Abstract
We have identified a gene located on chromosomes 21 that is expressed in normal and neoplastic prostate, and in normal testis, ovary, and placenta. We name this gene POTE (expressed in prostate, ovary, testis, and placenta). The POTE gene has 11 exons and 10 introns and spans ≈32 kb of chromosome 21q11.2 region. The 1.83-kb mRNA of POTE encodes a protein of 66 kDa. Ten paralogs of the gene have been found dispersed among eight different chromosomes (2, 8, 13, 14, 15, 18, 21, and 22) with preservation of ORFs and splice junctions. The synonymous:nonsynonymous ratio indicates that the genes were duplicated rather recently but are diverging at a rate faster than the average for other paralogous genes. In prostate and in testis, at least five different paralogs are expressed. In situ hybridization shows that POTE is expressed in basal and terminal cells of normal prostate epithelium. It is also expressed in some prostate cancers and in the LnCAP prostate cancer cell line. The POTE protein contains seven ankyrin repeats between amino acids 140 and 380. Expression of POTE in prostate cancer and its undetectable expression in normal essential tissues make POTE a candidate for the immunotherapy of prostate cancer. The existence of a large number of closely related but rapidly diverging members, their location on multiple chromosomes and their limited expression pattern suggest an important role for the POTE gene family in reproductive processes.
The publication of the human genome sequence has provided a new era for cancer research (1). With the help of bioinformatics and the EST and human genome sequence databases one can identify new genes that can be used as targets for cancer therapy or could be involved in essential biological pathways, which control growth, differentiation, or transformation of normal cells into cancer. Other experimental methods can be used to identify these genes, which include differential display (2), subtractive hybridization (3), serial analysis of gene expression (4), and microarray analysis (5). Our laboratory is interested in discovering genes that may be used for the therapy or diagnosis of prostate cancer. Although prostate cancer is one of the leading causes of death for men in the United States, little is known about the mechanisms and the genes involved. Moreover, identifying prostate-specific genes could aid in the early detection of prostate cancer. Over the past several years, we have developed a computer-based screening strategy to generate clusters of ESTs that are expressed in prostate cancer and/or in normal prostate but not in essential normal tissues (6). Using this approach we have identified several genes that are expressed in prostate cancer and in normal prostate but not in essential normal tissues (7–11). In this report, we describe the identification of a gene family POTE, which is selectively expressed in prostate, testis, ovary, and placenta, as well as in prostate cancer. The POTE family consists of at least 10 highly homologous genes located on chromosomes 2, 8, 13, 14, 15, 18, 21, and 22.
Materials and Methods
Primers.
Primers used in this study are as follows: T444, CAA TGC CAG GAA GAT GAA TGT GCG; T445, TCT CTG GCC GTC TGT CCA GAT AGAT; T455, GGT AGA CGC GAT CTG TTC GCT ACT; and T456, CCT AAG CTG TCC ACT GTA CTT AAA.
Dot Blot and Northern Blot Analysis.
Hybridizations on a human multiple tissue mRNA dot blot (RNA master blot, CLONTECH). The probe used is a PCR fragment generated by using primers T444 and T445 (Fig. 1) labeled with 32P by random primer extension (Lofstrand Laboratories, Gaithersburg, MD). POTE mRNA expression and transcript size were also examined on commercial multitissue Northern blot (MTN, CLONTECH). mRNA (2 μg per lane) was electrophoresed under denaturing conditions and subsequently transferred to a nylon membrane according to established procedures. Hybridizations were performed as described (11) by using the same probe used for the dot blot.
Fig. 1.
Schematic showing the genomic organization of a POTE gene. Two ESTs (BF675049 and BF676987), which match exons in the POTE gene, are from a prostate cDNA library. There are 11 exons (numbered 1–11) in the POTE gene.
Expression of POTE by PCR Analysis.
PCR was performed on cDNA from 16 different human tissues (Multiple Tissue cDNA panel from CLONTECH) following the manufacturer's instructions. The PCR conditions used are as follows: initial denaturation at 94°C for 35 sec, 30 cycles of denaturation at 94°C for 1 min, annealing at 65°C for 1 min, and elongation at 72°C for 2 min. The PCR primers used were T444 and T445 that should give a 400-bp fragment.
Cloning of the Full-Length cDNA.
Rapid amplification of cDNA ends (RACE) was performed on Marathon Ready prostate and testis cDNA (CLONTECH). Gene-specific primers T444 and T445 were used for the 3′- and 5′-RACE, respectively. Several individual clones from the RACE product were isolated and sequenced to establish the correct POTE sequence. Finally the full-length POTE was amplified from prostate cDNA by PCR by using primer pair T455 and T456. The PCR product was gel purified and cloned into the pCR2.1 TOPO vector (Invitrogen). The clones were identified by restriction digestion and sequenced by using Perkin–Elmer's rhodamine terminator sequencing kit (Perkin–Elmer).
