Abstract
Class II MHC molecules undergo conformational changes on shifts of the pH. As a consequence, low-affinity peptides tightly bound at pH 7.0 can be released at pH 5.0. The imidazole group of histidine is the only amino acid side chain affected within this range. At pH 5.0 the group is positively charged, polar, and hydrophilic, whereas at pH 7.4 it is neutral, apolar, and hydrophobic. In this study, we used soluble forms of HLA-DR and substituted conserved histidine residues with tyrosine, an isosteric analogue to the uncharged form of histidine. The goal of this substitution was to identify crucial His residues by an increase in pH stability of the ligand complex. HLA-DM-mediated release experiments revealed that substitution of His-33 in the α1 domain of the HLA-DR molecule almost doubled the half-life of HLA-DR1/class II-associated invariant-chain peptide complexes. The divergence in the off-rate of WT and H33Y mutated complex was strictly pH-dependent and correlated with the theoretical titration curve of the imidazole group. For both HLA-DR1 and HLA–DR4 molecules the mutation resulted in a shift of class II-associated invariant-chain peptide release curves by up to 0.5 pH units. His-33α1 is present in all HLA-DR and H-2E molecules. It connects the α1 and α2 domains in its noncharged form by hydrophobic interactions with residue Val-136α2. It is located in close proximity to the putative interface with HLA-DM and may function as a pH-sensitive “button,” which is closed at pH 7.0 but opens below pH 6.0 to allow conformational transitions necessary for ligand exchange.
The loading of class II MHC molecules with exogenous peptide ligands takes place in the endosomal MIIC compartment (1). Both the removal of the class II-associated invariant-chain peptide (CLIP) and the loading with exogenous peptides is assisted by HLA-DM (2–4). After generation of MIIC by the fusion of vesicles containing the components required for antigen processing (class II MHC, HLA-DM, proteases, etc.) with vesicles formed by endocytosis that contain the protein source, the endosome acquires a slightly acidic pH (≈pH 4.5). This decrease in pH initiates proteolytic degradation of the protein and a conformational change in the class II MHC molecule, which allows the release of CLIP and HLA-DM-catalysed ligand exchange reactions (4). During the transport of the MIIC endosome to the plasma membrane the pH is gradually raised to the neutral pH of the extracellular matrix (pH 7.4). This change in pH is accompanied by a decrease in HLA-DM activity and an increase in stability of the peptide/class II MHC complex. 8-Anilino-1-naphthalene sulfonate fluorescence experiments detected the conformational changes of the MHC/peptide complex during this transition, because two pH-dependent conformers seem to differ in the degree of surface hydrophobicity (5–7). More detailed information, however, including the structural basis for these transitions, is not available yet.
Conversions of pH-dependent conformers frequently are triggered by small molecular switches. These switches can consist of pairs or small clusters of amino acids, which on protonation/deprotonation rearrange bonds or produce subtle shifts in their relative position. Amplified by hinges and levers formed by secondary structure elements these shifts translate into major rearrangements in the tertiary or quaternary structure of the protein. One example is hemoglobin. The affinity of the protein to oxygen depends on CO2 concentration and blood pH, and His-146β seems to account primarily for this so-called “Bohr effect” by the pH-dependent breakage or formation of an intramolecular salt bridge with Asp-94β (8, 9).
Imidazole groups are particularly suitable elements for pH-sensitive molecular switches. Imidazole forms the side chain of histidine and its pK (≈6.0) is well within physiological pH ranges. Furthermore, the protonated and the nonprotonated forms of imidazole are chemically very different. The nonprotonated form has essentially a hydrophobic and aromatic character whereas the protonated form is hydrophilic and positively charged. Consequently the nature of chemical interactions differs significantly at pH above or below the pK. At pH 7.0 the nonprotonated form is dominant. It favors interactions with other hydrophobic groups. At pH 5.0 the imidazole group is protonated and prefers a hydrophilic environment. Thus, a pair of amino acids consisting of histidine and another hydrophobic residue could function as a pH-sensitive “His button.” It “closes” tightly at pH 7.0 but “opens” at pH 5.0, because hydrophobic amino acids are repelled by the charged form of histidine.
