Abstract
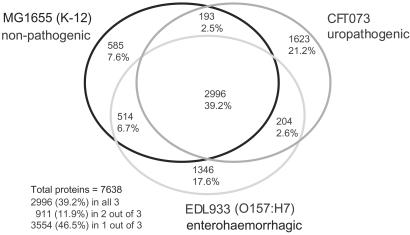
We present the complete genome sequence of uropathogenic Escherichia coli, strain CFT073. A three-way genome comparison of the CFT073, enterohemorrhagic E. coli EDL933, and laboratory strain MG1655 reveals that, amazingly, only 39.2% of their combined (nonredundant) set of proteins actually are common to all three strains. The pathogen genomes are as different from each other as each pathogen is from the benign strain. The difference in disease potential between O157:H7 and CFT073 is reflected in the absence of genes for type III secretion system or phage- and plasmid-encoded toxins found in some classes of diarrheagenic E. coli. The CFT073 genome is particularly rich in genes that encode potential fimbrial adhesins, autotransporters, iron-sequestration systems, and phase-switch recombinases. Striking differences exist between the large pathogenicity islands of CFT073 and two other well-studied uropathogenic E. coli strains, J96 and 536. Comparisons indicate that extraintestinal pathogenic E. coli arose independently from multiple clonal lineages. The different E. coli pathotypes have maintained a remarkable synteny of common, vertically evolved genes, whereas many islands interrupting this common backbone have been acquired by different horizontal transfer events in each strain.
The bacterium Escherichia coli is one of the best and most thoroughly studied free-living organisms. It is also a remarkably diverse species because some E. coli strains live as harmless commensals in animal intestines, whereas other distinct genotypes including the enteropathogenic, enterohemorrhagic, enteroinvasive, enterotoxigenic, and enteroaggregative E. coli cause significant morbidity and mortality as human intestinal pathogens. Extraintestinal E. coli are another varied group of life-threatening pathogens of this manifestly versatile species. This latter group of pathogens include distinct clonal groups responsible for neonatal meningitis/sepsis and urinary tract infections. The uropathogenic group is responsible for 70–90% of the 7 million cases of acute cystitis and 250,000 cases of pyelonephritis reported annually in the United States (1). The extraintestinal E. coli differ from the diarrheal pathogens because they can behave as either harmless human intestinal inhabitants or serious pathogens when they enter the urinary tract, bloodstream, or cerebrospinal fluid. To begin to understand the genetic bases for pathogenicity and the evolutionary diversity of E. coli, we present here the genome sequence of E. coli CFT073, a pathogenic strain isolated from the blood of a woman with acute pyelonephritis (2) and compare it with the genome sequences of enterohemorrhagic E. coli strain EDL933 and the nonpathogenic laboratory strain MG1655.
Materials and Methods
Clones and Sequencing.
CFT073 (serotype O6:H1:K?) was isolated at the University of Maryland Hospital (2). The sequence of the K-antigen regions is similar to the K5 type but is also consistent with K2 (not sequenced), which was assigned recently to CFT073 (3). The sequenced strain has been deposited at the American Type Culture Collection (accession no. 700928). Whole-genome libraries in M13Janus and pBluescript were prepared from genomic DNA as described (4, 5). Random clones were sequenced by using dye-terminator chemistry and data collected on Applied Biosystems ABI377 and 3700 automated sequencers. Sequence data were assembled by SEQMANII (DNASTAR, Madison, WI). Finishing used sequencing of opposite ends of linking clones, several PCR-based techniques, and primer walking. A whole-genome XhoI optical map permitted ordering of contigs and confirmation of contig structure during the assembly process as well as providing an independent physical map of the whole genome (6).
Sequence Analysis and Annotation.
The genome sequence was annotated in the multiuser, web-based annotation environment called magpie (7). This system used glimmer to define ORFs (8). Predicted proteins were searched against the nonredundant database by using BLAST (9). magpie assigned automatic annotations for all ORFs, which then were checked individually and corrected. These formed the basis for the GenBank submission. The island annotations contain unique identifiers in the form CI no., for islands of all sizes. Orthology was inferred when matches for CFT073 genes in either the MG1655 or EDL933 database exceeded 90% identity, alignments included at least 90% of both genes, and the MG1655 and EDL933 genes did not have an equivalent match elsewhere in the CFT073 genome. Genome comparisons were carried out by a modification of the method used to compare EDL933 and MG1655 (4).