In Vitro Transcription-Coupled Translation.
The in vitro translation of the POTE cDNA was examined in an in vitro transcription-coupled translation system (TNT, rabbit reticulocyte lysate system, Promega). 35S-Met (Amersham Pharmacia) was incorporated in the reaction for visualization of translated products. The reaction mixture was analyzed under reducing conditions on a polyacrylamide gel (4–20% Tris/Glycine, Bio-Rad) together with a prestained protein molecular weight marker (Bio-Rad). The gel was dried and subjected to autoradiography.
In Situ Hybridization.
Pretreatment of the tissue sections for in situ hybridization was performed as described (12). Biotinylated probes were prepared by using 1-kb 5′-end of POTE and U6 (250 bp) cDNA cloned in pBluescript II (+) plasmid. Biotinylated pBluescript II (+) with CD22 insert was used as a negative control. Probe labeling, hybridization, and washing conditions were similar to those described (11). Microscopic evaluation (bright-field) was performed by using a Nikon Eclipse 800 microscope.
Fluorescent In Situ Hybridization (FISH).
A bacterial artificial chromosome clone (RP11–279C9) containing POTE gene was purchased from Research Genetics (Huntsville, AL) and the DNA was prepared by Lofstrand Laboratories. The probe was labeled with biotin or digoxigenin (Random primed DNA labeling kit, Roche) and used for FISH of human metaphase chromosomes derived from methotrexate-synchronized normal peripheral lymphocytes. The posthybridization washing was carried under standard conditions. Digital-image acquisition, processing, and analysis as well as the procedure for direct visualization of fluorescent signals to banded chromosomes were done as described (13). Metaphases with specific hybridization signals were recorded, and slides were rehybridized for spectral karyotyping analysis (14) to unequivocally establish the identity of labeled chromosomes. The POTE signals were localized on look-up table (LUT)-inverted and contrast-enhanced digital images of 4′,6-diamidino-2-phenylindole-counterstained G-like banded chromosomes (13) with ≈400 bands resolution ideogram (ISHN1985).
Results
Computer Analysis and the Identification of a New Prostate-Specific EST Cluster, C34AG.
To identify prostate-specific genes by EST database mining, we have developed a computer-based screening strategy that has enabled us to identify a number of genes that are specifically expressed in normal prostate and prostate cancer (6). One such cluster, C34AG, consists of two ESTs (BF675049 and BF6987); both are from a prostate cDNA library (Fig. 1).
Expression Analysis of C34AG Cluster in Prostate and Other Normal Tissues.
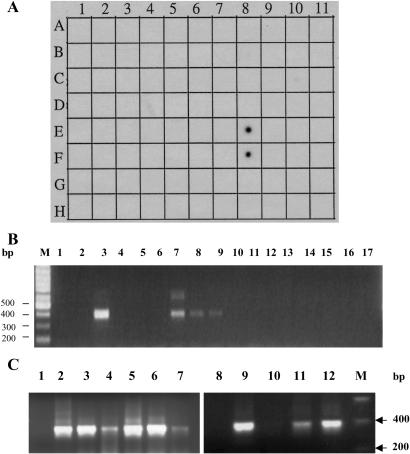
To investigate the expression of the C34AG cluster in different normal tissues, we performed a multitissue dot blot analysis by using a 32P-labeled PCR-generated DNA fragment as a probe. As shown in Fig. 2A, among the 76 different samples of normal and fetal tissue examined, C34AG is detected strongly in prostate (E8) and testis (F8) and weakly in placenta (B8) and in one leukemia cell line K-562 (C10). The expression of the C34AG cluster was not detected in any of the essential organs, including brain, heart, liver, and kidney. Dot blot analysis is not a very sensitive technique and can give a negative signal for genes, which are expressed at a very low level. To validate the dot blot result, we used a sensitive RT-PCR method on a panel of cDNAs isolated from many normal tissues including brain, heart, liver, lung, pancreas, and colon by using primers T444 and T445 shown in Fig. 1. As shown in Fig. 2B, a specific band ≈400 bp in size is detected in testis (lane 3), prostate (lane 7), and placenta (lane 8), and very weakly in ovary (lane 9). Because of its expression in prostate, ovary, testis, and placenta, we name this gene POTE.
Fig. 2.