Arginine and tyrosine are chemically the closest homologues of the charged and the noncharged forms of histidine, respectively. Their codon usage differs from histidine only in one base, which may suggest a close functional relation of these amino acids during the evolution of proteins. In contrast to histidine their side chains remain unchanged on pH changes between 5.0 and 7.0. The replacement of histidine by arginine should therefore stabilize the “protonated” position of pH-sensitive molecular switches whereas a tyrosine substitution should stabilize the “nonprotonated” position disregarding the surrounding pH. The primary objective of this study was to identify histidine residues that control the pH-dependent stability of HLA-DR/ligand complexes. We therefore substituted conserved histidine residues at strategic hinge positions with tyrosine and analyzed the influence of the substitution on the HLA-DM-mediated ligand release at various pH conditions.
Materials and Methods
Production of Substituted HLA-DR Molecules.
Tyrosine substitutions of three histidine residues, His-5α (5′-GGG CTA TCA AAG AAG AAT ATG TGA TCA TCC AGG CCG-3′), His-33α (5′-GAT GGT GAT GAG ATT TTC TAT GTG GAT ATG GCA AAG-3′), and His-143α (5′-CCT GCC CAG GGA AGA CTA TCT TTT CCG GAA GTT CC-3′), were introduced into a soluble HLA-DRA*0101 construct by using the QuikChange site-directed mutagenesis kit (Stratagene). The mutant DNA was cloned into the pMT-BIP vector (Invitrogen), an inducible expression vector for S2 Drosophila cells (Invitrogen). Mutated and WT forms of soluble HLA-DR1 (DRA*0101, DRB1*0101) (2) and HLA-DR4 (DRA*0101, DRB1*04011) (2) as well as soluble HLA-DM molecules (2) were expressed in S2 insect cells and produced as described (10–12).
HLA-DR Binding Assay with Immobilized Oligomeric CLIP-Peptide Ligands.
Polypeptide ligands were generated by producing linear peptide oligomers (13) containing four copies of the CLIP peptide (IC106–120: KMRMATPLLMQALPM) separated by a short spacer sequence (GGPG)3. The following oligonucleotides (encoding the CLIP peptide) were used for the construction: + strand, 5′-T AAA ATG CGT ATG GCT ACT CCG CTG CTG ATG CAG GCT CTG CCG ATG GG-3′; − strand, 5′-CAT CGG CAG AGC CTG CAT CAG CAG CGG AGT AGC CAT ACG CAT TTT ACC-3′. The peptide oligomers were produced in Escherichia coli bacteria as described (13). The oligomers were then linked via amine groups to N-hydroxysuccinimide (NHS)-agarose beads (taken from HiTrap NHS-activated columns, Amersham Pharmacia). The coupling was carried out according to the manufacturer's recommendation. The CLIP ligand beads were stored as a 1:10 suspension in PBS. For release experiments, 80 μl cell culture supernatants (containing between 1 and 10 μg/ml soluble HLA-DR molecules) were loaded overnight onto 4 μl of the bead suspension at 37°C. The release reaction was carried out for 4 h with 2 μM soluble HLA-DM at pH 5.0 at 37°C in the presence of 1 mg/ml high-affinity hemagglutinin (HA)306–318 peptide (PKYVKQNTLKLAT) to prevent rebinding of released HLA-DR to the beads. After release the amount of HLA-DR still bound to the beads was measured by fluorescence-activated cell sorting (FACS) with the phycoerythrin (PE)-labeled α-HLA-DR antibody L243 (Becton Dickinson). Cells were analyzed by using a FACScalibur flow cytometer (Becton Dickinson) and cellquest software.
SDS/PAGE Separation of Peptide/MHC Complexes.
Complexes were formed at pH 7.3 in a volume of 6 μl by incubating 0.5 μg/ml HLA-DR with 0.16 μg/ml peptide ligand in the presence of 5% ethanol (14) at 37°C for 48 h. SDS/PAGE separation was done on a 4–15% Tris-glycine gradient gel (Bio-Rad) at 4°C. Samples were loaded without prior boiling by using a nonreducing sample buffer. A protein standard of 25–150 kDa (Novagen) was used as a marker. Protein bands were visualized by Coomassie stain (Bio-Rad).
ELISA Release Experiments.
