Abstract
A strain of an unknown coryneform bacterium was repeatedly isolated in pure culture from the blood of a patient affected by endocarditis. Comparative 16S rRNA gene sequence analysis revealed that this isolate represented a new subline within the genus Corynebacterium. This new taxon can be identified by the presence of corynomycolic acids and its enzymatic activities and fermentation of sugars. Acid production from glucose and maltose, pyrazinamidase and alkaline phoshatase activities, and hippurate hydrolysis were the most characteristic phenotypic features of the bacterium. On the basis of both phenotypic and phylogenetic evidence, it is proposed that this isolate be classified as a novel species, Corynebacterium tuscaniae sp. nov. The type strain, ISS-5309, has been deposited in the American Type Culture Collection (ATCC BAA-1141) and in the Culture Collection of the University of Göteborg (CCUG 51321).
For many years, the pathogenic potential of coryneform bacteria other than Corynebacterium diphtheriae has been grossly underestimated. It is only in the last decade that both clinical microbiologists and physicians became more aware of the pathogenic potential of coryneform bacteria and of the great diversity of coryneform bacteria associated with human diseases (6).
The use of chemical markers (1, 2, 3, 11, 17), in combination with a phylogenetic approach based on 16S rRNA gene sequence analyses (14, 16) and improved phenotypic criteria (e.g., miniaturized tests), resulted in a much improved taxonomy of the genus Corynebacterium and proved invaluable for the delineation of novel taxa. During the past decade, a considerable number of novel corynebacterial species has been described. The vast majority of these newly described species have been isolated from human clinical and veterinary sources (7, 20, 21).
During the course of the characterization of medically relevant coryneform bacteria, we isolated a strain from blood cultures of a patient with infective endocarditis. By the use of chemotaxonomic methods, it was found that this isolate had the main characteristics of the genus Corynebacterium, but it could not be biochemically identified as a member of any previously described species. Further taxonomic and phylogenetic investigations indicated that it constitutes a distinct new species within the genus Corynebacterium. Therefore, it is proposed here that strain ISS-5309, deposited in the American Type Culture Collection (ATCC) as strain ATCC BAA-1141 and in the Culture Collection of the University of Göteborg (CCUG) as strain CCUG 51321, be classified as Corynebacterium tuscaniae sp. nov.
CASE REPORT
In June 2003, an 85-year-old woman was admitted to the Department of Internal Medicine of the Hospital of Lucca, Italy, because of fever of unknown origin. On admission, the patient complained of persistent fever (temperature, 37.5 to 38°C), anorexia, and weight loss (about 3 kg) during the previous month. The patient had undergone a gastrectomy due to gastric carcinoma at the age of 75 years. In the following years she suffered from protein-calorie malnutrition and osteoporosis.
Physical examination showed a poor general condition: a body weight of 41 kg, a height of 160 cm, a body mass index of 16 kg/m2, loss of muscle mass, expiratory crackles at the lungs, holosystolic murmur most prominent at the cardiac apex, diastolic murmur on the third-fourth interspaces, and a scar on the midline of the abdomen.
Laboratory investigations showed the following data: erythrocyte sedimentation rate, 25 mm; red blood cell count, 2,980,000/ml; hemoglobin concentration, 9.1 g/dl; mean corpuscular volume, 98.7 fl; white cell count, 3.500/ml; neutrophils, 77%; eosinophils, 0.80%; basophils, 0.40%; lymphocytes, 11.6%; monocytes, 7%; leukocytes, 3,2%; platelets, 80,000/ml; serum protein concentration, 5.1 g/dl; lactate dehydrogenase concentration, 495 IU/liter; C-reactive protein concentration, 6.4 mg/dl; and β2-microglobulin concentration, 6 mg/dl. The results of other routine laboratory tests, the Widal-Wright test and tests for carcinoembryonic antigen, Ca19-9, and Ca125, were within normal limits. Her electrocardiograph was normal.
Chest radiography revealed a diffuse interstitial thickness, hilar enlargement, and a left pleural effusion. Abdominal computed tomography showed an aortic aneurysm below the origin of the renal arteries with parietal thrombosis (false lumen, 5 cm; true lumen, 2 cm) and lymphadenopathies (3.5 cm) in the para-aortic seat as a result of splenic infarction. Transesophageal echocardiography detected left atrium enlargement (49 mm) with thrombotic formations, mitral valve regurgitation of 3+/4+, normal left ventricular function, and the aortic valve with a prolapse in the aorta during systole vegetation that caused aortic regurgitation of 3+/4+.