Specificity of POTE mRNA expression. (A) RNA hybridization of a multiple tissue dot blot containing mRNA from 76 normal human cell types or tissues by using a cDNA probe. Strong expression is observed in prostate (E8) and testis (F8). There is no detectable expression in brain (A1), heart (A4), kidney (A7), liver (A9), lung (A8), and colon (A6). (B) PCR on cDNAs from 16 different human tissues. The expected size of the PCR product is 400 bp. A specific 400-bp PCR product is detected in testis (lane 3), prostate (lane 7), placenta (lane 8), and ovary (lane 9). (C) RT-PCR analysis of RNAs from normal prostate, prostate cancer, and prostate cancer cell lines LnCAP and PC3. Lanes: 1 and 8, negative control for PCR; 2–4, three prostate cancer samples; 5–7 and 11, normal prostate samples; 9, LnCAP; 10, PC3; 12, testis. A specific band of 400 bp in size is detected in all lanes except negative controls and PC3.
Cloning of the Full-Length POTE cDNA.
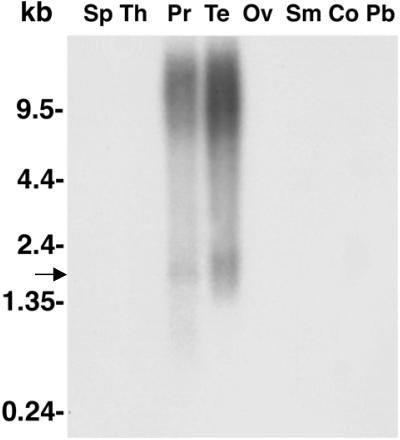
To determine the transcript size of POTE, we performed an analysis of a Northern blot containing mRNAs from many different tissues including prostate and testis with the same probe used for the dot blot analysis. As shown in Fig. 3, a band of ≈2.0 kb in size is detected in the prostate and testis lanes. The high molecular weight bands, which are detected by the probe in both prostate and testis, are frequently found in RNA extracted from tissues and are often incompletely spliced variants of the transcript (15). To isolate the full-length cDNA for POTE we used a 5′- and 3′-RACE PCR method and isolated a clone of 1,826 bp in size. Complete nucleotide sequence of the cDNA reveals that it has an ORF of 584 aa (Fig. 4). The nucleotide sequence analysis of the full-length cDNA indicates that it is a new gene. Amino acid sequence analysis of the predicted gene product by using the pfam database (16) reveals that it has an ankyrin repeat domain (see Discussion).
Fig. 3.
Northern blot analysis of POTE transcript in different normal tissues. Human multiple tissue Northern blot was probed with the same PCR probe used in the dot blot experiment. The only detectable signal is in prostate (Pr) and testis (Te) lanes. In both samples there is a weak band of ≈2 kb (arrow) in size and a smear at the high molecular weight region (which is probably the unprocessed nuclear RNA). There is no detectable signal in spleen (Sp), thymus (Th), ovary (Ov), small intestine (Sm), colon (Co), and peripheral blood leukocytes (Pb).
Fig. 4.
Amino acid sequence of POTE protein. The blue-and green-colored residues are the N-terminal three repeats of 37 residues each. The red colored residues are the seven ankyrin repeats. The prediction for the ankyrin repeat is weak for the first and the last members. The putative membrane-spanning region is underlined.
POTE Paralogs in Different Chromosomes.
Sequence alignment of the POTE cDNA with the human genome sequence by using the GoldenPath genome browser shows a 99.8% match with exons at chromosome 21q11.2. The gene is comprised of 11 exons and 10 introns with consensus splice donor-acceptor sequences at each intron–exon junction. Further analysis shows that there are at least nine more significant matches of POTE sequence in the human genome (Table 1). Four matches are in chromosome 2 (three at q22.2 and one at q14.3), two in chromosome 8 (p11.1 and q11.1), and one each in 14 (q11.2), 15 (q11.2), and 22 (q11.1). In most cases the exons and the splice sites are well conserved. When the DNA sequences are compared to that of POTE, there is a 90–98% sequence identity.
Table 1.