Preformed HLA-DR/CLIP peptide complexes were generated by incubating 10 μl HLA-DR (1 mg/ml) with 0.5 μl of biotinylated CLIP peptide (1 mg/ml) overnight at 37°C. The reaction mixture was then diluted 1:5 with PBS, and 4 μl was mixed with 4 μl of buffer (100 mM NaPO4/100 mM NaOAc/150 mM NaCl; the pH was adjusted between 5.0 and 7.4), 1 μl of indicated amounts of HLA-DM, and 1 μl of HA306–318 peptide (10 mg/ml). The reaction mixture was incubated for 3 h or indicated amounts of time at 37°C. The release was determined in an ELISA reaction, using an α-HLA-DR antibody as capture antibody (L243, American Type Culture Collection). The detection of stable HLA-DR/biotinylated CLIP complexes was carried out as described (14) with streptavidin-coupled peroxidase (Sigma) using TMB substrate (Kirkegaard & Perry Laboratories) or with Eu3+-labeled streptavidin (DELFIA, Wallac). The Eu3+ fluorescence was enhanced with 15 μM β-naphtoyltrifluoroacetone, 50 μM tri-n-octylphosphine oxide, 6.8 mM potassium hydrogen phtalate, 100 mM acetic acid, and 0.1% Triton X-100 (15) and measured with a Victor fluorescence reader (Wallac) by using the time-resolved mode at an excitation wavelength of 340 nm and emission wavelength of 614 nm.
Results and Discussion
The influence of pH on the stability of class II MHC/ligand complexes is a general phenomenon (4). Histidine residues responsible for this pH dependency therefore should be conserved in various class II MHC molecules. To identify a set of candidate residues an alignment was carried out in which the sequences of various allelic forms of class II MHC molecules were compared. This alignment included human HLA-DR, HLA-DP, and HLA-DQ molecules but also H-2E and H-2A molecules, the mouse homologues of HLA-DR and HLA-DQ, respectively. In addition to their presence in different class II MHC molecules the location within the MHC molecule was an important criterion. The histidine residues had to be located at strategic hinge regions. Also close interaction with hydrophobic residues, which potentially could form the counter amino acid of a His button, was another criterion to be considered.
By comparing an extended list of class II MHC sequences (16) and analyzing the crystal structures of several HLA-DR and HLA–DQ molecules (11, 17, 18) three putative His buttons (His-5α, His-143α, and His-33α) were identified (Fig. 1). The histidine residues of these buttons all were located on the α-chain of the class II MHC molecule, which contains three additional histidine residues at positions 149, 167, and 177 (Fig. 1). The latter three, however, were located at sites that either did not appear to be crucial for structural transitions or did not have hydrophobic counterparts. Of the three selected candidates His-5α is most conserved. This residue was found in α-chains of all MHC molecules analyzed (H5α corresponds to H7α of HLA-DQα and H8α of H-2Aα alleles). This residue interacts with the hydrophobic residue 91β, which is valine in HLA-DR and H-2E and leucine in HLA-DP and HLA–DQ (Table 1). The only exception was found in H-2A molecules where this position is occupied by a polar serine residue. The second most conserved residue is H143α. It is present in most class II MHC molecules (H143α in HLA-DR and H-2E, H146α in HLA-DQ, and H147α in H-2A) except for HLA-DPα and some allelic forms of H-2Aα where it is substituted by tyrosine. The hydrophobic counterpart is either I31β or F31β. The least conserved histidine is H33α. This histidine residue is missing in the α-chain of HLA-DP and HLA-DQ molecules and in mouse H-2A molecules but is present in all HLA-DRα and H-2Eα alleles. In contrast to histidine residues of the other putative buttons His-33α interacts directly with another residue on the α-chain, V136α. Analysis of the spatial structure of HLA-DR revealed that these two residues connect the two extracellular α-domains, with H33α located on the α1 domain and V136α located on the α2 domain.
Fig 1.
Alignment of α-chains of human and mouse class II MHC molecules. The amino acid sequences of human HLA-DRA1*0101, HLA-DPA1*0103, and HLA-DQA1*0101 and mouse H2-Abα, H2-Adα, and H2-Edα are aligned to the histidine residues of the HLA-DRA1*0101 molecule. Histidine residues and corresponding amino acids are indicated in bold.