Six blood cultures (three samples obtained on the first day of hospitalization and three samples obtained on the second day of hospitalization) were positive for a gram-positive coryneform bacterium.
The patient was treated with ampicillin-sulbactam at 3 g every 8 h intravenously and gentamicin at 80 mg/day intravenously for 4 weeks. The fever disappeared, and repeated blood cultures were negative; but the patient kept on complaining of severe anorexia.
No further investigation to check the para-aortic lymphadenopathies was performed because of the poor condition of the patient and as requested by her family. The patient died at home 2 months later.
MATERIALS AND METHODS
Strain and culture conditions.
Strain ISS-5309 (strains ATCC BAA-1141 and CCUG 51321) was isolated from blood after 2.5 days of incubation in the nonvented aerobic culture bottles of the blood BacT/Alert FA with the Bact/Alert 3D Microbial detection system (bioMèrieux). The growth from the blood culture was subcultured onto chocolate agar with IsoVitalex and Columbia agar (Merck) supplemented with 5% sheep blood and incubated at 37°C in 6% CO2. After 24 h of incubation, growth was visible on the medium used.
Biochemical tests.
The strain was grown on Columbia agar supplemented with 5% sheep blood aerobically at 37°C. Smears were stained by the Gram and Ziehl-Neelsen methods. The API Coryne system was used, according to the manufacturer's instruction (API bioMèrieux). The CAMP and reverse CAMP tests were performed with Staphylococcus aureus ATCC 25923, according to the standard procedure. The lipid requirement was determined by comparison of the growth of cultures in Trypticase soy broth (Difco) and in the same medium supplemented with 1% (vol/vol) Tween 80 (Merck) after 24 h at 37°C. Tween esterase activity was detected on Trypticase soy agar supplemented with 1% Tween 80, and the presence of this enzyme was determined by the detection of opacity surrounding the colonies. Urease production was tested in Christensen's medium (Difco), and nitrate reductase production was tested by using brain heart broth (Difco) supplemented with 2 g/liter of NaNO3.
Antimicrobial susceptibility patterns.
The susceptibility of the isolate to 11 antimicrobials agents commonly used for the treatment of infections caused by gram-positive rods was determined by the disk diffusion method by use of the methodology of the CLSI (formerly the National Committee for Clinical Laboratory Standards) and interpreted according to the criteria for streptococci established by CLSI (13), although it is acknowledged that the CLSI has not explicitly expressed susceptibility criteria for coryneform bacteria.
The MICs of penicillin G, amoxicillin, cefotaxime, imipenem, vancomycin, and ciprofloxacin were determined by Etest, according to the recommendations of the manufacturer (AB biodisk, Solna, Sweden).
Cell wall analysis.
A previously described reverse-phase high-pressure liquid chromatography method was used for the determination of mycolic acids (4).
DNA-DNA hybridization.
DNA was extracted and purified as described previously (15). Hybridization of the labeled DNA of isolate ISS-5309 and the fragmented DNA preparation of Corynebacterium appendicis type strain CIP 107643 (DSM 44531) was carried out at 60°C for 16 h in 0.42 M NaCl by the S1 nuclease-trichloroacetic acid method (15).
PCR amplification and 16S rRNA gene sequencing.
A genomic DNA preparation was made by suspending a loop of overnight colonies in a tube containing 1 ml water and extracted with Chelex by using the InstaGene matrix (Bio-Rad Laboratories), according to the manufacturer's protocol described for the preparation of genomic DNA from bacteria.
An aliquot of the supernatant (10 μl) was used as the template in a final PCR mixture volume of 50 μl, which contained 1× PCR buffer, 2 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 400 nM each primer, and 0.5 U Taq DNA polymerase (Life Technologies) for mediated amplification of the 16S rRNA gene. Previously described universal primer sequences were used (5). The samples were amplified on a DNA thermal cycler (MJ Research, Inc.) by heating for 3 min at 96°C, followed by 30 cycles of 95°C for 60 s, 57°C for 60 s, and 72°C for 60 s and concluding with a cycle of 72°C for 10 min.
The PCR products were purified by using a Qiaquick PCR purification kit (QIAGEN), according to the manufacturer's instructions, and both strands were automatically sequenced (M-Medical) by using universal and de novo-synthesized primers.
Phylogenetic analysis.