Results
CFT073 Genome Organization Relative to Other E. coli.
The assembly of DNA sequences from a shotgun library of CFT073 DNA fragments, combined with PCR strategies and primer walking experiments for finishing, resulted in a circular, 5,231,428-bp chromosomal sequence with seven times coverage. An XhoI restriction fragment optical map confirmed the circular assembly of the genome. The principal features of the CFT073 genome are summarized in Table 1 and Fig. 1. The beginning of the sequence corresponds to minute 0 on the E. coli K-12 MG1655 genetic map, with the origin and terminus of replication corresponding to those identified in MG1655 (5). Although virulence plasmids are common to many E. coli isolates, they are not usually associated with uropathogenic strains, and none were found in CFT073. There are five cryptic prophage genomes in the CFT073 chromosome, none with sufficient genetic information to produce viable phage.
Table 1.
Genome contents
| Genome length | 5,231,428 bp |
| Plasmids | None |
| Protein-coding genes | 5,533 |
| tRNAs | 89 [1 pseudo, 1 novel (Arg), 1 extra tandem (Arg), 3 phage-encoded] |
| rRNAs | 22 genes in 7 operons |
| Miscellaneous RNAs annotated | 11 |
| G + C% | 50.47% |
| Backbone genes | 3,190 |
| Island genes | 1,827 |
| Backbone regions | 359 (3,925,047 bp; 75.02%) |
| CFT073-specific islands | 247 (1,306,391 bp; 24.98%) |
| Cryptic prophage | 5 |
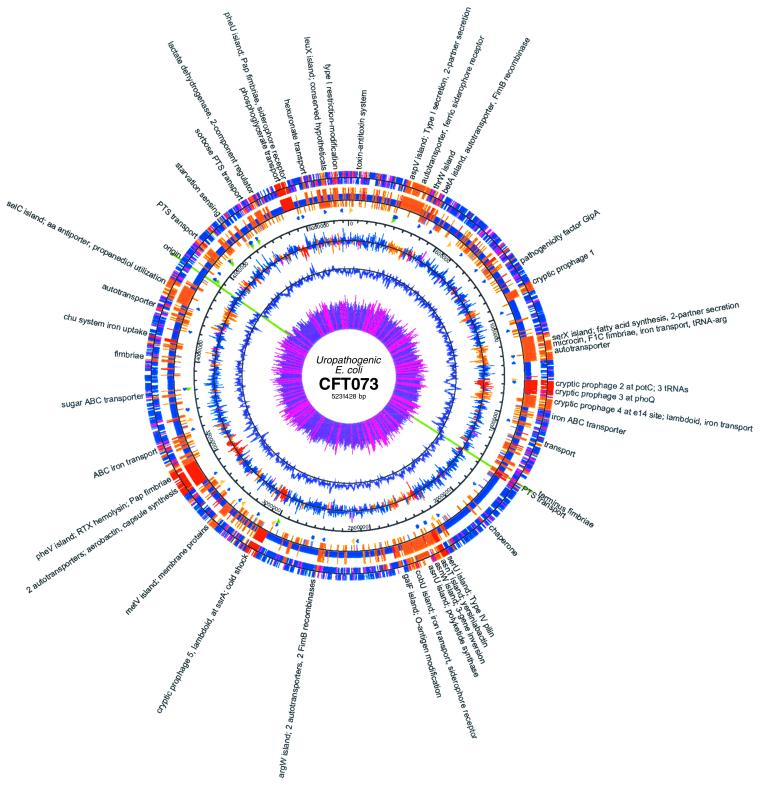
Fig 1.
Map of the CFT073 genome and comparison with K-12 strain MG1655. The outer circle shows ORFs, colored according to the K-12 comparison in the second circle, where DNA regions are shown: blue, backbone, i.e., E. coli near match to MG1655; red, CFT073 islands (insertions); orange, islands (substitutions replacing K-12 segments); violet, K-12 islands. ORFs in the outer ring that span island–backbone junctions are pink. Third circle, RNAs: green, rRNA operons; blue, tRNAs; gold, miscellaneous RNAs. Fourth circle, scale in bp. Fifth circle, GC skew calculated for each ORF >100 aa, colored according to the same scheme of the ORF circle and plotted around the mean. Sixth circle, GC skew calculated over the whole sequence (window, 10 kb) plotted around the mean. Seventh circle, codon-adaptation index CAI (inverse, 1-CAI is plotted); pink rays indicate CAI values <0.2; purple rays, values >0.2. The pink rays can be seen to correspond with islands. A detailed linear map with ORF annotations is available at www.genome.wisc.edu. Maps were created by GENVISION from DNASTAR.