Alignment of the POTE sequence at different chromosomal locations
| Chro
|
Strand
|
Chromosome | Score
|
CDNA | Identity (%)
|
||
|---|---|---|---|---|---|---|---|
| Start | End | Start | End | ||||
| 22 | + | 11645620 | 11677028 | 1808 | 0 | 1826 | 99.8 |
| 15 | + | 18715046 | 18746092 | 1738 | 0 | 1826 | 98.1 |
| 2 | + | 140937343 | 141259911 | 1487 | 0 | 1779 | 92.7 |
| 14 | − | 17309857 | 17339582 | 1348 | 0 | 1585 | 93.2 |
| 2 | + | 141927000 | 141961449 | 1346 | 0 | 1743 | 92.5 |
| 2 | − | 124217411 | 124260895 | 1344 | 0 | 1585 | 93.0 |
| 8 | − | 43962699 | 44033444 | 1068 | 10 | 1825 | 89.3 |
| 22 | − | 13206523 | 13232594 | 976 | 571 | 1743 | 92.7 |
| 2 | − | 140822569 | 140835195 | 928 | 0 | 1188 | 93.8 |
| 8 | + | 47053750 | 47094566 | 433 | 1107 | 1825 | 89.1 |
The alignments were generated by using blat (17) and the ucsc genome assembly (“GoldenPath” at http://genome.ucsc.edu/index.html; ref. 18). Each line is a hit to a chromosomal location. The chromosome number, the direction of the alignment, and the absolute address of the hitting region are indicated in the first through the fourth columns. The fifth column gives the blat alignment score. In many cases, not all of the 1,826 bp of the cDNA align to the genome sequence. The beginning and ending base pair of the POTE cDNA that align are indicated in the sixth and seventh columns. The last column gives the percent identity of the matches.
Localization of Dispersed POTE Sequences in the Human Genome by FISH.
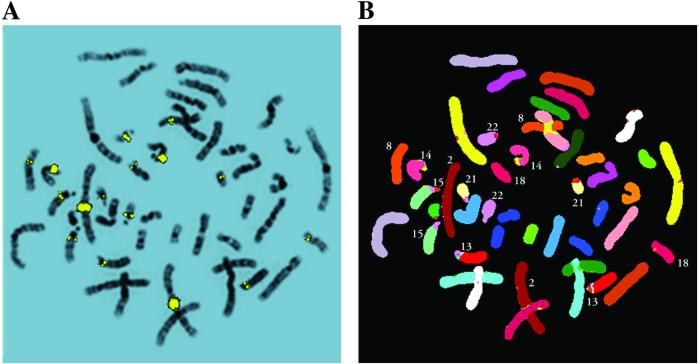
Because the human genome sequence is still incomplete and there are some gaps in most of the chromosomes, we validated our computer analysis data experimentally by performing FISH with a biotin-labeled bacterial artificial chromosome DNA from chromosome 2 corresponding to the genomic region of the POTE. A total of eight sites of hybridization were detected by FISH on chromosomes 2, 8, 13, 14, 15, 18, 21, and 22 with a bacterial artificial chromosome probe labeled with biotin and under standard hybridization conditions. Thirty randomly selected metaphases from two FISH experiments with cells derived from different peripheral leukocytes cultures, showed a fluorescent signal on pairs of the same chromosomes. From 30 spreads with informative signal, 15 were selected and examined sequentially by spectral karyotyping, for individual chromosome identification, and by G-banding for signal localization. All metaphases had signals on chromosomes 2, 14, and 22; >70% on chromosomes 13, 15 and 21; >40% exhibited signal on chromosome 18, and >30% on chromosome 8. The signal on chromosome 8 was the weakest one. The most intense signals were observed on chromosomes 2, 14, and 22, weaker and smaller on chromosomes 13, 15, 18, and 21, whereas chromosome 8 showed the weakest signal. With the exception of chromosome 2 where a strong signal spans region 2q13-q22, and chromosome 18 where a signal is located at 18p11.2, all other signals were localized at pericentromeic regions 13q11, 14q11, 15q11, 21q11, and 22q11 where we assigned POTE sequences on human chromosomes. FISH and spectral karyotyping hybridizations of the same representative metaphase with signals on all eight sites are shown in Fig. 5.
Fig. 5.
FISH analysis of POTE gene. (A) Digital images of normal human chromosomes hybridized with a biotinylated bacterial artificial chromosome probe for POTE gene counterstained with 4′,6-diamidino-2-phenylindole. Double symmetrical fluorescent signals are distributed on seven chromosome pairs. The signal is localized on G-banded chromosomes generated by contrast enhancement and LUT inversion of the 4′,6-diamidino-2-phenylindole-stained chromosomes. (B) Spectral karyotype of the same metaphase as in A for precise identification of chromosomes with hybridization signal.
POTE Paralogs from Different Chromosomes Are Expressed in Prostate.
Because there are so many paralogs of the POTE gene in different chromosomes, we investigated how many are expressed in prostate and testis. Using primer pair T455 and T445, we cloned and sequenced a 1.0-kb region of POTE from prostate and testis cDNA by using primers that would amplify all of the forms of POTE. Analysis of >24 individual clones and subsequent alignment with the human genome sequence indicates that POTE RNAs are transcribed from chromosomes 2, 14, 15, 21, and 22 in both prostate and testis.