Table 1.
Three putative histidine buttons in human and mouse class II MHC molecules
| Molecule | His-5 button | His-143 button | His-33 button |
|---|---|---|---|
| HLA-DR (all alleles) | H-5α:V-91β | H-143α:I/F-31β | H-33α:V-136α |
| H-2E (all alleles) | H-5α:V-91β | H-143α:I/F-31β | H-33α:V-136α |
| HLA-DP (all alleles) | H-5α:L-91β | (Y-143α, I-29β) | (Y-33α, L-136α) |
| HLA-DQ (all alleles) | H-7α:L-91β | H-146α:I-31β | (Y-36α, S-139α) |
| H-2A (f,d,s,q) | (H-8α, S-91β) | H-147α:I-31β | (Y-37α, S-140α) |
| H-2A (k,b,u) | (H-8α, S-91β) | (Y-147α, I-31β) | (Y-37α, S-140α) |
The amino acids located at the position of pairs forming three putative His buttons, His-5α, His-143α, and His-33α, are listed for human HLA-DR, HLA-DP, and HLA-DQ and for mouse H-2A and H-2E molecules (complete amino acid sequences are listed in ref. 16). Amino acids are printed in single letter code; origin of the chain (α or β) and the position of the amino acid are indicated (shifts in the relative numbering of amino acids in HLA-DQα and H-2Aα and in the HLA-DPβ chain are caused by insertions or deletions, respectively). Matching pairs that could potentially form pH-sensitive His buttons are in bold and connected by a colon. Pairs that cannot form such a button are displayed in parentheses and separated by a comma.
In all cases, where one of the three putative button-forming histidines was missing, it was replaced by tyrosine, the isosteric analogue of the noncharged form (Fig. 1). For our studies similar tyrosine substitutions were introduced at positions 5α, 33α, and 143α of a soluble form of HLA-DRA*0101 that lacks the intracellular domain and the transmembrane region by site-directed mutagenesis. Single mutants as well as mutants with combinations of these substitutions were expressed together with a similarly truncated β-chain of HLA-DRB1*0101 to produce soluble HLA-DR1 molecules. Insect cells were used to obtain HLA-DR1 molecules essentially free of endogenously bound peptide ligands (10–12).
A sensitive assay was established to screen these constructs by using as little as 80 μl of HLA-DR1 containing cell culture supernatant (Fig. 2). In this assay soluble HLA-DR molecules were captured with CLIP ligands covalently linked to small agarose beads [instead of single CLIP peptides, oligomers consisting of four copies of the CLIP epitope (13) were coupled to the agarose beads to reduce steric hindrance of the binding region and to provide sufficient distance to the bead surface]. Detection and quantitation of bead-bound HLA-DR was done by FACS analysis after staining with a PE-labeled anti-HLA-DR antibody. As shown in Fig. 2A the assay allows the detection of less than 2 μg/ml HLA-DR1 with a linear correlation of the median of fluorescence to the HLA-DR1 concentration. With this assay the stability of HLA-DR1/CLIP complexes at pH 5.0 could be determined by using supernatant of transiently transfected insect cells. In this experiment the HLA-DM-mediated CLIP release from HLA-DR1 molecules containing the WT HLA-DR α-chain (WT) or α-chains with single (H5αY, H143αY, and H33αY), double (5/33 and 33/143), or triple tyrosine substitutions (5/33/143) of the histidine residues was compared. Although no difference from WT HLA-DR1 was detected with constructs containing a single tyrosine substitution at positions 5 or 143, a significant reduction of released HLA-DR1 was observed for the H33αY construct. The amount of HLA-DR1 molecules released within 5 h decreased from 51% (WT) to 33% (H33αY). A similar effect was also seen for the double and triple mutants containing the H33αY substitution (Fig. 2B).
Fig 2.