The CFT073 genome is 590,209 bp longer than MG1655 and similar in size to EDL933. When the CFT073 genome sequence was compared with the reference MG1655 genome (3), 247 CFT073-specific DNA segments >50 bp were found inserted or substituted into a conserved backbone sequence of 3.92 Mb. Sixty unique segments >4.0 kb encode known or potential virulence genes. The CFT073-specific islands total 1.303 Mb. Conversely, the MG1655-specific sequences, absent in CFT073, amount to 715.7 kb. A similar pattern was observed when the enterohemorrhagic E. coli O157:H7 EDL933 genome sequence was compared with MG1655 (4). Comparisons revealed that >70% of the ORFs previously identified as unique to either MG1655 or EDL933 are replaced with new genes specific to the uropathogenic isolate (Fig. 2). A search for disrupted ORFs of previously characterized E. coli genes resulted in detection of only 62 pseudogenes.
Fig 2.
Shared E. coli proteins. Comparison of the predicted proteins of the three E. coli strains shows the number of orthologs in each shared category and numbers of strain-specific proteins. Hypervariable proteins and proteins spanning island–backbone junctions were excluded from the analysis. Number of proteins counted: K-12, 4,288; CFT073, 5,016; EDL933, 5,063. In the totals for the three strains, orthologous proteins are counted only once. Orthologous proteins meet the same match criteria used for designation of backbone (see Materials and Methods).
Islands and Horizontal Transfer.
Distinctive codon usage is considered to be a hallmark of lateral gene transfer. In CFT073, codon usage analysis was performed to test the hypothesis that different patterns of usage occur between the backbone and island genes. When frequency distributions for each codon were examined, 52 of 61 codons in island ORFs had frequency distributions significantly different from those in backbone with >95% confidence values, measured by Student's t test. In contrast, the codon usage pattern in EDL933 backbone ORFs was indistinguishable from CFT073 backbone in the same tests. The average amino acid sequence identity of backbone ORFs is >98% for each pairwise comparison between the three strains. We also observed a bias for rare codons in island genes, with AUA (Ile), AGA (Arg), and AGG (Arg) occurring at frequencies 3.1, 4.0, and 4.5 times higher, respectively, in island ORFs than in backbone.
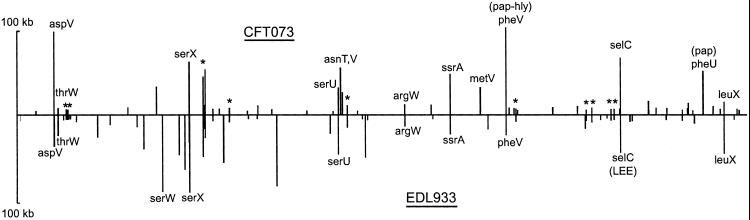
The CFT073-specific islands contain 2,004 genes, of which only 204 also occur among the EDL933-specific genes. Two-thirds of these island genes shared by EDL933 and CFT073 have unknown functions or are associated with phage or insertion sequence elements (10). The remaining shared genes encode putative iron-uptake systems, a complex set of potential fatty acid biosynthetic enzymes, several adhesins, and phosphotransferase system and ATP-binding cassette (ABC)-type transport systems. CFT073 and EDL933 contain, respectively, 60 and 57 islands >4 kb in length. The locations and sizes of these are shown in Fig. 3. Many island locations are at the same relative backbone position in the two pathogens although the island contents are unrelated. Thirteen CFT073 and 10 EDL933 islands are closely associated with known tRNA genes (nine are at the same tRNA in both genomes). Ten other locations also are occupied by unrelated islands in both strains.
Fig 3.
