Abstract
Circulating human natural killer (NK) lymphocytes have been functionally defined by their ability to exert cytotoxic activity against MHC class I-negative target cell lines, including K562. Therefore, it was proposed that NK cells recognized the “missing self.” We show here that the Ig-like CD160 receptor expressed by circulating CD56dim+ NK cells or IL-2-deprived NK cell lines is mainly involved in their cytotoxic activity against K562 target cells. Further, we report that HLA-C molecules that are constitutively expressed by K562 trigger NK cell lysis through CD160 receptor engagement. In addition, we demonstrate, with recombinant soluble HLA-Cw3 and CD160 proteins, direct interaction of these molecules. We also find that CD158b inhibitory receptors partially interfere with CD160-mediated cytotoxicity, whereas CD94/CD159a and CD85j have no effect on engagement with their respective ligands. Thus, CD160/HLA-C interaction constitutes a unique pathway to trigger NK cell cytotoxic activity.
Keywords: activation, NK receptors, HLA class I
Human circulating natural killer (NK) lymphocytes can be divided into two subsets based on their cell-surface density of CD56 and -16, namely the major CD56dim+CD16+ and the minor CD56bright+CD16− subpopulations. Only the former NK cell subset was shown to exert effector cytotoxic function (1). During the past years, several receptors have been identified on NK lymphocytes for their ability to switch NK cells “off” or “on” (2). Among the inhibitory receptors are the killer cell Ig-like receptors (KIR), the Ig-like transcript receptors (ILT), and the lectin-like complex receptor CD94/CD159a (NKG2A) recognizing classical and/or nonclassical MHC class I molecules (3). These receptors possess immunoreceptor tyrosine-based inhibition motifs (ITIM) in their cytoplasmic tail (3). In contrast, besides the clonally expressed CD94/NKG2C that binds to HLA-E (4) and KIR immunoreceptor tyrosine-based activation motif (ITAM)-bearing receptors that bind to HLA class Ia, the receptors that provide an “on” signal to NK cell cytotoxicity are mostly NKG2D and the natural cytotoxicity receptors (NCR), which do not recognize constitutive MHC class I molecules (2, 5). NKG2D is a lectin-like molecule associated with the protein DAP10 containing the cytoplasmic YxxM motif capable of recruiting the p85 subunit of the (PI)-3 kinase (5). NKG2D is expressed on all NK cells, T cell antigen receptor (TCR)-γδ+, and CD8+ T cells, and binds the stress-inducible MIC-A and -B or ULBP proteins (5). The NCR group mainly consists of NKp46, NKp30, and NKp44 (2). Only NKp46 and -30 are expressed by circulating NK cells, whereas the NKp44 is selectively expressed by IL-2-activated NK cells, which may explain the high efficiency of NK cells cultured in IL-2 in killing of tumor cells. The cellular ligands for NCR are unknown except for NKp46, which could be hemagglutinins present on virus-infected cells (6).
We previously found that human CD56+dimCD16+ NK lymphocytes expressed the Ig-like glycosyl phosphatidyl inositol (GPI)-anchored CD160 receptor (7). Further, using the original anti-CD160 mAb BY55 to separate circulating CD160+ NK lymphocytes, we demonstrated that the latter cells corresponded to the effector cytotoxic population (8, 9). However, because the BY55 anti-CD160 mAb did not inhibit cytotoxicity of the separated peripheral blood NK (PB-NK) lymphocytes, we were unable to assess whether the structure recognized by BY55 mAb was involved in the target cell recognition (8, 9). In the present study, we investigated the function of CD160 expressed on NK cells by using another anti-CD160 mAb, CL1-R2, which inhibits the binding of CD160 receptor to its MHC class I ligands (10). We report that CL1-R2 mAb inhibits fresh CD160+ PB-NK cell cytotoxicity. Further, using selective combination of effector NK cell lines and target tumor cell lines, our results show for the first time that a CD160 molecule triggers NK cell cytotoxicity after its engagement with an HLA-C molecule. Further direct interaction of CD160 with HLA-C was demonstrated using soluble recombinant molecules. Interestingly, we found that mAb-mediated cross-linking of CD85j or CD94/CD159a inhibits the redirected cell lysis induced by anti-CD160 mAb. In contrast, neither CD85j nor CD94/CD159a stimulated by their respective physiological ligands provided significant inhibition to the CD160-mediated triggering signal, whereas coengagement of both CD160 and -158b reduced but did not abolish it. Thus, these data suggest that more than one KIR should be engaged to “turn off” circulating NK cell activity mediated through CD160/HLA-C interaction.
Materials and Methods
Antibodies and Flow Cytometry.
mAbs used included W6/32 (anti-HLA class I), B1.23.2 (anti-HLA-B, -C, referred to as “anti-HLA-C”), L31 (anti-free HLA-C heavy chain, a gift from A Beretta, Hôpital Saint-Joseph, Paris; ref. 11) and 87G (anti-HLA-G, a gift from D. Geraghty, Fred Hutchinson Cancer Research Center, Seattle). Anti-CD158a (EB6), anti-CD158b (GL183), anti-CD94 (HP-3B1), anti-CD159a (Z199), and anti-CD244 (C.1.7.1) mAbs were from Beckman Coulter. The following mAbs were produced in our laboratories: anti-CD160 (CL1-R2, IgG1, and BY55, IgM), anti-CD16 (CRET16), anti-CD56 (CRET56), anti-CD59 (P282), anti-HLA-B/C (B1.23.2), anti-HLA-G (MEM-G9), and anti-HLA-E (MEM-E/06). The mAbs anti-CD158e (DX9 and Z27), anti-CD158i (PAX180), anti-CD159a (p25), anti-NKG2A/C (Z270, IgG1), anti-CD85j (VMP55), and anti-CD244 (pp35) were obtained through the exchanges of the 7th International Workshop on Leukocyte Antigens (12). For flow cytometry analysis, cells were stained with the appropriate mAbs followed by phycoerythrin (PE)-conjugated goat anti-mouse Ig (Caltag, South San Francisco, CA). Samples were analyzed on a Coulter Epics Elite flow cytometer.