Detection of soluble HLA-DR molecules with immobilized CLIP ligands by FACS. Agarose beads carrying covalently attached CLIP oligomers (linear polypeptides containing four copies of the CLIP ligand) were used to capture soluble HLA-DR molecules in cell supernatants and quantified by FACS. The beads were stained with PE-labeled α-HLA-DR antibody. (A) Sensitivity and linearity of the assay. The beads were incubated with 80 μl of insect cell medium containing the indicated amounts of purified HLA-DR1. (Left) A dot plot of the FACS analysis. Beads were incubated with four different concentrations of soluble HLA-DR1 (the data were recorded in linear mode but are displayed in a log scale, resulting in some loss of resolution at lower values). (Right) The linear correlation of fluorescence median and amount of HLA-DR in the medium is evident in a plot of fluorescence median vs. HLA-DR1 concentration. The graph shows the data obtained with PE-labeled anti-HLA-DR antibodies (○) and PE-labeled isotype-matched control antibodies (•). (B) HLA-DM-mediated release of mutated and WT HLA-DR1 molecules from CLIP beads at pH 5.0. Supernatants of S2 cells expressing soluble WT or mutated HLA-DR1 molecules were incubated overnight with CLIP beads. Constructs with single tyrosine substitutions (H5αY, H143αY, and H33αY), dual substitutions (5/33 and 33/143), and one triple substitution (5/33/143) of the HLA-DR α-chain were tested. After loading, the beads were transferred into a pH 5.0 buffer and incubated with soluble HLA-DM (2 μM) and high-affinity HA306–318 peptide for 5 h at 37°C. The amount of HLA-DR still remaining on the beads was determined by FACS. The bars indicate the percentage of HLA-DR molecules released from the beads. Little or no release was determined at pH 7.4 or in the absence of HLA-DM for both WT and mutant HLA- DR1 (data not shown).
The previous experiment suggested that H33α has some important function in the pH-dependent control of the ligand release. For more detailed experiments soluble HLA-DR1 molecules containing H33αY or the WT α-chain were expressed on a large scale and purified by affinity chromatography using anti-HLA-DR antibodies and gel filtration (12). As expected SDS/PAGE analysis did not reveal any significant differences between the two HLA-DR1 molecules (Fig. 3A). Both WT HLA-DR1 and H33Y HLA-DR1 were able to form a stable complex with the HA306–318 peptide, and the gel did not reveal any significant differences in the peptide complex formed. In accordance with the bead assay (Fig. 2B), however, differences were noticed when the purified HLA-DR molecules were used to test the stability of the CLIP complexes at pH 5.0. ELISA experiments with biotinylated peptide ligands revealed a half-life of the H33Y HLA-DR1/CLIP complex that was almost doubled compared with the complex formed with the WT HLA-DR1 (Fig. 3B). In the presence of HLA-DM a 50% release was observed for the H33Y complex after 79 min whereas for the WT complex the 50% release point was 42 min.
Fig 3.
Comparison of purified HLA-DR1 molecules containing WT or H33Y substituted α-chains. (A) SDS/PAGE analysis of empty and peptide-loaded soluble HLA-DR1 molecules. Purified HLA-DR1 molecules containing WT (lanes 1 and 2) or H33Y α-chains (lanes 3 and 4) were loaded with HA306–318 peptide (lanes 2 and 4) or separated without prior loading (lanes 1 and 3) under nonreducing conditions. After SDS/PAGE the bands were visualized by Coomassie staining. The position of separate α- and β-chains and the peptide/MHC complex are indicated. (B) Kinetics of the HLA-DM-mediated CLIP release at pH 5.0. HLA-DR1 molecules containing the WT α-chain (•) or the H33Y α-chain (▪) were loaded overnight with biotinylated CLIP peptide. The complex was then incubated for the indicated amounts of time at pH 5.0 with soluble HLA-DM (0.4 μM) in the presence of HA306–318 peptide. The amount of stable HLA-DR1/biotinylated CLIP complex was determined by ELISA using anti-HLA-DR capture antibodies (L243) and peroxidase-labeled streptavidin.