The 16S rRNA gene sequence of strain ISS-5309 was determined in this study and was aligned with other corynebacterial sequences by the use of the program ClustalW and default gap penalties. Unrooted trees were constructed by sequential use of the programs SEQBOOT, DNADIST or DNAPARS, and CONSENSE (implemented in the Phylogeny Inference Package [PHYLIP], version 3.6) to construct a consensus tree based on 100 bootstrap replications of the original alignment. The evolutionary distances between pairs of DNA sequences were calculated by the Jukes-Cantor one-parameter method, which assumes an equality of substitution rates between all nucleotides (10).
Nucleotide sequence accession number.
The nucleotide sequence of the 16S rRNA gene of strain ISS-5309 was deposited in the GenBank database under accession number AY677186.
RESULTS AND DISCUSSION
Strain ISS-5309 showed good growth within 24 h on Columbia blood agar, forming nonhemolytic white rounded colonies. Cells stained gram positive and were non-acid fast. The cells were nonmotile and non-spore forming and showed a typical coryneform morphology. No visible growth was detected under anaerobic conditions. The strain was nonlipophilic, and no Tween esterase was detected on Trypticase soy agar supplemented with 1% Tween 80. The API Coryne system provided the numerical code of 2100124 for this species.
The microorganism possessed the following biochemical reactions: positivity for catalase; the presence of pyrazinamidase and alkaline phosphatase activities; the absence of pyrrolidonyl arylamidase, nitrate reductase, beta-glucuronidase, beta-galactosidase, alpha-glucosidase, beta-N-acetyl-glucosaminidase, and urease activities; the production of acids from glucose and maltose but not from ribose, xylose, mannitol, lactose, sucrose, or glycogen; positivity for hydrolysis of hippurate; and negativity for hydrolysis of tyrosine, esculin, and gelatin. The results of both the CAMP test and the reverse CAMP test were negative.
Nevertheless, these features did not allow us to readily distinguish our isolate from some other nonlipophilic species of the genus Corynebacterium (8). The biochemical characteristics that differentiated our isolate from other Corynebacterium species (8, 9, 20, 21) are given in Table 1. Phenotypically, the isolate could be confused with Corynebacterium minutissimum, but the latter species is not able to hydrolyze hippurate, unlike the isolate studied. Corynebacterium amycolatum displays variable responses to routine biochemical tests and exhibits typical colonies with irregular edges, while our isolate showed rounded and regular colonies. Furthermore, C. amycolatum lacks mycolic acids.
TABLE 1.
Characteristics that differentiate C. tuscaniae sp. nov. from other biochemically related Corynebacterium spp. encountered in clinical specimensa
| Organism | Pyrazinamidase activity | Urease activity | Alkaline phosphatase activity | Acid produced from:
|
Other traits | ||
|---|---|---|---|---|---|---|---|
| Glucose | Maltose | Sucrose | |||||
| C. tuscaniae | + | − | + | + | + | − | Hippurate positive, tyrosine negative, contains mycolic acids, CAMP |
| reaction negative | |||||||
| C. amycolatum | + | − | + | + | v | v | Lacks mycolic acids |
| C. argentoratense | + | − | v | + | − | − | |
| C. atypicum | − | − | − | + | + | + | Lacks mycolic acids |
| C. aurimucosum | + | − | − | + | + | + | |
| C. diphtheriae | − | − | − | + | + | − | |
| C. coyleae | + | − | + | + | − | − | |
| C. imitans | w | − | + | + | + | v | Tyrosine negative, CAMP |
| reaction positive | |||||||
| C. glaucum | − | − | + | + | − | + | |
| C. minutissimum | + | − | + | + | + | v | Hippurate negative, tyrosine positive |
| C. sanguinis | + | − | + | w | − | − | |
| C. mucifaciens | + | − | + | + | − | v | Mucoid colonies |
| C. afermentans | + | − | + | − | − | − | |
| C. riegelii | v | + | v | − | w | − | |
| C. sundsvallense | v | + | v | + | + | + | |
| C. thomsseni | + | + | + | + | + | + | |
| C. appendicis | + | + | + | + | + | − | |
Abbreviations: w, weakly positive; v, variable; +, positive; −, negative.
C. tuscaniae sp. nov. may be easily differentiated from the more phylogenetically related species by use of the following characteristics: (i) it does not hydrolyze urea, unlike C. appendicis, Corynebacterium riegelii, Corynebacterium sundsvallense, and Corynebacterium thomsseni; (ii) it produces acid from glucose, unlike Corynebacterium afermentans; and (iii) it produces acid from maltose, which is not the case for Corynebacterium glaucum.
