perforin-dependent mechanism, because perforin-deficient mice (Pfp−/− mice; ref. 31) also showed impaired trophoblast clearance yet did not show any change in pulmonary NK cell number (2.3 ± 0.3% of total lung cells; n = 6) compared with B6 mice.
In support of these in vivo assays, we found that both trophoblast stem cells as well as differentiated giant cells could also be killed in vitro by IL-2-activated NK cells [which generates LAKs (Fig. 3E), as shown previously with primary mouse trophoblasts (32)]. Interestingly, naive anti-DX5-purified NK cells from whole splenocytes showed only moderate trophoblast killing activity in the standard 51Cr release assay (data not shown), which is similar to previous results (9). This relative resistance toward NK cell-mediated killing in vitro despite rapid clearance in vivo is reminiscent of similar findings from the early studies of “natural resistance,” where it was noted that several allogeneic tumor cell lines rapidly cleared in vivo by NK cells were simultaneously poor targets for NK cell-mediated cytolysis in vitro (29, 30). Thus, taken together, our in vitro and in vivo killing experiments show that both trophoblast stem cells as well as differentiated giant cells are susceptible to direct NK cell-mediated attack, with their relative resistance in vitro likely caused by the limitations of in vitro NK killing assays.
Rejection of Wild-Type Trophoblasts in Pregnant Allogeneic Mice.
Lastly, we used our lung clearance assay to ask whether the systemic state of pregnancy influenced the clearance of ectopic trophoblast stem cells, which might occur through an effect either on the activity of the immune system or on the susceptibility of the trophoblasts. As shown in Fig. 3F, pregnant wild-type mice showed a mild and inconsistent increase in trophoblast retention (i.e., a decrease in clearance) in early gestation compared with nonpregnant mice and a moderate and consistent increase in retention by midgestation (E11.5–16.5). We could not rule out the possibility that this increase had an immunological component, because the retention in pregnant Rag2−/−γc−/− mice at midgestation did not show an obvious parallel increase over nonpregnant Rag2−/−γc−/− mice, although the wide variance in trophoblast retention during pregnancy prevented a definitive statistical analysis. In wild-type mice, the number of pulmonary NK cells at midgestation was not different from the number in nonpregnant mice [1.9 ± 0.4% of total viable lung cells in E12.5–16.5 pregnant mice (n = 14) vs. 1.8 ± 0.7% in nonpregnant mice (n = 29)]; however, partially purified splenic NK cells from E14.5–15.5 pregnant mice showed a reproducible and statistically significant 25% decrease in lytic activity toward YAC-1 targets (Fig. 6, which is published as supporting information on the PNAS web site), which agrees with the general depression in NK cell activity during pregnancy previously noted with peripheral human blood and whole mouse splenocytes (33, 34). Thus, the moderate decrease in trophoblast clearance during midgestation, if it is in fact an immune phenomenon, was likely caused by a systemic impairment of NK cell activity on a per cell basis rather than an effect of pregnancy hormones on trophoblasts themselves. Nonetheless, the significantly greater trophoblast clearance throughout gestation in wild-type mice compared with both pregnant and nonpregnant Rag2−/−γc−/− mice showed that trophoblasts could still be rejected, even during pregnancy.
Discussion
Immune-Mediated Rejection of Ectopic Trophoblasts in Vivo.
Because trophoblasts are not rejected by the maternal immune system despite their direct contact with maternal tissue, it has been tempting historically to speculate that they are intrinsically resistant to immune rejection. We have taken advantage of recently developed techniques for the continuous culture of mouse trophoblast stem cells (14) to readdress this basic question of reproductive immunology in an in vivo system. Surprisingly, we found that the choriocarcinoma-like TSras2 cell line was rejected in wild-type allogeneic B6 mice yet formed tumors in completely alymphoid Rag2−/−γc−/− mice on a nearly identical strain background. Similarly, wild-type trophoblasts showed impaired lung clearance in Rag2−/−γc−/− as well as in perforin-deficient mice, which have a relatively subtle immune defect expected only to reduce lymphocyte cytotoxicity (31). Thus, the differential clearance of wild-type trophoblasts in immunodeficient vs. immunocompetent mice reflected immune rejection as well. It was also unlikely that the rejection of wild-type trophoblasts was an artifact of their in vitro culture, because trophoblast stem cells show no irreversible developmental restriction and retain the capacity to contribute to all trophoblast lineages after injection into blastocysts (14). Collectively, our results show that both transformed and nontransformed trophoblasts are not intrinsically resistant to immune rejection but rather are efficiently killed in vivo after introduction into nonpregnant allogeneic mice at extrauterine sites.