Locations and sizes of CFT073 and EDL933 islands. Island size, vertical axis; position in colinear backbone, horizontal axis. All islands >4 kb are shown. Islands located at tRNAs are indicated by tRNA labels. One tmRNA (ssrA) is also an insertion target. *, CFT073 and EDL933 islands in the same backbone location but not near tRNAs.
Variation Among E. coli Uropathogenic Strains.
The differences in disease potential between enterohemorhaggic and uropathogenic E. coli are reflected specifically in the absence of genes in CFT073 for type III secretion system and phage- and plasmid-encoded virulence genes common to E. coli O157:H7 isolates. In CFT073, the strain-specific regions contain genes that encode specific fimbrial adhesins, secreted autotransporters, and phase-switch recombinases. The chromosomal locations, gene order, and composition of the large pathogenicity islands encoding these potential virulence genes in CFT073 are different when compared with two other well-studied uropathogenic E. coli strains, 536 and J96 (11–13). We compared previously described CFT073 islands with those identified in this project. Based on the genome sequence, the two pap pilus operon-containing pathogenicity islands in CFT073 (14, 15) actually are located at pheV and pheU and their gene maps have been revised. Numerous insertion sequence elements including multiple copies of an IS629-like sequence probably contributed to errors in the original screening and ordering of genes from cosmid clones. Interestingly, pheV and pheU sites are identical to the insertion sites for the two large, pap-associated islands in J96 (13). In CFT073 and J96, the pap-associated islands at pheV also encode hemolysin, hlyCABD, but the order of the pap and hlyCABD genes relative to the K-12 backbone is different. Other known virulence genes located on the two islands are also different. The J96 pheV island contains hra (heat-resistant hemagglutinin) and cnf-1 (cytotoxic necrotizing factor), but not aerobactin biosynthesis genes, whereas the CFT073 pheV island has neither hra nor cnf-1 but does have the aerobactin genes (13). In strain 536, the third well-studied uropathogenic E. coli, the large pap- and hly-associated islands are located at selC and leuX sites. In addition, 536 possesses an S-type pilus operon on a 25-kb island associated with the thrW site (PAI III536) whereas the thrW site in CFT073 is intact, and a related S type, Foc pilus operon, is present on a 47-kb island close to serX. The remaining large 536 island described by Hacker's laboratory encodes part of the “high pathogenicity island” of Yersinia pestis and is located at asnT (11, 16). The yersinabactin genes are also found in CFT073 on a island in the asnT region. This finding suggests that the introduction of the high pathogenicity island may have been one of the earliest events in the evolution of extraintestinal E. coli.
Potential for New Niches and Different Pathogenic Mechanisms.
The ability to inhabit the different niches during an ascending urinary tract infection and cause particular pathologies at each site resides largely in the island genes specific to uropathogenic E. coli. The CFT073 genome sequence has revealed many possible factors that may contribute to colonization of the urinary tract tissues and the disease. The most important examples are mentioned here.
Surface structures known as fimbriae or pili mediate specificity for and attachment to host cells, an essential event for host colonization. We found genes encoding 12 distinct, putative fimbriae in the genome of CFT073, 10 fimbriae of the chaperone–usher family, and two type IV pili. Two pap operons (pylonephritis-associated pilus) encode P fimbriae with PapGII adhesins (17), located in islands at pheV and pheU. These are specific to uropathogens but are not the sole adhesins in CFT073 that are important for virulence. The foc operon encoding F1C fimbria and a chaperone–usher family operon with two chaperone genes both have been linked to urinary tract infections (18). Several of the chaperone–usher pilus operons are common to CFT073, EDL933, and MG1655, including the yad fimbriae (19) and the type I fimbrial operon, which plays an essential role in the pathogenesis of urinary tract infection (20). Type 1 fimbriae are ubiquitous, but they are not all identical. Also common to all three sequenced strains is a pilus similar to Stf of Salmonella enterica serotypes Typhimurium and Typhi and to the Mrp pilus of Proteus mirabilis, a confirmed urovirulence determinant (21). In CFT073, these proteins are highly divergent from those in MG1655 and EDL933, with amino acid sequence identities ranging from 53% to 81%, suggesting that the selective pressure on the expression of this pilus has varied among E. coli lineages. Four other fimbrial operons are shared by two of the three strains or by S. enterica. These have similarly variable amino acid sequence identity. Presumably, the variable sequences of the shared operons allows for the specificity of each adhesin to its individual target tissue.