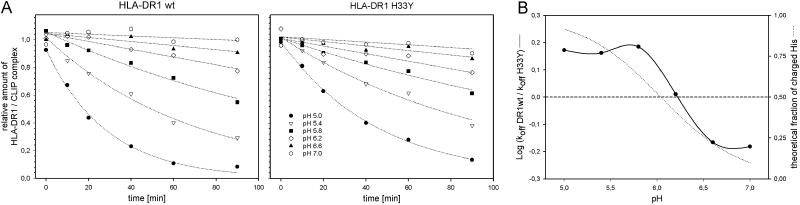
To investigate the influence of pH on the dissociation rates or WT and H33Y mutated complexes in more detail kinetic analysis of the HLA-DM-mediated CLIP release was carried out at pH 5.0, 5.4, 5.8, 6.2, 6.6, and 7.0 (Fig. 4A). The pH-specific dissociation rates of WT and mutant complex differed most at pH 5.0, where the CLIP release from the mutant complex was significantly slower (koff WT: −3.23e-2 min−1; koff H33Y: −2.17e-2 min−1). With increasing pH, the difference decreased and was almost zero at pH 6.2 (koff WT: −2.80e-3 min−1; koff H33Y: −2.73e-3 min−1). Above pH 6.2 the CLIP release from the mutant complex was even slightly higher compared with the WT complex.
Fig 4.
pH dependency of the HLA-DM-mediated CLIP release from WT and H33Y-substituted HLA-DR1. (A) Kinetic analysis of the CLIP release. CLIP-loaded complexes of WT HLA-DR1 (Left) and H33Y mutant HLA-DR1 (Right) were incubated at indicated amounts of time as described in Fig. 3B with 1.2 μM HLA-DM at a pH of 5.0 (•), 5.4 (▿), 5.8 (▪), 6.2 (◊), 6.6 (▴), or 7.0 (open octagon). The amount of nondissociated complex was measured by ELISA using Eu3+-labeled streptavidin. Off-rates were determined with a graphic analysis software (sigmaplot) using a nonlinear regression [f = a exp(−bx)]. The analysis revealed the following koff [min−1] for the WT and the H33Y-substituted HLA-DR1 complex, respectively: −3.23e-2; −2.17e-2 (pH 5.0), −1.45e-2; −9.98e-3 (pH 5.4), −7.90e-3; −5.16e-3 (pH 5.8), −2.80e-3; −2.73e-3 (pH 6.2), −1.10e-3; −1.61e-3 (pH 6.6), −6.40e-4; −9.72e-4 (pH 7.0). (B) Correlation of pH-dependent off-rates with the theoretical fraction of charged imidazole groups. The logarithmic values of the ratio of koff HLA-DR1 WT and koff HLA-DR1 H33Y (see Fig. 4A) were calculated and plotted vs. the pH (straight line). The experimental curve is shown together with plot representing the theoretical fraction of charged imidazole group at the respective pH (dotted line). The calculation was done by using the Henderson–Hasselbalch equation with a pKs = 6.04.
As expected the effect of pH on the off-rates was not linear but rather correlated with the titration curve of imidazole. This is illustrated in Fig. 4B where the influence of pH on the off-rates and the putative degree of the His-33 protonation are compared. In this graph the experimental curve of the logarithmic values of koff ratios [log(koff WT/koff H33Y)] revealed a good correlation with the theoretical titration curve of imidazole. This finding indicates that the pH controls the dissociation rate of HLA-DR1 ligand complexes at least in part by effecting the charged/noncharged state of His-33 of the α-chain.
To test whether the results are also valid for other alleleic forms of class II MHC molecules release experiments were carried out with H33Y-mutated HLA-DR4 (Fig. 5). The HLA-DM-mediated CLIP release of WT and H33Y mutated forms of HLA-DR1 (Fig. 5 Left) and HLA-DR4 molecules (Fig. 5 Right) was carried out at different pH (5.0–6.0). Under these conditions the pH for the 50% release point for WT forms of HLA-DR1 and HLA-DR4 was around 6.0, whereas the 50% release point for the H33Y forms was below 5.6. Thus, the influence of H33α resulted in a shift of ≈0.5 pH units and was evident for both HLA-DR molecules investigated. This finding suggests that H33α participates in general in the control of pH-dependent transitions in HLA-DR molecules.
Fig 5.
pH-titration curves of the HLA-DM-mediated dissociation of CLIP peptide from WT and H33Y HLA-DR1 and HLA-DR4 molecules. Purified HLA-DR molecules containing the WT α-chain (○) or the mutated H33Y α-chain (•) were preloaded with biotinylated CLIP peptide and incubated for 1.5 h with soluble HLA-DM (2 μM) in buffers with indicated pH. The experiment was carried out with soluble HLA-DR1 (Left) as well as with soluble HLA-DR4 (Right) in the presence of HA306–318 peptide. After the release the amount of stable HLA-DR/biotinylated CLIP was determined by ELISA as described in Fig. 3B.