The inability of trophoblasts to resist rejection in turn implies that the intrinsic characteristics of these cells, at least in the mouse, are insufficient to explain how the fetus and placenta escape rejection during pregnancy. In the present study, we have shown that two trophoblast subtypes in particular can be rejected: trophoblast stem cells, which in rodents such as mice, rats, and hamsters are directly exposed early in gestation (e.g., E7.5) to the maternal blood that bathes their presumed location within the porous ectoplacental cone (35–37), and differentiated trophoblast giant cells, which are exposed to the uterine stroma throughout gestation as they invade the uterus and ultimately form the external shell around the mature placenta. Thus, these exposures represent important points of vulnerability where nontrophoblastic mechanisms must be invoked to explain the survival of the fetus and placenta. Testing the immunological susceptibility of labyrinthine trophoblasts, another trophoblast subtype in extensive contact with maternal blood, must await the development of methods for their pure isolation.
Broadly, the nontrophoblastic components of pregnancy that might protect the fetus and placenta from rejection are systemic pregnancy hormones (such as progesterone) and the uterine microenvironment. Certainly, neither alternative alone sufficiently explains the special immune status of the fetus and placenta, because pregnant mammals do not accept allografts (5), and the nonpregnant uterus is not an immune-privileged site (3, 4). Furthermore, it is unlikely that systemic pregnancy hormones directly induce trophoblasts to become resistant to the maternal immune system, because we have shown that pregnant mice also immunologically clear ectopic trophoblasts. The possibility that pregnancy hormones and the uterus act in concert to create an ad hoc immune-privileged site, however, has not been thoroughly explored. In particular, the uterine decidua, a progesterone-dependent structure that develops in response to the implanting blastocyst, might function not only to attenuate immune cell activity locally (1, 2), but also to establish the unusual kind of systemic immune tolerance that has been observed toward placental antigens during pregnancy, at least in the mouse (2). Systemic tolerance induced by the decidua, if inappropriately maintained, might explain how a gestational choriocarcinoma originally arising in the pregnant uterus is able to persist and metastasize after completion of the antecedent pregnancy. In addition, it remains possible that the pregnant uterus might produce local factors that secondarily induce trophoblasts to become resistant to rejection.
Involvement of NK Cells in the Killing of Ectopic Trophoblasts.
By using a modified lung clearance assay on mouse strains with identical or nearly identical strain backgrounds, we found impaired clearance of allogeneic trophoblast stem cells in Rag2−/−γc−/− mice (which selectively lack B, T, NKT, and NK cells) and NKD mice (which have a selective NK cell deficiency) but not Rag2−/− mice (which lack B, T, and NKT cells, but retain NK cells) or wild-type mice. This pattern of rejection shows both directly and indirectly that NK cells are required for trophoblast rejection in vivo, a conclusion further supported by our demonstration of trophoblast stem cell and trophoblast giant cell killing by activated NK cells in vitro. Interestingly, Rag2−/− mice showed increased trophoblast clearance compared with wild-type mice, which is consistent with the increased numbers of NK cells in Rag2−/− mice and the similar increase in NK cell activity previously noted in T cell-deficient nu/nu mice (28). Our finding that efficient trophoblast clearance also requires host perforin expression indicates that NK cells likely kill trophoblasts in vivo by a direct mechanism, although this does not rule out the possibility that NK cells might indirectly cause trophoblast death through the activation of secondary effector cells, such as macrophages, as well.