Type IV pili are assembled by the type II general secretory pathway. They occur in a wide range of species and frequently are associated with diseases. Genes encoding a putative type IV pilin and tip adhesin were found in CFT073. In all three strains, ppdD and hofBC genes may encode type IV prepilin. Although there is no evidence for its expression in MG1655, PpdD can be incorporated into a type IV pilus in a suitable host (22). Genes encoding the putative secretin, a nucleotide-binding protein required for twitching motility, and other type IV pilin-like proteins also are present in all three strains. The type II general secretory pathway secreton for chitinase (23) is found in CFT073 and K-12 in the backbone region between rpsJ and tufA but is absent from the EDL933 genome, although the large plasmid pO157 carries a functional type II secretion system (24).
FimE and FimB recombinases control expression of the fimbriae encoded by the widespread fim operon in a phase-switch system that involves site-specific inversion of a small, 314-bp DNA element. Five different copies of fimBE-like genes were found in the CFT073 genome. Two copies are associated with the type I fimbrial locus present in the same place as other E. coli genomes. There are two divergently transcribed copies linked to the d-serine deaminase locus near argW and a fifth linked to the osmoregulatory choline–glycine betaine locus, betABIT.
E. coli CFT073 encodes at least seven putative autotransporters, proteins that export a large passenger-domain cleavage fragment across the outer membrane via a β-barrel pore formed by the C terminus of the same protein. The secreted polypeptides often confer virulence (25). For example, in CFT073, Sat, a serine protease, elicits cytopathic effects on bladder and kidney epithelial cells (26). Unique examples in the CFT073 genome are similar to hemagglutinin or diffuse adherence (AIDA)-like adhesins. One is a homologue of Pic, a mucinase of enteroaggregative E. coli and Shigella flexneri, that contains within its sequence on the opposite strand two ORFs >95% identical to SetA and SetB, the AB subunit enterotoxin (ShET-1) in Shigella (27).
The well-characterized hemolysin genes (hlyCABD) at the pheV island encode a cytolytic toxin and its secretion apparatus (28). An additional member of the type I RTX-like secretion family, upxBDA, is found in the 100-kb island at aspV. The gene order for this member is atypical when compared with the originally characterized RTX determinants, the B and D secretion genes preceding the A gene. It also lacks a C-like gene that typically encodes a fatty acid modification enzyme. There are no notable UpxA sequence features that indicate that it is a member of one of the known RTX family branches (i.e., pore-forming toxin, protease, or lipase). This finding suggests that this locus encodes a unique class of RTX-like secreted protein.
Discussion
Both pathogenic and nonpathogenic types of E. coli have evolved through a complex process. The ancestral backbone genes that define E. coli have undergone slow accumulation of vertically acquired sequence changes, but genes in the remainder of the chromosome are, in a relative sense, newly introduced via numerous, independent horizontal gene-transfer events at many discrete sites, some serving as universal insertion targets used independently in separate lineages. The codon usage analysis supports the conclusion that there are a set of backbone E. coli genes that have a shared codon bias that is not seen in the genes unique in each of the three genomes. The net result is a mosaic genome structure in which newly acquired genes in each of the E. coli types are placed into a framework made of genes that distinguishes E. coli from its closer relatives such as S. enterica.
For uropathogenic strains of E. coli, island acquisition resulted in the capability to infect the urinary tract and bloodstream and evade host defenses without compromising the ability to harmlessly colonize the intestine. For the different intestinal pathogens, acquired genes promote the colonization of specific regions of the intestine and new modes of interaction with the host tissue that produce clinically distinct variations of gastrointestinal disease. Each type of E. coli possesses combinations of island genes that confer its characteristic lifestyle or disease-causing traits. Hacker and colleagues (11) elaborated the pathogenicity island concept based on the genetic behavior, virulence gene linkage relationships, and location of unique inserts near several tRNA genes. Pathogenicity-associated islands were designated based on the presumption that pathogenic traits are present in all inserts and with the assumption that each unique DNA segment has some unifying physical features and similar genetic history and behavior. Our sequence comparisons show that this is not true even for similar uropathogenic strains that have two islands containing some similar genes inserted at the same tRNA site. Comparisons of CFT073 islands with those of other extraintestinal E. coli isolates indicate that similar virulence genes may come into play, but their linkage relationships and chromosomal locations vary considerably. Our observation provides evidence that extraintestinal E. coli may be oligoclonal despite the apparent linkage relationships of a handful of virulence genes and suggests that the uropathogenic E. coli may be as diverse as the intestinal strains. Recent epidemiological analyses lend support to the proposal that specific subsets of genes are characteristic of each of the E. coli uropathogenic subtypes: cystitis, pyelonephritis, and urosepsis (14, 15).