Cells.
Effector cells were the human NKL, NK92, and NK3.3 (13–15) cell lines, cultured without IL-2 for 4 days, as well as fresh purified human PB-NK cells derived from normal donors by using a magnetic-activated cell sorter NK cell isolation kit (Miltenyi Biotec, Auburn, CA). Populations were between 80% and 99% CD3−CD56+CD16+. Such freshly isolated NK cells were used immediately or after 3 days of culture in the presence of IL-2 (200 units/ml). In some experiments, when the purified PB-NK cell population contained at least 20% CD3−CD56bright+CD16−CD160− we further separated by flow cytometry CD160+ from CD160− populations by using BY55 mAb. The following target cells were used: P815 (FcγR+ murine mastocytoma), human erythroleukemia K562 and K562-HLA-G1 (K562-G) transfectant (gift of A. Horuszko, Institute of Molecular Medicine and Genetics, Augusta, GA), and JAR and JAR-HLA-Cw3 transfected with HLA-Cw0304 cDNA (gift of B. van den Eynde, Ludwig Institute for Cancer Research, Brussels). Jurkat and Jurkat-CD160 transfectant (gift of G. Freeman, Dana-Farber Cancer Institute, Boston) were also used.
NK Cell Cytotoxicity.
NK cell cytotoxicity and redirected cell lysis against P815 were tested in a 51Cr-release assay in the absence or presence of various mAbs or isotypic control Ig. The concentrations of these mAbs, as well as Ig-isotype controls, were 10 μg/ml for the masking experiments and 2 μg/ml for the redirected killing experiments, unless otherwise specified.
Soluble CD160-Flag and HLA-Cw3 Proteins.
cDNA encoding a C-terminal, Flag-tagged soluble CD160 was generated by PCR amplification of the aa 1–160 region. The resulting PCR product was ligated into the pcDNA3 expression vector (Invitrogen), sequenced, and transfected in Jurkat cells. Positive clones were identified by immunoprecipitation of soluble CD160 from culture supernatants followed by anti-Flag immunoblotting. Flow cytometry binding experiments were performed using culture supernatant from a soluble CD160 transfectant cultured in serum-free medium, followed by incubation with anti-Flag mAb (Sigma) and PE-labeled conjugate. Using HLA-Cw0304 cDNA as a template, extracellular domains were amplified by PCR primers: 5′-GGATTCCATATGGGTTCCCATTCCATGCGTTATTTCTACACCGCC-3′ and 5′-AGCTGGGATCCCGGCTCCCATCTCAGGGTGAGGGG-3′ and cloned into a PET24a vector derivative containing a BirA recognition and biotinylation site in frame at the COOH terminus (Avidity, Denver). The NH2-terminal primer contained several synonymous nucleotide substitutions designed to optimize protein expression from an Escherichia coli BL21 pLys strain. HLA-Cw3 monomers were refolded as described (16). HLA-Cw3 recombinant and human β2-microglobulin, produced in E. coli, were solubilized in urea and injected together with IAIIPSKKL synthetic peptide (17). Soluble HLA-Cw3 was finally purified by gel filtration on a Sephacryl S-200 column (Amersham Pharmacia). Flow cytometry analysis was performed using soluble HLA-Cw3 cross-linked with B1.23.2 mAb, followed by incubation with PE-conjugated streptavidin.
Results
CD160 Engagement Triggers Cytotoxic Activity in Peripheral Blood NK Lymphocytes and in IL-2-Deprived NK Cell Lines.