The synonymous (Ks) and nonsynonymous (Ka) nucleotide substitution rates between each pair of POTE and three of its paralogous sequences in chromosomes 2, 14, and 15 were calculated by using the Pamilo-Bianchi-Li method (19, 20). These values are listed in Table 2. Excluding the exceptionally conserved chromosome 15 and 21 pair, the Ks values are nearly the same and average to 0.1. Assuming an average rate of 2.5 synonymous substitutions per billion years for humans (21), the Ks value indicates that the duplication of these genes occurred ≈20 million years ago. The average Ka/Ks ratio is 0.60 (excluding the 21–15 pair, for which the Ka/Ks ratio is unreliable because of the small number of substitutions). The fact that Ka is less than Ks indicates that “purifying” selection is operating to conserve the protein sequence of the original gene. However, the ratio is substantially larger than the average ratio of 0.44 or 0.45 reported for mammalian paralogs (21, 22), suggesting that these genes are evolving faster than most other paralogs.
Table 2.
The Ka (upper right triangle) and Ks (lower left triangle) substitutions per site for each pair of the four POTE paralogs at the chromosomes indicated
| Chr. 2 | Chr. 14 | Chr. 15 | Chr. 21 | |
|---|---|---|---|---|
| Chr. 2 | 0.04 | 0.06 | 0.07 | |
| Chr. 14 | 0.08 | 0.06 | 0.07 | |
| Chr. 15 | 0.12 | 0.10 | 0.02 | |
| Chr. 21 | 0.11 | 0.09 | 0.01 |
Chr., chromosome.
POTE mRNA Is Expressed in Prostate Cancer and Is Localized in Epithelium of Normal Prostate and Prostate Cancer.
To determine whether the POTE gene is expressed in prostate cancer, we performed an RT-PCR by using RNAs isolated from normal prostate and prostate cancers. As shown in Fig. 2C, all three prostate cancer samples tested have POTE expression. It should be noted that the prostate cancer samples are often associated with normal prostate tissues. To determine the expression of POTE in prostate cancer cell lines, we performed an RT-PCR analysis by using RNA from LnCAP and PC3 cells. A specific band of 400 bp was detected in the androgen-responsive LnCAP cell line but not in the androgen-unresponsive PC3 (Fig. 2C).
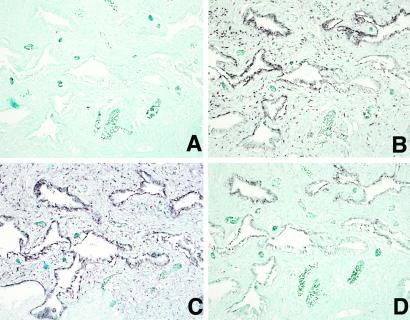
The cell types that express the POTE mRNA in normal prostate were determined by in situ hybridization with biotin-labeled POTE cDNA. As shown in Fig. 6D, POTE mRNA is expressed in the basal and terminal epithelial cells of normal prostate epithelium and in prostate cancer cells. There is no detectable signal in cells of the stromal compartment of the tissue. In comparison, U6 RNA is found in both epithelial cells and stromal cells of the prostate (Fig. 6B) whereas TARP is only in the epithelial cells (Fig. 6C).
Fig. 6.
In situ hybridization of prostate tissue sections probed with: CD22 control probe (A), note absence of signal; U6 (B); TARP (C); and POTE (D).
In Vitro Transcription and Translation of the POTE cDNA.
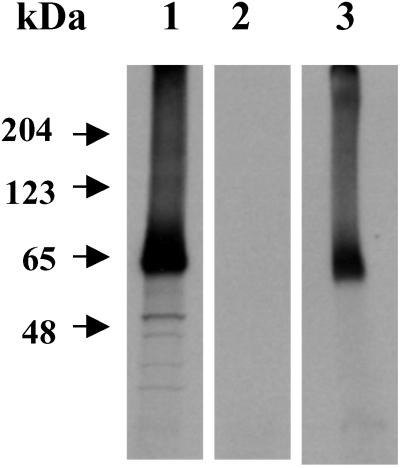
The POTE cDNA isolated from prostate has a predicted ORF of 584 aa with a calculated molecular mass of 66.4 kDa. To determine the actual size of the protein encoded by this cDNA, in vitro transcription and translation was performed. SDS/PAGE analysis and fluorography of the translated product show that the POTE cDNA encodes a protein product of ≈66 kDa in size (Fig. 7, lane 3) and that the size of the protein product is in agreement with the predicted ORFs of the cDNA.