Many biochemical characteristics of strain ISS-5309 were identical to those of C. appendicis, the species that is the most phylogenetically related to strain ISS-5309 when enzymatic activities (the presence of pyrazinamidase and alkaline phosphatase activities but a lack of nitrate reductase, pyrrolidonyl arylamidase, glucuronidase, galactosidase, and glucosidase activities) and the fermentation of sugars (acid from glucose and maltose but not from sucrose or mannitol) are considered. Nevertheless, no degradation of urea occurred for strain ISS-5309 with the API Coryne system or Christensen's medium, unlike in C. appendicis, as it was given in the original description (20) and determined by us by study of the type strain.
By the disk diffusion method, the isolate was susceptible to amoxicillin, cefotaxime, pristinamycin, ciprofloxacin, tetracycline, vancomycin, rifampin, gentamicin, and erythromycin, while it was resistant to ceftazidime and fosfomycin. The MICs, as estimated by Etest, were as follows: penicillin, 0.064 mg/liter; amoxicillin, 0.064 mg/liter; cefotaxime, 0.125 mg/liter; ciprofloxacin, 0.125 mg/liter; vancomycin, 0.5 mg/liter; and imipenem, 0.023 mg/liter. Overall, the antimicrobial susceptibility pattern corresponded to those of most nonlipophilic Corynebacterium species.
The strain ISS-5309 cell wall contains mycolic acid of short chain lengths (C26 to C36), typical of the genus Corynebacterium, while C. amycolatum, which exhibits biochemical characteristics similar to those of our isolate, has no mycolic acids.
In order to identify this isolate accurately, we investigated its genomic similarities to the nearest phylogenetically recognized species by using quantitative DNA-DNA hybridization. DNA-DNA similarity experiments, according to a stringent nuclease S1 method described previously (15), showed only 16% DNA relatedness at 60°C with C. appendicis type strain CIP 107643T (DSM 44531T). Thus, this isolate was genetically confirmed to be distinct from the species C. appendicis, as the genetic definition of a species is more than 70% DNA similarity (19).
The almost complete 16S rRNA gene sequence of strain ISS-5309 was determined in this study and was found to comprise 1,502 nucleotides. The 16S rRNA gene sequence comparison by using both the FASTA and the BLASTN programs showed that strain ISS-5309 displayed high sequence similarity (95 to 97%) to previously described members of the genus Corynebacterium (Table 2). The highest sequence similarity (≥97%) was shown to C. appendicis, Corynebacterium mucifaciens, C. riegelii, and Corynebacterium coyleae and, to a lesser extent, to C. glaucum, Corynebacterium lipophiloflavum, Corynebacterium imitans, C. afermentans subsp. lipophilum, Corynebacterium accolens, Corynebacterium sundvallense, C. afermentans subsp. afermentans, C. thomssenii, and Corynebacterium mycetoides. All these species are members of the same cluster within the family Corynebacteriaceae (20).
TABLE 2.
16S rRNA sequence similarities between C. tuscaniae sp. nov. and other Corynebacterium spp.
| Species (GenBank accession no.) | Straina | 16S rRNA sequence similarity (%) |
|---|---|---|
| C. appendicis (AJ314919) | DSM 44531T | 97.4 |
| C. mucifaciens (Y11200) | DMMZ 2278 | 97.2 |
| C. riegelii (AF537602) | CCUG 38180 | 97.2 |
| C. afermentans subsp. lipophilum (X82055) | CIP 103500T | 97.2 |
| C. coyleae (X96497) | DSM 44184T | 97.0 |
| C. glaucum (AJ431634) | IMMIB R-5091T | 96.8 |
| C. lipophiloflavum (Y09045) | DMMZ 1944 | 96.6 |
| C. imitans (Y09044) | NCTC 13015 | 96.6 |
| C. afermentans subsp. afermentans (X82054) | CIP 103499T | 96.6 |
| C. accolens (AJ439346) | CIP 104783T | 96.3 |
| C. sundsvallense (Y09655) | CCUG 36622 | 96.2 |
| C. mycetoides (X84241) | NCTC 9864 | 96.2 |
| C. thomssenii (AF010474) | DSM 44276 | 95.6 |
DSM, German Collection of Microorganism, Braunschweig, Germany; DMMZ, Department of Medical Microbiology, University of Zürich, Switzerland; CIP, Collection of Institute Pasteur, Paris, France; IMMIB, Institute of Medical Microbiology and Immunology, University of Bonn, Bonn, Germany; NCTC, National Collection of Type Cultures, London, United Kingdom.