The position of H33α and the putative His-33α button within the spatial structure of HLA-DR1 is illustrated in Fig. 6. H33α and its counterpart V136α are located at the interface of the α1 and α2 domains. They are in close proximity to E40α and F51α, two residues recently identified to interact with HLA-DM (19). Another recent study showed that HLA-DM tethered by a linker to a bound peptide on its N-terminal end (but not at the C-terminal end) accelerates the rate of peptide exchange (20). Both of those papers suggest the face of the HLA-DR protein with which HLA-DM interacts, i.e., the side where the N terminus of the peptide ligand extrudes from the binding groove. The histidine button described here is located on that face at the beginning of the long strand that connects the β-sheet platform to the α1-helix in class II MHC proteins. HLA-DM may trigger the release of peptide ligands by affecting pocket 1 of the HLA-DR molecule, presumably by redirecting important H-bonds (2, 21–23). The disruption of the connection formed by the H33α1/V136α2 button between the two extracellular domains of the HLA-DR α-chain could be a crucial step for the release of the bound peptide. As illustrated in Fig. 6 the opening of the His-33α button could allow the shift of the α2 domain, for instance, after a “push” by the HLA-DM molecule, which is transferred via the short interdomain connection to the α1 domain. This, in turn, could relax the grip of the alpha1 α-helix, which together with the beta1 α-helix holds the peptide ligand like a clamp.
Fig 6.
Position of H33α in the HLA-DR1 molecule and the putative role of the His-33 button in the pH-dependent ligand release by HLA-DM. The figure was adapted from an x-ray crystallographic study of an HLA-DR1/ligand complex (18). The HLA-DRA*0101 α-chain is presented as a white ribbon, the HLA-DRB1*0101 β-chain as a gray ribbon (the relative positions of the α1, α2, β1, and β2 domains are indicated). The peptide ligand is shown in red and the N terminus of the peptide is indicated in cyan. The amino acid side chains forming the His-33α button, H33α (dark green or orange) of the α1 domain, and V136α (green) of the α2 domain, as well as HLA-DM contact residues E40α and F51α (both yellow) (19), are displayed in space-filling mode. (Left) The stable pH 7.3 conformation is shown with the closed His-33 button (deprotonated H33α in dark green). (Right) A hypothetical pH 5.0 conformation with an open His-33α button (charged H33α in orange). The pH-dependent opening of the button promotes a break of the connection between the α1 and α2 domains that could allow HLA-DM to shift the α2 domain of the HLA-DR molecule toward the C-terminal end of the peptide. This shift leads to a lift of the α-helix of the α1 domain, which provides sufficient space for the release of the peptide ligand.
A recent study showed that small molecular compounds with H-bond donor function can significantly accelerate the ligand exchange of HLA-DR molecules (14). These molecules act as carriers of “immobilized” protons and pH-sensitive switches, such as the His-33α button, that may be the immediate target affected by these molecules. In summary, these results indicate that pH-sensitive molecular switches control the stability of HLA-DR ligand complexes, in a similar way as described for hemoglobin. In both cases the molecular basis is the protonation/deprotonation of imidazole groups of histidine residues. In this study H33α could be identified as a pH sensor in HLA-DR molecules that seems to form a His button with V136α. The incomplete inhibition of the pH dependency after tyrosine substitution, however, suggests that this button is only one of several pH-sensitive elements that control the conformational transitions that influence the stability of HLA-DR/ligand complexes.
Acknowledgments
We thank E. Stratikos and D. Wiley for HLA-DM and H. Finley for excellent technical assistance. The work was supported by National Institutes of Health Grant 5R35-CA47554.