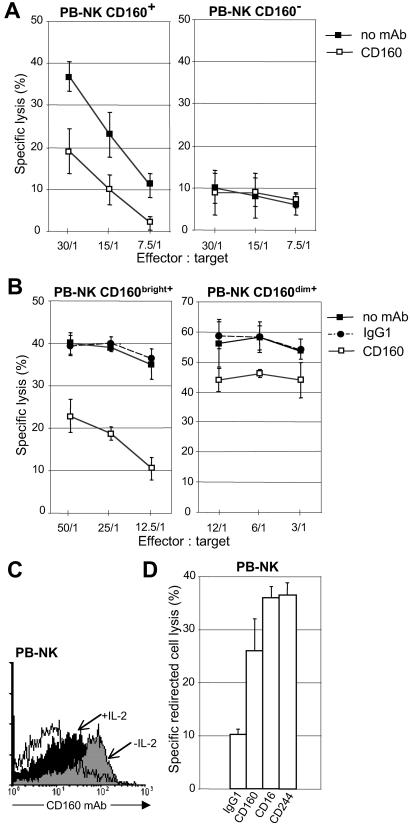
Freshly purified PB-NK cells from normal individual were sorted in CD160+ and -160− effector cells by using BY55 mAb labeling, and analyzed for their ability to lyse K562 target cells in the absence or presence of anti-CD160 (CL1-R2) mAb. We found that, in contrast to CD160+ cells, which exhibited significant NK activity (Fig. 1A Left), CD160− cells were poorly efficient as effector cytotoxic cells (Fig. 1A Right). CL1-R2 mAb inhibited CD160+ cells cytotoxicity, whereas it had no effect on CD160− cells (Fig. 1A). Further, when we purified PB-NK cells containing >95% CD160+ cells, lysis of K562 was sharply inhibited in the presence of soluble CL1-R2 mAb, whereas isotype-matched IgG1 alone had no effect (Fig. 1B Left). When the same fresh CD160+ (CD56dim+CD16+) NK cell population was cultured ex vivo for a few days in the presence of exogenous IL-2, the NK cell lysis of K562 was strongly enhanced, and we found that CL1-R2 mAb only marginally blocked the IL-2-activated NK cell-mediated cytotoxicity (Fig. 1B Right). Interestingly, this low blocking effect was associated with a significant diminishment of CD160 cell-surface expression in IL-2-activated NK cells (Fig. 1C). These results suggested that CD160 receptor expressed by circulating NK cells was involved in the lysis of K562. We tested whether cross-linked anti-CD160 mAb was capable of triggering PB-NK CD160+ cytotoxic effector function. Using the standard redirected killing assay with P815, we found that CL1-R2 mAb-mediated cross-linking of CD160 resulted in the induction of NK-mediated cytotoxicity, almost as efficiently as anti-CD16 or anti-CD244 mAbs-triggering NK cytotoxicity (Fig. 1D). Taken together, these results showed that on specific engagement, the CD160 receptor triggers human PB-NK cell-mediated cytotoxicity.
Fig 1.
Activation of fresh peripheral blood NK cell-mediated cytolysis by CD160. (A) Cytotoxic activity of freshly isolated human PB-NK separated in CD160+ and -160− populations against K562 target cell in the absence of mAb (no mAb) or in the presence of mAb CL1-R2 (CD160). (B) Cytotoxic activity of PB-NK either untreated (CD160bright+) or cultured for 3 days with IL-2 (CD160dim+) against K562 target cell in the absence of mAb (no mAb) or in the presence of mAb CL1-R2 or Ig-isotype control (IgG1). (C) CD160 expression in fresh PB-NK cells either untreated (−IL-2) or cultured for 3 days with IL-2 (+IL-2) was analyzed by flow cytometry using FITC-labeled BY55 mAb. Data are representative of several independent experiments. Open profile is Ig-isotype control staining. (D) Redirected lysis assay of P815 using fresh human CD160+ PB-NK in the presence of mAbs specific for the indicated molecules or control IgG1. The effector-to-target cell (E:T) ratio used was 25. Results of A, B, and D are mean ± SD of triplicates and are representative of three independent experiments.
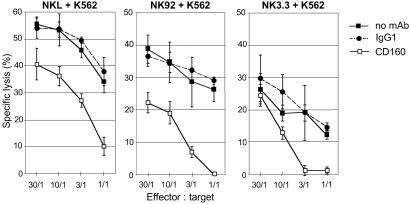
To assess that CD160 receptor was involved in clonal NK cell population-mediated cytotoxicity, we studied the effect of anti-CD160 mAb in three different well defined functional NK cell lines, namely NKL, NK92, and NK3.3, that were found to express significant levels of CD160 at their cell surface when cultured for 4 days in the absence of IL-2 (data not shown). Such NK lines only express a limited number of inhibitory receptors (Table 1) and therefore represent a good tool to study the specificity and the activatory function of CD160. Further, we used the target cell line K562 because it appeared to be an appropriate target cell to avoid dominant NCR effector cell triggering (18). We observed that addition of CL1-R2 mAb resulted in a strong inhibition of the IL-2-deprived NKL-, NK92-, and NK3.3-mediated cytotoxicity against K562 target cells (Fig. 2). It should be emphasized that, similar to circulating NK lymphocytes (Fig. 1B), when these cell lines were cultured in the presence of IL-2 before functional assay, their cytotoxic activity was only partially inhibited by CL1-R2 mAb (data not shown). These experiments performed with NK cell lines further demonstrate that CD160 is a unique receptor that triggers NK lysis mainly in an IL-2-free environment and suggest that the CD160 ligand was present in K562.
Table 1.
NK92, NKL, and NK3.3 markers/phenotype
| mAb
|
Ligand
|
NK cell lines | ||
|---|---|---|---|---|
| NK92 | NKL | NK3.3 | ||
| BY55, CL1-R2 | CD160 | ++ | ++ | ++ |
| EB6 | CD158a (KIR2DL1/S1) | − | − | − |
| GL183 | CD158b (KIR2DL2/S1) | − | − | ++ |
| DX9, Z27 | CD158e1/2 (KIR3DL1/S1) | − | − | − |
| PAX180 | CD158i (KIR2DS4) | − | − | − |
| HP-3B1 | CD94 | + | ++ | ++ |
| p25 | CD159a (NKG2A) | ++ | ++ | ++ |
| Z270 | NKG2A/C | ++ | ++ | ++ |
| VMP55 | CD85j (ILT2) | ++ | ++ | ++ |
| CRET56 | CD56 | ++ | − | +/− |
| CRET16 | CD16 | − | + | + |
| C.1.7.1, pp35 | CD244 (2B4) | ++ | ++ | ++ |
Flow cytometry analysis using indirect immunofluorescence labeling. −, <5%; +/−, <20%; +, <50%; ++, 100%.