Because the NK cell defects of NKD mice are incomplete, this strain could not be used in long-term tumorigenesis experiments. Nonetheless, we found that the MHC class I-positive TSras2 cell line formed tumors in Rag2−/−γc−/− but not Rag2−/− or wild-type mice, and that tumor growth of the MHC class I-negative TSras1 cell line in syngeneic mice led to reexpression of MHC class I molecules, likely as a result of a selection for resistance to NK cell killing. Together, these data provide strong indirect evidence that NK cells are also involved in the rejection of transformed allogeneic trophoblasts. However, as is also the case for nontransformed trophoblasts, the experiments here only show that B, T, and NKT cells are not absolutely required for trophoblast rejection in vivo; they do not comment on whether these cell types can kill ectopic trophoblasts in the absence of NK cells.
Supplementary Material
Acknowledgments
We thank J. Rossant, R. DePinho, N. Sharpless, and W. Yokoyama for valuable reagents; D. Zhang, K. Sigrist, L. Kangaloo, and C. McCall for technical and administrative assistance; A. Singer, M. Oukka, E. Jaeckel, and M. Grusby for critical review of the manuscript; and J. Hunt and members of the Glimcher lab for advice and discussions. This work was supported by National Institutes of Health Grants AI01650 and AI54370 and a gift from The G. Harold and Leila Y. Mathers Charitable Foundation.
Abbreviations
NK, natural killer
FGF4, fibroblast growth factor 4
EFCM, embryonic fibroblast conditioned media
LAK, lymphokine-activated killer
References
- 1.Clark D. A. (1999) Crit. Rev. Immunol. 19, 509-539. [PubMed] [Google Scholar]
- 2.Mellor A. L. & Munn, D. H. (2000) Annu. Rev. Immunol. 18, 367-391. [DOI] [PubMed] [Google Scholar]
- 3.Beer A. E. & Billingham, R. E. (1974) J. Reprod. Fertil. Suppl. 21, 59-88. [Google Scholar]
- 4.Schlesinger M. (1962) J. Natl. Cancer Inst. 28, 927-945. [PubMed] [Google Scholar]
- 5.Billingham R. E. (1964) N. Engl. J. Med. 270, 667. [DOI] [PubMed] [Google Scholar]
- 6.Head J. R. (1989) Am. J. Reprod. Immunol. 20, 100-105. [DOI] [PubMed] [Google Scholar]
- 7.Baergen R. N. (1997) Gen. Diagn. Pathol. 143, 127-141. [PubMed] [Google Scholar]
- 8.Simmons R. L. & Russell, P. S. (1962) Ann. N.Y. Acad. Sci. 99, 717-732. [DOI] [PubMed] [Google Scholar]
- 9.Zuckermann F. A. & Head, J. R. (1988) Cell. Immunol. 116, 274-286. [DOI] [PubMed] [Google Scholar]
- 10.Zuckermann F. A. & Head, J. R. (1987) J. Immunol. 139, 2856-2864. [PubMed] [Google Scholar]
- 11.Saji F., Kameda, T., Koyama, M., Matsuzaki, N., Negoro, T. & Tanizawa, O. (1989) Am. J. Reprod. Immunol. 19, 108-113. [DOI] [PubMed] [Google Scholar]
- 12.King A., Birkby, C. & Loke, Y. W. (1989) Cell. Immunol. 118, 337-344. [DOI] [PubMed] [Google Scholar]
- 13.Kovats S., Main, E. K., Librach, C., Stubblebine, M., Fisher, S. J. & DeMars, R. (1990) Science 248, 220-223. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka S., Kunath, T., Hadjantonakis, A. K., Nagy, A. & Rossant, J. (1998) Science 282, 2072-2075. [DOI] [PubMed] [Google Scholar]