Abbreviations
CLIP, class II-associated invariant-chain peptide
HA, hemagglutinin
PE, phycoerythrin
FACS, fluorescence-activated cell sorting
References
- 1.Busch R., Doebele, R. C., Patil, N. S., Pashine, A. & Mellins, E. D. (2000) Curr. Opin. Immunol. 12, 99-106. [DOI] [PubMed] [Google Scholar]
- 2.Sloan V. S., Cameron, P., Porter, G., Gammon, M., Amaya, M., Mellins, E. & Zaller, D. M. (1995) Nature 375, 802-806. [DOI] [PubMed] [Google Scholar]
- 3.Busch R., Reich, Z., Zaller, D. M., Sloan, V. & Mellins, E. D. (1998) J. Biol. Chem. 273, 27557-27564. [DOI] [PubMed] [Google Scholar]
- 4.Jensen P. E., Weber, D. A., Thayer, W. P., Westerman, L. E. & Dao, C. T. (1999) Immunol. Rev. 172, 229-238. [DOI] [PubMed] [Google Scholar]
- 5.Runnels H. A., Moore, J. C. & Jensen, P. E. (1996) J. Exp. Med. 183, 127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ullrich H. J., Doring, K., Gruneberg, U., Jahnig, F., Trowsdale, J. & van Ham, S. M. (1997) Proc. Natl. Acad. Sci. USA 94, 13163-13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boniface J. J., Lyons, D. S., Wettstein, D. A., Allbritton, N. L. & Davis, M. M. (1996) J. Exp. Med. 183, 119-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perutz M. F., Kilmartin, J. V., Nishikura, K., Fogg, J. H., Butler, P. J. & Rollema, H. S. (1980) J. Mol. Biol. 138, 649-668. [DOI] [PubMed] [Google Scholar]
- 9.Perutz M. F., Gronenborn, A. M., Clore, G. M., Fogg, J. H. & Shih, D. T. (1985) J. Mol. Biol. 183, 491-498. [DOI] [PubMed] [Google Scholar]
- 10.Stern L. J. & Wiley, D. C. (1992) Cell 68, 465-477. [DOI] [PubMed] [Google Scholar]
- 11.Dessen A., Lawrence, C. M., Cupo, S., Zaller, D. M. & Wiley, D. C. (1997) Immunity 7, 473-481. [DOI] [PubMed] [Google Scholar]
- 12.Rotzschke O., Falk, K., Mack, J., Lau, J. M., Jung, G. & Strominger, J. L. (1999) Proc. Natl. Acad. Sci. USA 96, 7445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rotzschke O., Falk, K. & Strominger, J. L. (1997) Proc. Natl. Acad. Sci. USA 94, 14642-14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falk K., Lau, J., Santambrogio, L., Marin Esteban, V., Puentes, F., Rotzschke, O. & Strominger, J. L. (2002) J. Biol. Chem. 277, 2709-2715. [DOI] [PubMed] [Google Scholar]
- 15.Malle E., Munscher, G., Muller, T., Vermeer, H. & Ibovnik, A. (1995) J. Immunol. Methods 182, 131-144. [DOI] [PubMed] [Google Scholar]
- 16.Rammensee H.-G., Bachmann, J. & Stevanovic, S., (1997) MHC Ligands and Peptide Motifs (Springer, Heidelberg). [DOI] [PubMed]
- 17.Lee K. H., Wucherpfennig, K. W. & Wiley, D. C. (2001) Nat. Immunol. 2, 501-507. [DOI] [PubMed] [Google Scholar]
- 18.Stern L. J., Brown, J. H., Jardetzky, T. S., Gorga, J. C., Urban, R. G., Strominger, J. L. & Wiley, D. C. (1994) Nature 368, 215-221. [DOI] [PubMed] [Google Scholar]
- 19.Doebele C. R., Busch, R., Scott, M. H., Pashine, A. & Mellins, D. E. (2000) Immunity 13, 517-527. [DOI] [PubMed] [Google Scholar]
- 20.Stratikos E., Mosyak, L., Zaller, D. M. & Wiley, D. C. (2002) J. Exp. Med. 196, 173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosyak L., Zaller, D. M. & Wiley, D. C. (1998) Immunity 9, 377-383. [DOI] [PubMed] [Google Scholar]
- 22.Roche P. A. (1996) Science 274, 526-527. [DOI] [PubMed] [Google Scholar]
- 23.Weber D. A., Evavold, B. D. & Jensen, P. E. (1996) Science 274, 618-620. [DOI] [PubMed] [Google Scholar]