Fig. 7.
Analysis of the protein encoded by POTE mRNA. POTE cDNA was transcribed in vitro with T7 RNA polymerase, and the RNA was translated with rabbit reticulocyte lysate in the presence of [35S]methionine. The translated products were analyzed by SDS/PAGE and fluorography. Lane 1, luciferase cDNA as positive control; lane 2, empty vector control; and lane 3, POTE cDNA.
Discussion
Using the EST database and the human genome browser as a guide, we identified a previously uncharacterized gene, POTE, which is expressed in prostate (both normal and cancer), testis, ovary, and placenta. Full-length POTE, a member of this gene family, encodes a protein of 66.4 kDa. There are several closely related POTE genes that are located on different chromosomes and are actively transcribed in both prostate and testis.
POTE Encodes a New Protein with Seven Ankyrin Repeats.
The longest ORF of POTE cDNA encodes a 66.4-kDa protein with 584 aa. A blastp (23) run against the ncbi-nr database resulted in many hits, of which the first two (XP•063686 and XP•063387) are predicted proteins from genomic sequences, which coincide with the regions of chromosomes 15 and 14, given in Table 1. In addition, there were hits for ankyrin repeat containing proteins. An analysis of the query sequence against the pfam database (16, http://pfam.wustl.edu/hmmsearch.shtml) indicates that the protein contains five to seven ankyrin repeats (residues 138–369, Fig. 4). Ankyrin repeats are tandemly repeated modules of ≈33 aa each. They have been identified in >600 different, functionally diverse proteins and have been implicated in mediating protein–protein interactions. A blastp run against the ncbi-pdb database lists bcl-3 (1k1a) as the top hit, which is an NF-κB-binding protein that contains seven ankyrin repeats. Other high scoring hits were the ankyrin repeat domains of the transcription regulator GA-binding protein α/β (1awc), cyclin-dependent kinase inhibitor protein P19ink4d (1bd8), and a p53-binding protein (1ycs), all of which contain four ankyrin repeats. Two transmembrane prediction programs (24, 25) predict a transmembrane region for the 19 residues (307–325) with a high score. This region is in between, and partly overlaps with, the fifth and the sixth predicted ankyrin repeats.
The C-terminal domain (residues 370–584) is predicted to be mainly α-helical. Three different fold-recognition programs (26–28), gave different lists of predicted protein structures, but all of them belonged to the “all α” class according to the structural classification of proteins (SCOP) classification (29). One of these, 1cun, the common hit from the bioinbgu (28) and 3d-pssm (26) programs, belongs to the α-spectrin family. Spectrin α/β heterodimers are the main constituents of the cytoskeleton, which is anchored to the plasma membrane by means of protein complexes that include ankyrins (30). At the N-terminal end of the protein, there are three repeats of 37 peptide units at residues 20–56, 57–93, and 94–130. There are clustered cysteines and histidines in the repeats, but the arrangement of these residues does not conform to any of the known Zn-finger domains.
Paralogs of POTE That Are Expressed from Different Chromosomes Might Evolve by Gene Duplication.
The POTE cDNA sequence we determined aligns almost 100% to human chromosome 21q11.2. It also aligns to chromosomes 2, 8, 14, 15, and 22 with a significant homology (90–98%). Our FISH analysis confirms the presence of POTE variants in the above mentioned chromosomes. In addition FISH analysis identified two more chromosomes (13 and 18) where POTE variants are located.
POTE is a new and interesting example of gene duplication and dispersion in evolution. All seven sites of hybridization at 2q13-q22, 13q11, 14q11.2, 15q11.218, p11.2, 21q11.1, and 22q11.1 correspond with the location of the same family SF2 of α satellite DNA (31, 32). A similar distribution has been demonstrated for keratinocyte growth factor (KGF) gene (33). The site on chromosome 2 in this study is the only one outside pericentromeric regions. Human chromosome 2 is the result of ancestral telomeric fusion of chromosomes 11 and 12 (in gorilla) or 12 and 13 (in chimpanzee) and this accounts for the chromosome number reduction from 24 pairs in great apes to 23 in humans (34, 35). The fusion occurred at human band 2q13, and resulted in loss of distal heterochromatic segments, accompanied or followed by inactivation or elimination of one of the ancestral centromeres that carried alphoid sequences at human band 2q21 (36). POTE sequences are localized within this region of chromosome 2. α-Satellite DNA, highly repetitive short DNA segments arranged in tandem arrays, has been implicated in genomic rearrangements (31) as well as chromosome translocations in cancer cells (31, 37).