Fig 2.
CD160 engagement triggers cytolytic activity in NKL, NK92, and NK3.3 cell lines. Shown is the cytotoxicity of NKL, NK92, and NK3.3 effector cells cultured without IL-2 against K562 in the absence of mAb (no mAb) or in the presence of mAb CL1-R2 (CD160) or control IgG1. Results are mean ± SD of triplicates and are representative of three independent experiments.
CD160 Triggers NK Cell-Mediated Cytotoxicity Through HLA-C Recognition.
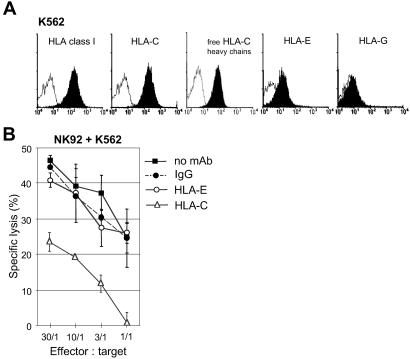
We previously reported (10) that CD160 receptor had a broad specificity for MHC class Ia/Ib ligands. Thus, although generally considered as HLA class I negative, we investigated whether some MHC class I molecules constitutively expressed on the K562 target cell surface could be responsible for CD160 triggering of NK cell-mediated cytotoxicity. Using flow cytometry analysis, we observed that W6/32 mAb constantly bound to K562 (Fig. 3A), confirming previous observations (19). We then examined the binding of several HLA class I locus-specific mAbs. We found that the B1.23.2 mAb clearly bound to K562 (Fig. 3A). Knowing that K562 did not transcribe HLA-A and -B (19), and that L31-specific anti-HLA-C mAb bound to K562, we demonstrated that only HLA-C molecules were expressed on this cell line. Further, genotype analysis revealed that HLA-Cw3 and -Cw5 were present in K562 cells (20). To a lesser extent, anti-HLA-E mAb also stained K562, whereas anti-HLA-G mAb did not (Fig. 3A). These data demonstrate that K562 cells constitutively express a significant amount of cell-surface HLA-C and -E.
Fig 3.
Engagement of CD160 by HLA-C, constitutively expressed by K562, triggers NK cell cytotoxicity. (A) K562 cells were analyzed by flow cytometry for surface expression of HLA class I using broad anti-HLA-class I (mAb W6/32), anti-HLA-C (mAb B1.23.2), anti-HLA-C free heavy chains (mAb L31), anti-HLA-E (mAb MEM-E/06), or anti-HLA-G (mAb 87G) (black profiles) followed by PE-labeled conjugate. Open profiles are Ig-isotype control stainings. Data are representative of several independent experiments. (B) Cytotoxicity of NK92 cultured without IL-2 against K562 in the absence of mAb (no mAb) or in the presence of mAb MEM-E/06 (HLA-E, 5 μg/ml), mAb B1.23.2 (HLA-C, 10 μg/ml), or Ig-isotype control (IgG). Results are mean ± SD of triplicates and are representative of several independent experiments.
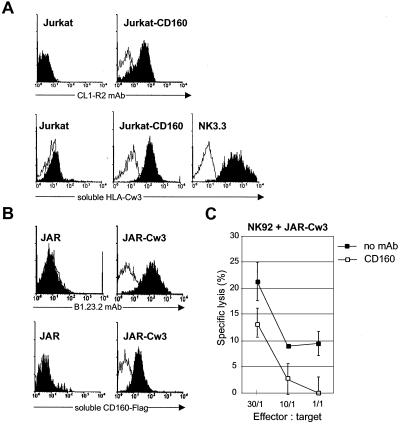
We then investigated whether MHC class I molecules present on K562 could trigger cytotoxic activity through CD160 interaction in IL-2-deprived NK cell lines. We demonstrated that incubation of K562 with the mAb B1.23.2 led to a drastic inhibition of cytotoxicity by the NK92 line, whereas the anti-HLA-E mAb had no effect (Fig. 3B). Similar results were obtained with NK3.3 (see Fig. 5D) and NKL (data not shown). These results indicate that HLA-C, but not HLA-E, expressed at the cell surface of K562 triggers CD160-induced NK cell cytotoxic function. The direct interaction of CD160 with HLA-C was further demonstrated with soluble recombinant HLA-Cw3 and CD160 molecules. First, we showed that soluble HLA-Cw3 specifically bound to Jurkat-CD160 transfectant, as well as to NK3.3 cells, which express high levels of CD160 (Table 1), but not to untransfected Jurkat (Fig. 4A). Second, we found that a soluble CD160-Flag protein specifically bound to HLA-Cw3-transfected JAR trophoblast cells but not to the parental JAR (Fig. 4B), which is HLA class I-negative (21). The functional consequences of the HLA-C/CD160 interaction was demonstrated by the finding that addition of CL1-R2 mAb resulted in a strong inhibition of the NK92-mediated cytotoxicity against JAR-Cw3 transfectant (Fig. 4C), whereas it did not modify the NK92-NCR-mediated lysis against JAR (ref. 22 and data not shown). Taken together these results demonstrate that CD160 receptor bound to HLA-Cw3 and that this ligation triggered cytotoxic activity.