The presence of three extra homologues of the fimBE-like recombinases suggests that the DNA segment-inversion mechanism of genetic phase variation may operate at regions other than the type I fimbrial adhesin in CFT073. The extent of genotypic differences from other E. coli, on a scale larger than previously known, is not altogether surprising given the complexity in the lifestyle of this pathogen, where it colonizes distinct niches including the intestine, perineum, urethra, bladder, and kidney of humans as well as these sites in other mammals such as dogs (29).
The common core chromosome of the E. coli genomes has been preserved throughout its vertical evolution, with very limited intragenomic rearrangement, resulting in the conserved synteny apparent today. The backbone also provides a large, core set of markers for this group, including genes of nutrient synthesis and others that form the signature of Escherichia physiology. No extensive genome reductions have taken place to take advantage of nutrients available in the intestinal environment, and this presumably has remained true despite millions of years of a commensal lifestyle in animals. The presence of “black holes,” i.e., deletions that remove genes detrimental to the uropathogenic lifestyle, is difficult to assess at this time because of the large number of genetic differences already observed, the absence of Shigella spp., and additional E. coli genome sequences needed for comparisons (30). The detection of only a relatively small number of pseudogenes in CFT073 stands in contrast to the numbers that have been recently observed, 204 in S. enterica serovar Typhi and 149 in Y. pestis (31, 32). In this respect, CFT073 parallels the broad host range and varied lifestyle of S. enterica serovar Typhimurium (39 pseudogenes) than the more restricted lifestyles of Typhi or Y. pestis. However, the sheer amount of unique DNA in each E. coli strain that can be explained by the frequent gain and loss of accessory genes suggests that careful reconsideration is due for defining species by a few phenotypic traits and low-resolution mapping. The CFT073 and EDL933 genome sequences enable us to design far more discriminating tools for diagnosis of particular E. coli pathotypes that cause such a wide range of intestinal and extraintestinal diseases.
Acknowledgments
We thank Bob Mau for software and help with genome comparisons. We thank the technical staff of the University of Wisconsin Bacterial Pathogen Genome Initiative for their excellent cloning and sequencing work. This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Awards AI44387 (to F.R.B.) and AI39000 (to R.A.W.) and National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Award DK49720 (to M.S.D. and H.L.T.M.). This is paper no. 3599 from the Laboratory of Genetics.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AE014075).
References
- 1.Hooton T. M. & Stamm, W. E. (1997) Infect. Dis. Clin. North Am. 11, 551-581. [DOI] [PubMed] [Google Scholar]
- 2.Mobley H. L., Green, D. M., Trifillis, A. L., Johnson, D. E., Chippendale, G. R., Lockatell, C. V., Jones, B. D. & Warren, J. W. (1990) Infect. Immunol. 58, 1281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manges A. R., Johnson, J. R., Foxman, B., O'Bryan, T. T., Fullerton, K. E. & Riley, L. W. (2001) N. Engl. J. Med. 345, 1007-1013. [DOI] [PubMed] [Google Scholar]
- 4.Perna N. T., Plunkett, G., III, Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A., et al. (2001) Nature 409, 529-533. [DOI] [PubMed] [Google Scholar]
- 5.Blattner F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Lim A., Dimalanta, E. T., Potamousis, K. D., Yen, G., Apodoca, J., Tao, C., Lin, J., Qi, R., Skiadas, J., Ramanathan, A., et al. (2001) Genome Res. 11, 1584-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaasterland T. & Sensen, C. W. (1996) Trends Genet. 12, 76-78. [DOI] [PubMed] [Google Scholar]
- 8.Salzberg S. L., Delcher, A. L., Kasif, S. & White, O. (1998) Nucleic Acids Res. 26, 544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 10.Mahillon J. & Chandler, M. (1998) Microbiol. Mol. Biol. Rev. 62, 725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacker J., Blum-Oehler, G., Janke, B., Nagy, G. & Goebel, W., (1999) Pathogenicity Islands of Extraintestinal Escherichia coli (Am. Soc. Microbiol., Washington, DC).