The initial sequencing and analysis of the human genome revealed that there are (numerous) large blocks of genome sequence that share a high degree of sequence identity (>90%). These blocks range in size from a few kilobase to hundreds of kilobase and can include exonic and intronic sequences (38). More recently, Eichler and coworkers (39, 40) have systematically analyzed the human genome for blocks of highly similar sequences that are duplicated across multiple chromosomes. They have identified 169 large regions with highly similar duplications of which 24 are associated with genetic diseases. In addition, their analysis suggests that these duplications in the pericentromeric regions have emerged relatively recently ≈40 million years of human evolution.
POTE, a Potential Candidate for Prostate Cancer Immunotherapy.
The specificity of POTE expression and the fact that it is expressed in prostate cancer samples from different individuals makes POTE an attractive candidate for targeted therapy for prostate cancer. At this point it is not clear where the POTE protein is localized in the cell. Analysis of the POTE amino acid sequence indicates that it has short potentially membrane-spanning regions but no signal sequence is present to help insert the protein into the membrane. These findings indicate POTE could be a valuable diagnostic marker as well as a target for Ab- or vaccine-based prostate cancer therapies.
Other Prostate-Specific Transcripts in the POTE Locus.
We have identified another prostate-specific noncoding cDNA, C34, which is localized at 22q11 (also known as the Cat-Eye Syndrome region) and 14q11, at the same position as POTE paralogs but transcribed from the opposite direction. Therefore, C34 might play a regulatory role in expression of POTE paralogs from this region.
Conclusion
The finding that there are many POTE paralogs on different chromosomes that encode RNAs with nearly identical ORFs indicates that a strong selective pressure must maintain expression of these POTE genes. This, together with their selective expression in reproductive organs, suggests an important role for POTE in reproductive processes.
Acknowledgments
We thank Dr. Evan E. Eichler for reviewing the manuscript, Dr. Igor Rogozin for the calculation of the Ka and Ks values, and Ms. Anna Mazzuca for editorial assistance.
Abbreviations
FISH, fluorescence in situ hybridization
POTE, gene expressed in prostate, ovary, testis, and placenta
Ks, synonymous
Ka, nonsynonymous
References
- 1.International Human Sequencing Consortium (2001) Nature 409, 860-920.11237011 [Google Scholar]
- 2.Liang P. & Pardee, A. B. (1992) Science 257, 967-971. [DOI] [PubMed] [Google Scholar]
- 3.Hara T., Harada, N., Mitsui, H., Miura, T., Ishizaka, T. & Miyajima, A. (1994) Blood 84, 189-199. [PubMed] [Google Scholar]
- 4.Velculescu V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 276, 1268-1272. [DOI] [PubMed] [Google Scholar]
- 5.Schena M., Shalon, D., Davis, R. W. & Brown, P. O. (1995) Science 270, 467-470. [DOI] [PubMed] [Google Scholar]
- 6.Vasmatzis G., Essand, M., Brinkmann, U., Lee, B. & Pastan, I. (1998) Proc. Natl. Acad. Sci. USA 95, 300-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkmann U., Vasmatzis, G., Lee, B., Yerushalmi, N., Essand, M. & Pastan, I. (1998) Proc. Natl. Acad. Sci. USA 95, 10757-10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Essand M., Vasmatzis, G., Brinkmann, U., Duray, P., Lee, B. & Pastan, I. (1999) Proc. Natl. Acad. Sci. USA 96, 9287-9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolfgang C. D., Essand, M., Vincent, J. J., Lee, B. & Pastan, I. (2000) Proc. Natl. Acad. Sci. USA 97, 9437-9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsson P., Bera, T. K., Essand, M., Kumar, V., Duray, P., Vincent, J., Lee, B. K. & Pastan, I. (2001) Prostate 48, 231-241. [DOI] [PubMed] [Google Scholar]