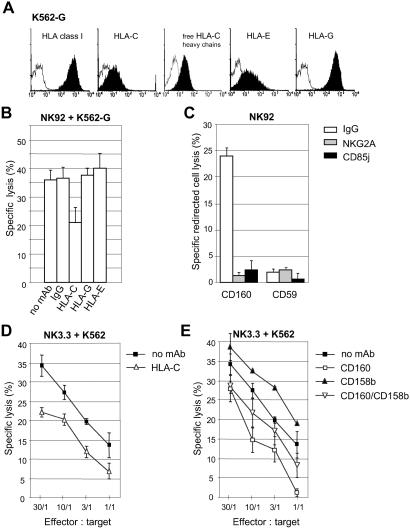
Fig 5.
CD160-induced NK92 cytotoxicity is not blocked by inhibitory receptors specific for HLA-E and -G, whereas HLA-C, through its interaction with CD158b, alters CD160-induced NK3.3 cytotoxicity. (A) Surface expression of HLA class I on K562-G transfectants was monitored with W6/32 (broad HLA class I), B1.23.2 (HLA-C), L31 (free HLA-C heavy chains), MEM-E/06 (HLA-E), or 87G (HLA-G) mAbs (black profiles). Open profiles are Ig-isotype control stainings. Data are representative of several independent experiments. (B) Cytotoxicity of NK92 cultured without IL-2 against K562-G target cells, either in the absence of mAb (no mAb) or in the presence of isotype control (IgG), mAb B1.23.2 (HLA-C), mAb 87G (HLA-G), or mAb MEM-E/06 (HLA-E). The effector-to-target cell (E:T) ratio was 10. Results are mean ± SD of triplicates and are representative of several independent experiments. (C) Redirected lysis assay of P815, using NK92 cells in the presence of anti-CD160 or -CD59 mAbs mixed with anti-CD159a or -CD85j mAb or with control IgG. The E:T ratio used was 25. Results are mean ± SD of triplicates and are representative of three independent experiments. (D) Cytotoxicity of NK3.3 cultured without IL-2 against K562 in the absence of mAb (no mAb) or in the presence of mAb B1.23.2 (HLA-C), or (E) mAb CL1-R2 (CD160), anti-CD158b (CD158b) mAb, or anti-CD158b together with anti-CD160 (CD160/CD158b) mAbs. Results are mean ± SD of triplicates and are representative of three independent experiments.
Fig 4.
Soluble recombinant HLA-Cw3 and CD160 proteins bind to JAR-Cw3 and Jurkat-CD160, respectively. (A Upper) By flow cytometry, mAb CL1-R2 stained Jurkat-CD160 but not Jurkat cells (black profiles). Open profiles are Ig-isotype control stainings. (A Lower) flow cytometry binding of soluble HLA-Cw3 cross-linked with B1.23.2 mAb, followed by incubation with PE-streptavidin on Jurkat-CD160 and NK.3.3 cells but not Jurkat (black profiles). Open profiles are control staining with B1.23.2 mAb and streptavidin-PE. (B) Flow cytometry showing binding of mAb B1.23.2 on JAR-Cw3 but not JAR (Upper, open profiles are Ig-isotype control staining), and of soluble CD160-Flag, after incubation with anti-Flag mAb followed by PE-labeled conjugate on Jurkat-CD160 and NK.3.3 cells but not Jurkat (Lower, open profiles are staining after incubation with control Jurkat supernatant). (C) Cytotoxicity of NK92 cultured without IL-2 against JAR-Cw3 target cells in the absence of mAb (no mAb) or in the presence of mAb CL1-R2 (CD160). Results are mean ± SD of triplicates and are representative of three independent experiments.
CD160/HLA-C-Mediated-NK Cytotoxicity Is Partially Controlled by Engagement of CD158b but Not of CD94/CD159a and CD85j Inhibitory Receptors with Their Respective Physiological Ligands.
We investigated whether different types of inhibitory NK receptors could interfere with CD160-mediated cytotoxicity. We analyzed the engagement of the Ig-like transcript CD85j and the lectin-like receptor complex CD94/CD159a, both present on the NK92 cell line (Table 1), by their respective ligands, namely HLA-G or HLA-C and -E (23). We used K562 transfected with HLA-G (K562-G) as target cells. Flow cytometry analysis revealed that, in addition to HLA-G, this transfectant also expressed low levels of both endogenous HLA-C and -E (Fig. 5A). We found that K562-G target cell line was as efficiently killed as the parental K562 cell line by NK92 (Fig. 5B), although the level of HLA-C was much lower on the former cell line surface. Moreover, such cytotoxic activity was strongly inhibited in the presence of soluble anti-CD160 mAb (data not shown). To determine which MHC class I molecule(s) was involved, we performed a cytolytic assay in the presence of different anti-HLA class I-specific mAbs. The results clearly showed that only the use of anti-HLA-C mAb resulted in a consistent inhibition of IL-2-deprived, NK92-mediated killing of K562-G (Fig. 5B). In contrast, no blocking effect was detected in the presence of mAb directed to HLA-G or -E (Fig. 5B), even at different E:T ratios (data not shown), suggesting that neither HLA-G nor HLA-E triggered CD160-induced cytotoxic activity. Importantly, as none of the mAbs specific for HLA-G or -E were capable of enhancing the cytotoxic activity mediated by NK92, these results further indicated that none of the MHC class Ib molecules, on interaction with their respective receptors, was able to provide sufficient negative signal to inhibit CD160-induced cytotoxicity. To demonstrate that both inhibitory receptors CD94/CD159a and CD85j were functional in the NK92 cell line, we performed a redirected cell lysis assay using P815 in the presence of low doses of anti-CD160 together with anti-CD85j, anti-CD159a, or isotype-matched mAbs. We observed in such experimental conditions that mAb-mediated cross-linking of CD85j or CD159a inhibited the redirected cell lysis induced by anti-CD160 mAb, whereas the isotype-matched antibody used in the same conditions did not modify the induced cytotoxicity (Fig. 5C). Note that as a control, mAb against the glycosyl phosphatidyl inositol (GPI)-linked CD59 molecule failed to trigger NK92 cytotoxic activity (Fig. 5C). Altogether, these data demonstrate that both CD85j and CD94/CD159a inhibitory receptors were functional. However, triggering with their respective physiological ligands did not interfere with CD160-mediated NK cell lysis.