- 15.Kim S., Iizuka, K., Aguila, H. L., Weissman, I. L. & Yokoyama, W. M. (2000) Proc. Natl. Acad. Sci. USA 97, 2731-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capon D. J., Chen, E. Y., Levinson, A. D., Seeburg, P. H. & Goeddel, D. V. (1983) Nature 302, 33-37. [DOI] [PubMed] [Google Scholar]
- 17.Iwasaki A. & Kelsall, B. L. (1999) J. Exp. Med. 190, 229-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunji Y., Vujanovic, N. L., Hiserodt, J. C., Herberman, R. B. & Gorelik, E. (1989) J. Immunol. 142, 1748-1754. [PubMed] [Google Scholar]
- 19.Serrano M., Lee, H., Chin, L., Cordon-Cardo, C., Beach, D. & DePinho, R. A. (1996) Cell 85, 27-37. [DOI] [PubMed] [Google Scholar]
- 20.Serrano M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. (1997) Cell 88, 593-602. [DOI] [PubMed] [Google Scholar]
- 21.Yan J., Tanaka, S., Oda, M., Makino, T., Ohgane, J. & Shiota, K. (2001) Dev. Biol. 235, 422-432. [DOI] [PubMed] [Google Scholar]
- 22.Athanassakis I., Aifantis, Y., Ranella, A. & Vassiliadis, S. (1996) Am. J. Reprod. Immunol. 36, 111-117. [DOI] [PubMed] [Google Scholar]
- 23.Bowen J. A. & Hunt, J. S. (1999) Biol. Reprod. 60, 428-434. [DOI] [PubMed] [Google Scholar]
- 24.Lohmann S., Wollscheid, U., Huber, C. & Seliger, B. (1996) Scand. J. Immunol. 43, 537-544. [DOI] [PubMed] [Google Scholar]
- 25.Bardawil W. A. & Toy, B. L. (1959) Ann. N.Y. Acad. Sci. 80, 197-261. [DOI] [PubMed] [Google Scholar]
- 26.Shinkai Y., Rathbun, G., Lam, K. P., Oltz, E. M., Stewart, V., Mendelsohn, M., Charron, J., Datta, M., Young, F., Stall, A. M. & Alt, F. W. (1992) Cell 68, 855-867. [DOI] [PubMed] [Google Scholar]
- 27.Cao X., Shores, E. W., Hu-Li, J., Anver, M. R., Kelsall, B. L., Russell, S. M., Drago, J., Noguchi, M., Grinberg, A., Bloom, E. T., et al. (1995) Immunity 2, 223-238. [DOI] [PubMed] [Google Scholar]
- 28.Karre K., Klein, G. O., Kiessling, R., Klein, G. & Roder, J. C. (1980) Int. J. Cancer 26, 789-797. [DOI] [PubMed] [Google Scholar]
- 29.Riccardi C., Puccetti, P., Santoni, A. & Herberman, R. B. (1979) J. Natl. Cancer Inst. 63, 1041-1045. [PubMed] [Google Scholar]
- 30.Riccardi C., Santoni, A., Barlozzari, T., Puccetti, P. & Herberman, R. B. (1980) Int. J. Cancer 25, 475-486. [DOI] [PubMed] [Google Scholar]
- 31.Kagi D., Ledermann, B., Burki, K., Seiler, P., Odermatt, B., Olsen, K. J., Podack, E. R., Zinkernagel, R. M. & Hengartner, H. (1994) Nature 369, 31-37. [DOI] [PubMed] [Google Scholar]
- 32.Drake B. L. & Head, J. R. (1989) J. Immunol. 143, 9-14. [PubMed] [Google Scholar]
- 33.Baley J. E. & Schacter, B. Z. (1985) J. Immunol. 134, 3042-3048. [PubMed] [Google Scholar]
- 34.Furukawa K., Itoh, K., Okamura, K., Kumagai, K. & Suzuki, M. (1984) J. Reprod. Immunol. 6, 353-363. [DOI] [PubMed] [Google Scholar]
- 35.Carpenter S. J. (1972) Am. J. Anat. 135, 445-476. [DOI] [PubMed] [Google Scholar]
- 36.Mehrotra P. K. (1988) Biol. Struct. Morphog. 1, 63-68. [PubMed] [Google Scholar]
- 37.Welsh A. O. & Enders, A. C. (1991) Am. J. Anat. 192, 347-365. [DOI] [PubMed] [Google Scholar]
- 38.Karlhofer F. M., Ribaudo, R. K. & Yokoyama, W. M. (1992) Nature 358, 66-70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.