- 12.Dobrindt U., Blum-Oehler, G., Hartsch, T., Gottschalk, G., Ron, E. Z., Funfstuck, R. & Hacker, J. (2001) Infect. Immunol. 69, 4248-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swenson D. L., Bukanov, N. O., Berg, D. E. & Welch, R. A. (1996) Infect. Immunol. 64, 3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasko D. A., Phillips, J. A., Li, X. & Mobley, H. L. (2001) J. Infect. Dis. 184, 1041-1049. [DOI] [PubMed] [Google Scholar]
- 15.Guyer D. M., Kao, J. S. & Mobley, H. L. (1998) Infect. Immunol. 66, 4411-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schubert S., Cuenca, S., Fischer, D. & Heesemann, J. (2000) J. Infect. Dis. 182, 1268-1271. [DOI] [PubMed] [Google Scholar]
- 17.Mobley H. L., Jarvis, K. G., Elwood, J. P., Whittle, D. I., Lockatell, C. V., Russell, R. G., Johnson, D. E., Donnenberg, M. S. & Warren, J. W. (1993) Mol. Microbiol. 10, 143-155. [DOI] [PubMed] [Google Scholar]
- 18.Pere A., Leinonen, M., Vaisanen-Rhen, V., Rhen, M. & Korhonen, T. K. (1985) J. Gen. Microbiol. 131, 1705-1711. [DOI] [PubMed] [Google Scholar]
- 19.Raina S., Missiakas, D., Baird, L., Kumar, S. & Georgopoulos, C. (1993) J. Bacteriol. 175, 5009-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connell H., Agace, W., Klemm, P., Schembri, M., Mårild, S. & Svanborg, C. (1996) Proc. Natl. Acad. Sci. USA 93, 9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahrani F. K., Massad, G., Lockatell, C. V., Johnson, D. E., Russell, R. G., Warren, J. W. & Mobley, H. L. (1994) Infect. Immunol. 62, 3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauvonnet N., Gounon, P. & Pugsley, A. P. (2000) J. Bacteriol. 182, 848-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francetic O., Belin, D., Badaut, C. & Pugsley, A. P. (2000) EMBO J. 19, 6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lathem W. W., Grys, T. E., Witowski, S. E., Torres, A. G., Kaper, J. B., Tarr, P. I. & Welch, R. A. (2002) Mol. Microbiol. 45, 277-288. [DOI] [PubMed] [Google Scholar]
- 25.Henderson I. R. & Nataro, J. P. (2001) Infect. Immunol. 69, 1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guyer D. M., Henderson, I. R., Nataro, J. P. & Mobley, H. L. (2000) Mol. Microbiol. 38, 53-66. [DOI] [PubMed] [Google Scholar]
- 27.Fasano A., Noriega, F. R., Maneval, D. R., Chanasongcram, S., Russell, R., Guandalini, S. & Levine, M. M. (1995) J. Clin. Invest. 95, 2853-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welch R. A. (2001) Curr. Top. Microbiol. Immunol. 257, 85-111. [DOI] [PubMed] [Google Scholar]
- 29.Johnson J. R., Stell, A. L., Delavari, P., Murray, A. C., Kuskowski, M. & Gaastra, W. (2001) J. Infect. Dis. 183, 897-906. [DOI] [PubMed] [Google Scholar]
- 30.Maurelli A. T., Fernandez, R. E., Bloch, C. A., Rode, C. K. & Fasano, A. (1998) Proc. Natl. Acad. Sci. USA 95, 3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkhill J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T., Prentice, M. B., Sebaihia, M., James, K. D., Churcher, C., Mungall, K. L., et al. (2001) Nature 413, 523-527. [DOI] [PubMed] [Google Scholar]
- 32.Parkhill J., Dougan, G., James, K. D., Thomson, N. R., Pickard, D., Wain, J., Churcher, C., Mungall, K. L., Bentley, S. D., Holden, M. T., et al. (2001) Nature 413, 848-852. [DOI] [PubMed] [Google Scholar]