To determine whether an HLA-Cw3-specific Ig-like inhibitory receptor could modulate CD160-induced cell cytotoxicity, we used the IL-2-deprived NK3.3 cell line, which expressed high amount of CD158b, in addition to CD85j, CD94/CD159a, and CD160 (Table 1), as well as CD158d (24). The results indicated that a NK3.3 cell line cultured without IL-2 for 4 days was capable of killing K562 expressing endogenous HLA-C molecules and that an anti-HLA-C mAb partially inhibited this cell-mediated cytotoxicity (Fig. 5D). Thus, in the NK3.3 cell, overall HLA-Cw3 molecules appear to trigger cytotoxicity through interaction with CD160. Failure of CD158b molecules to strongly inhibit cytotoxicity of NK3.3 cells against K562 target cells was not related to the expression of HLA-C molecules, because similar results were obtained with IFN-γ/TNF-α-treated K562 cells that expressed a high amount of HLA-C molecules (data not shown). We found that soluble anti-CD158b mAb was capable of inducing a slight but reproducible enhancement of NK3.3 cytotoxic activity against K562 (Fig. 5E). Further, addition of both anti-CD160 and -CD158b seemed to reduce the NK3.3 cytotoxicity inhibition obtained with anti-CD160 mAb alone (Fig. 5E). Importantly, we found that various concentrations of anti-CD158b mAb were able to inhibit NK3.3 anti-CD160 mAb-induced cytotoxicity in a redirected cell lysis assay (data not shown). Thus, these results demonstrate that CD158b molecules function as KIR2DL2 in NK3.3 cells by slightly reducing the HLA-Cw3-dependent NK3.3 cytotoxic activity mediated through CD160 molecules. It should be emphasized that even if the CD158b receptor is functional in NK3.3 cells, its inhibitory property is not sufficient to completely abrogate the triggering NK activity mediated through the engagement of CD160 molecules by HLA-C.
Taken together, these data demonstrate with physiological ligands that, in a cytokine-free environment, CD160 receptor/HLA-C interaction is capable of overriding inhibitory signals provided by specific MHC class Ib molecules through two structurally representative inhibitory receptors, namely CD94/CD159a and CD85j, and to partially override inhibitory signals provided by the HLA-C molecule through CD158b.
Discussion
In the present work, we used the CD56dim+CD16+CD160+ PB-NK or NK cell lines NK92, NKL, and NK3.3 cultured in the absence of IL-2 and the NK-sensitive target cell K562 expressing endogenous HLA-C and -E molecules and lacking other MHC class Ia and Ib alleles to demonstrate that HLA-C/CD160 interaction corresponded to a NK cell cytotoxicity-triggering pathway. The latter NK cell lines represented an important tool to investigate the function of CD160 NK receptor, because its expression is hardly detectable on long-termed NK cells cultured ex vivo in the presence of exogenous IL-2 (10). Further, such an effector/target cell system was found to be appropriate for study of the CD160-induced NK cell lysis and its regulation as it corresponded to the main pathway compared with NCR or NKG2D triggering pathways also used with the same effector NK cell lines but with other target cells (18). It is generally admitted that NK cell function is regulated primarily by inhibitory receptors that prevent NK cell lysis of target cells expressing MHC class I molecules. This view has now changed with the discovery of triggering NK receptors required for NK cell-mediated rejection of tumors and control of viral infection. Several MHC class I-independent activatory receptors have been described to date, including NCR and NKG2D (2, 5). Here, we described a MHC class I-dependent activating NK receptor expressed by all of the CD56dim+CD16+ PB-NK exhibiting spontaneous cytotoxic activity prior to any cytokine activation. CD160 receptor thus differs from the other already characterized MHC class I-specific triggering receptors, namely CD158c, -g, -h, -i, -j, and -e2, as well as CD94/NKG2C, which are clonally distributed within the circulating NK cell population (25). Further, CD160 exhibits several other unique properties, including its location on human chromosome 1 (26) and its restricted expression on resting NK cells (10). This is in contrast to the other NK cell activatory receptors, such as NKp44, which is absent from fresh NK cells and needs IL-2 or -15 stimulation to be expressed (27), and NKG2D, which is up-regulated by IL-15 (28). Finally, the Ig-like CD160 receptor is a glycosyl phosphatidyl inositol (GPI)-anchored cell surface molecule, whereas all of the other NK receptors described to date are linked to the cell membrane via transmembrane domains. Therefore, signaling through CD160 should be different from the lectin-like triggering receptors that use DAP12 or -10 (4). Preliminary results indicated that CD160 receptors associated with lck and LAT (unpublished data), similar to other receptors found in the lipid raft (29).