- 11.Bera T. K., Maitra, R., Iavarone, C., Salvatore, G., Kumar, V., Vincent, J. J., Sathyanarayana, B. K., Duray, P., Lee, B. K. & Pastan, I. (2002) Proc. Natl. Acad. Sci. USA 99, 3058-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar V. & Collins, F. H. (1994) Insect Mol. Biol. 3, 41-47. [DOI] [PubMed] [Google Scholar]
- 13.Zimonjic D. B, Rezanka, L. & Popescu, N. C. (1995) Cancer Genet. Cytogenet. 80, 100-102. [DOI] [PubMed] [Google Scholar]
- 14.Zimonjic D. B., Keck-Waggoner, C. L. & Popescu, N. C. (2001) Leukemia 15, 1582-1588. [DOI] [PubMed] [Google Scholar]
- 15.Yerushalmi N., Keppler-Hafkemeyer, A., Vasmatzis, G., Liu, X. F., Olsson, P., Bera, T. K., Duray, P., Lee, B. K. & Pastan, I. (2001) Gene 265, 55-60. [DOI] [PubMed] [Google Scholar]
- 16.Bateman A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M. & Sonnhammer, E. L. (2002) Nucleic Acids Res. 30, 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent W. J. (2002) Genome Res. 12, 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M. & Haussler, D. (2002) Genome Res. 12, 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pamilo P. & Bianchi, N. O. (1993) Mol. Biol. Evol. 10, 271-281. [DOI] [PubMed] [Google Scholar]
- 20.Li W. H. (1993) J. Mol. Evol. 36, 96-99. [DOI] [PubMed] [Google Scholar]
- 21.Lynch M. & Corney, J. S. (2000) Science 290, 1151-1155. [DOI] [PubMed] [Google Scholar]
- 22.Kondrashov F. A., Rogozin, I. B., Wolf, Y. I. & Koonin, E. V. (2002) Genome Biol. 3, research 0008.1-0008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claros M. G. & von Heijne, G. (1994) Comput. Appl. Biosci. 10, 685-686. [DOI] [PubMed] [Google Scholar]
- 25.Cserzo M., Wallin, E., Simon, I., von Heijne, G. & Elofsson, A. (1997) Protein Eng. 10, 673-676. [DOI] [PubMed] [Google Scholar]
- 26.Kelley L. A., MacCullum, R. M. & Sternberg, M. J. (2000) J. Mol. Biol. 299, 499-520. [DOI] [PubMed] [Google Scholar]
- 27.Jones D. T. (1999) J. Mol. Biol. 287, 797-815. [DOI] [PubMed] [Google Scholar]
- 28.Fisher D. (2000) in Proceedings of the Fifth Symposium on Bio-Computing, Honolulu, Hawaii, 4–9 January, 2000, eds. Altman, R. B., Dunker, A. K., Lauderdale, K. & Klein, T. E. (World Scientific, Teaneck, NJ), pp. 116–127.
- 29.Murzin A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995) J. Mol. Biol. 247, 536-540. [DOI] [PubMed] [Google Scholar]
- 30.Alberts B., Bray, D., Lewis, J., Raff, M., Roberts, K. & Watson, J. D., (1994) Molecular Biology of the Cell (Garland, New York), pp. 491–493.
- 31.Alexandrov I. A., Mitkevich, S. P. & Yurov, Y. B. (1988) Chromosoma 96, 443-453. [DOI] [PubMed] [Google Scholar]
- 32.Alexandrov I. A., Mashkova, T. D., Akopian, T. A., Medvedev, L. I., Kisselev, L. L., Mitkevich, S. P. & Yurov, Y. B. (1991) Genomics 11, 15-23. [DOI] [PubMed] [Google Scholar]
- 33.Zimonjic D. B., Kelley, M. J., Rubin, J. S., Aaronson, S. A. & Popescu, N. C. (1997) Proc. Natl. Acad. Sci. USA 94, 11461-11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jauch A., Wienberg, J., Stanyon, R., Arnold, N., Tofanelli, S., Ishida, T. & Cremer, T. (1992) Proc. Natl. Acad. Sci. USA 89, 8611-8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ijdo J. W., Baldini, A., Ward, D. C., Reeders, S. T. & Wells, R. A. (1991) Proc. Natl. Acad. Sci. USA 88, 9051-9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baldini A., Ried, T., Shridhar, V., Ogura, K., D'Aiuto, L., Rocchi, M. & Ward, D. C. (1993) Hum. Genet. 90, 577-583. [DOI] [PubMed] [Google Scholar]
- 37.Wong N., Lai, P., Pang, E., Leung, T. W., Lau, J. W. & Johnson, P. J. (2000) Hepatology 32, 1060-1068. [DOI] [PubMed] [Google Scholar]
- 38.Eichler E. E. (2001) Trends Genet. 17, 661-669. [DOI] [PubMed] [Google Scholar]
- 39.Samonte R. V. & Eichler, E. E. (2002) Nat. Genet. 3, 65-72. [DOI] [PubMed] [Google Scholar]
- 40.Bailey J. A., Gu, Z., Clark, R. A., Reinert, K., Samonte, R. V., Schwartz, S., Adams, M. D., Myers, E. W., Li, P. W. & Eichler, E. E. (2002) Science 297, 1003-1007. [DOI] [PubMed] [Google Scholar]