We have previously reported that CD160 bound to several MHC class Ia and Ib molecules (10). However, in the present study, we found that neither HLA-E nor HLA-G triggered efficient NK92 cell lysis through CD160 interaction because only the anti-HLA-C blocking mAb B1.23.2 inhibited cytotoxicity against K562 or K562-G transfected cells. Thus, CD160 triggers NK cell-mediated cytotoxicity through HLA-C recognition. This was confirmed by the use of soluble HLA-Cw3 molecules, which bound specifically to CD160 transfectants. Further, we demonstrated that soluble CD160 molecule bound to JAR-Cw3 transfectant, whereas it failed to bind the parental JAR cells, which lack expression of endogenous HLA class Ia and Ib. We found that NK92-mediated JAR-Cw3 cell lysis was inhibited in the presence of anti-CD160 mAb, whereas the same anti-CD160 mAb failed to block the NKG2-mediated JAR cell lysis. This clearly indicated that CD160 function could be achieved only when engaged by HLA-C and that in the absence of exogenous IL-2, only NKp46, expressed by NK92 (22), could be involved in the killing of JAR (22). The use of mAb-mediated cross-linking of CD85j or CD94/CD159a demonstrated that the CD160 triggering pathway is under the control of inhibitory receptors. However, we found that CD160-induced cell cytotoxicity is not turned off by CD85j and CD94/CD159a on ligation with HLA-G and -E, respectively, because none of the specific mAbs against these MHC class I molecules enhanced the CD160-induced cytotoxicity. It should be mentioned that expression of CD158d by a NK92 cell line recently reported (24) is very unlikely to trigger inhibitory signal after HLA-G interaction because the blocking anti-HLA-G mAb 87G did not modify CD160-induced cytotoxicity. The absence of detectable NK cell lysis inhibitory signaling mediated by CD94/CD159a or CD85j could not be attributed to the level of their respective ligand because neither up-regulation of HLA-E in IFNγ/TNFα-treated K562 (data not shown) nor the high amount of HLA-G in HLA-G-transfected cells resulted in the inhibition of CD160-induced cytotoxicity. In contrast to these MHC class Ib-specific receptors, the HLA-C-specific inhibitory receptor CD158b provided significant but reduced inhibitory signal to CD160 engagement in the CD158b-expressing NK3.3 cell line because soluble anti-CD158b slightly enhanced the level of cytotoxicity that is inhibited by anti-CD160 mAb. Further, the combination of both mAbs decreased the level of inhibition obtained with anti-CD160 alone. These results suggest that CD158b alone is not sufficient to prevent CD160-mediated NK cell cytotoxicity in vivo. Therefore, it is likely that additional inhibitory signals mediated by other MHC class I-specific receptors, such as CD158e1, CD158a, and CD158k (30), are required to avoid normal autologous cells from NK cell destruction.
The newly identified CD160/HLA-C interaction triggering activatory signal to circulating NK lymphocytes is likely to play a crucial role in the innate immunity against virally infected and malignant cells. It is striking that after HIV or cytomegalovirus infection, HLA-C, in contrast to HLA-A and -B, is not down-modulated by viral proteins (31). Thus, it appears that CD160 ligand (HLA-C) is still present in such virally infected cells, whereas the potential ligands for the KIR3D inhibitory receptors (HLA-A and -B) are diminished.
In conclusion, we report a triggering pathway of NK cell cytotoxicity, depending on HLA-C recognition by the CD160 receptor expressed in all circulating cytotoxic CD56dim+CD16+ cells. Further studies are needed to evaluate how and when this distinct triggering pathway is selected in vivo and whether this can be initiated independently of the NKG2D, NCR, and immunoreceptor tyrosine-based activation motif (ITAM)-bearing receptors.
Acknowledgments
We thank V. Horejsi for stimulating discussions. This work was supported by grants from Institut National de la Santé et de la Recherche Médicale, Association pour la Recherche sur le Cancer, Etablissement Français des Greffes, Universités Paul Sabatier and Paris XII, and Groupement des Entreprises Françaises dans la Lutte Contre le Cancer Paris Ile de France.
Abbreviations
KIR, killer cell Ig-like receptor
NCR, natural cytotoxicity receptor
NK, natural killer
PB-NK, peripheral blood NK
PE, phycoerythrin
References
- 1.Lanier L. L., Le, A. M., Civin, C. I., Loken, M. R. & Phillips, J. H. (1986) J. Immunol. 136, 4480-4486. [PubMed] [Google Scholar]
- 2.Moretta A., Bottino, C., Mingari, M. C., Biassoni, R. & Moretta, L. (2002) Nat. Immunol. 3, 6-8. [DOI] [PubMed] [Google Scholar]
- 3.Borrego F., Kabat, J., Kim, D. K., Lieto, L., Maasho, K., Pena, J., Solana, R. & Coligan, J. E. (2002) Mol. Immunol. 38, 637-660. [DOI] [PubMed] [Google Scholar]
- 4.Lanier L. L. (2001) Nat. Immunol. 2, 23-27. [DOI] [PubMed] [Google Scholar]
- 5.Cerwenka A. & Lanier, L. L. (2001) Immunol. Rev. 181, 158-169. [DOI] [PubMed] [Google Scholar]
- 6.Mandelboim O., Lieberman, N., Lev, M., Paul, L., Arnon, T. I., Bushkin, Y., Davis, D. M., Strominger, J. L., Yewdell, J. W. & Porgador, A. (2001) Nature 409, 1055-1060. [DOI] [PubMed] [Google Scholar]
- 7.Anumantha A., Bensussan, A., Boumsell, L., Christ, A., Blumberg, R., Voss, S., Patel, A., Robertson, M., Nadler, L. & Freeman, G. (1998) J. Immunol. 161, 2780-2790. [PubMed] [Google Scholar]
- 8.Maiza H., Leca, G., Mansur, I. G., Schiavon, V., Boumsell, L. & Bensussan, A. (1993) J. Exp. Med. 178, 1121-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bensussan A., Gluckman, E., el Marsafy, S., Schiavon, V., Mansur, I. G., Dausset, J., Boumsell, L. & Carosella, E. (1994) Proc. Natl. Acad. Sci. USA 91, 9136-9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal S., Marquet, J., Freeman, G. J., Tawab, A., Le Bouteiller, P., Roth, P., Bolton, W., Ogg, G., Boumsell, L. & Bensussan, A. (1999) J. Immunol. 162, 1223-1226. [PubMed] [Google Scholar]
- 11.Grassi F., Meneveri, R., Gullberg, M., Lopalco, L., Rossi, G. B., Lanza, P., De Santis, C., Brattsand, G., Butto, S., Ginelli, E., et al. (1991) J. Exp. Med. 174, 53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.André P., Vivier, E., et al. (2002) in Leucocyte Typing VII: White Cell Differentiation Antigens, eds. Mason, D., André, P., Bensussan, A., Buckley, C., Civin, C., Clark, E., de Haas, M., Goyert, S., Hadam, M. & Hart, D. (Oxford Univ. Press, New York), pp. 407–411.
- 13.Marti F., Xu, C. W., Selvakumar, A., Brent, R., Dupont, B. & King, P. D. (1998) Proc. Natl. Acad. Sci. USA 95, 11810-11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson M. J., Cochran, K. J., Cameron, C., Le, J. M., Tantravahi, R. & Ritz, J. (1996) Exp. Hematol. 24, 406-415. [PubMed] [Google Scholar]
- 15.Maki G., Klingemann, H. G., Martinson, J. A. & Tam, Y. K. (2001) J. Hematother. Stem Cell Res. 10, 369-383. [DOI] [PubMed] [Google Scholar]
- 16.Altman J. D., Moss, P. A. H., Goulder, P. J. R., Barouch, D. H., McHeyzer-Williams, M. G., Bell, J. I., McMichael, A. J. & Davis, M. M. (1996) Science 274, 94-96., and erratum (1998) 280, 1821. [DOI] [PubMed] [Google Scholar]
- 17.Zappacosta F., Borrego, F., Brooks, A. G., Parker, K. C. & Coligan, J. E. (1997) Proc. Natl. Acad. Sci. USA 94, 6313-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sivori S., Pende, D., Bottino, C., Marcenaro, E., Pessino, A., Biassoni, R., Moretta, L. & Moretta, A. (1999) Eur. J. Immunol. 29, 1656-1666. [DOI] [PubMed] [Google Scholar]
- 19.Johnson D. (2000) Hum. Immunol. 61, 389-396. [DOI] [PubMed] [Google Scholar]
- 20.Van den Elsen P. J., van der Stoep, N., Vietor, H. E., Wilson, L., van Zutphen, M. & Gobin, S. J. (2000) Hum. Immunol. 61, 850-862. [DOI] [PubMed] [Google Scholar]
- 21.Boucraut J., Hakem, R., Gauthier, A., Fauchet, R. & Le Bouteiller, P. (1991) Tissue Antigens 37, 84-89. [DOI] [PubMed] [Google Scholar]
- 22.Sivori S., Parolini, S., Marcenaro, E., Millo, R., Bottino, C. & Moretta, A. (2000) Hum. Immunol. 61, 1055-1058. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Botet M., Llano, M., Navarro, F. & Bellon, T. (2000) Semin. Immunol. 12, 109-119. [DOI] [PubMed] [Google Scholar]
- 24.Rajagopalan S., Fu, J. & Long, E. O. (2001) J. Immunol. 167, 1877-1881. [DOI] [PubMed] [Google Scholar]
- 25.Long E. & Rajagopalan, S. (2000) Semin. Immunol. 12, 101-108. [DOI] [PubMed] [Google Scholar]
- 26.Bensussan A. (2000) Protein Rev. Web 1, 72-73. [Google Scholar]
- 27.Vitale M., Bottino, C., Sivori, S., Sanseverino, L., Castriconi, R., Marcenaro, E., Augugliaro, R., Moretta, L. & Moretta, A. (1998) J. Exp. Med. 187, 2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts A. I., Lee, L., Schwarz, E., Groh, V., Spies, T., Ebert, E. C. & Jabri, B. (2001) J. Immunol. 167, 5527-5530. [DOI] [PubMed] [Google Scholar]
- 29.Horejsi V., Drbal, K., Cebecauer, M., Cerny, J., Brdicka, T., Angelisova, P. & Stockinger, H. (1999) Immunol. Today 20, 356-361. [DOI] [PubMed] [Google Scholar]
- 30.Moretta A., Biassoni, R., Bottino, C. & Moretta, L. (2000) Semin. Immunol. 12, 129-138. [DOI] [PubMed] [Google Scholar]
- 31.Tortorella D., Gewurz, B., Furman, M., Schust, D. & Ploegh, H. (2000) Annu. Rev. Immunol. 18, 861-926. [DOI] [PubMed] [Google Scholar]