The sequence divergence observed between isolate ISS-5309 and the corynebacterial species mentioned above suggests that it is a separate species. In fact, it is generally recognized that a divergence value of about 3% is significant (18). However, the 97% limit is not always fulfilled in the genus Corynebacterium, and additional distinctive characters are generally needed to allow the definition of a novel species (14, 16).
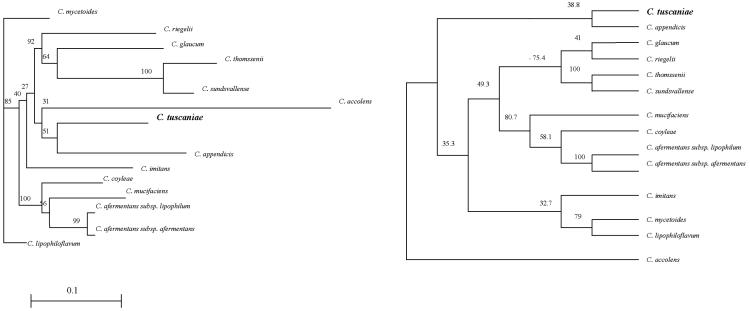
The phylogenetic analysis corroborated the affiliation of strain ISS-5309 within the genus Corynebacterium and its independent radiation from the members of the same cluster to which strain ISS-5309 belongs. The 16S rRNA gene sequence-based trees showed that strain ISS-5309 forms a distinct subline loosely associated with but distinct from C. appendicis (Fig. 1).
FIG. 1.
Distance matrix tree (left) and parsimony tree (right) inferred from comparison of the 16S rRNA gene sequences of C. tuscaniae sp. nov. (strains ATCC BAA-1141 and CCUG 51321) and some close relatives. Bootstrap values, expressed as a percentage of 100 replications, are given at the branch points. The bar represents the average number of substitutions per sequence position.
Bootstrap values of ≥95% were observed in both the distance matrix and the parsimony trees only for nodes that grouped closely related species like C. sundsvallense, C. thomssenii, C. afermentans subsp. afermentans, and C. afermentans subsp. lipophilum. The phylogenetic trees presented bootstrap values that were not significant for the other nodes and only a partially superimposable clustering of species (Fig. 1). This difficulty with assessment of the phylogenetic positions of some corynebacterial species by using 16S rRNA gene sequence phylogenies has been reported previously (12).
Nevertheless, the evidence that the new isolate has a separate placement proved that strain ISS-5309 represents a novel taxon (C. tuscaniae) that is phylogenetically distinct from the previously known corynebacterial taxa, as expected from the biochemical and microbiological distinctness of the isolate. However, the affiliation of C. tuscaniae sp. nov. remains to be further investigated, as the alternative phylogenetic reconstruction methods used (the distance matrix and maximum-parsimony methods) placed the novel taxon with C. appendicis, but this was not significantly supported (51 to 38.8% bootstrap confidence values for the distance matrix and parsimony analyses, respectively) (Fig. 1).
Further data on the pathogenic role of this strain need to be collected. However, in general, the clinical significance of corynebacteria other than C. diphtheriae is strengthened by the presence of these organisms in the direct Gram stain or their isolation from a normally sterile body site, such as blood, provided that multiple cultures of blood become positive (8). In the case reported here, the isolate was obtained from six blood cultures and no other bacteria were isolated. According to the clinical and biological data, the case of endocarditis described here can be attributed to this unrecognized Corynebacterium strain. Commensal organisms of the skin or inhabitants of the upper respiratory tract cause most cases of endocarditis due to corynebacteria. The possible source of infection of this patient remains unknown. The patient had predisposing factors, such as a neoplasm and a poor general condition; and the endocarditis occurred with a native valve, while most Corynebacterium-related cases of endocarditis occur with prosthetic valves.
Although it is, in general, not desirable to describe a new bacterial species on the basis of the detection of a single strain, we believe that it is more likely that other microbiologists will also identify this new species once its characteristics have been described. Therefore, the description and delineation of this strain as a new species should be useful for further studies, including evaluation of its prevalence among normal flora and its clinical implication.
Corynebacterium tuscaniae sp. nov.
Corynebacterium tuscaniae (tuscaniae, Latin name of the Italian region, Tuscany, where the strain was isolated). The cells are gram positive, non-spore forming, nonmotile, and non-acid-alcohol fast. Colonies are whitish and circular with entire edges, are nonhemolytic, and grow aerobically and in the presence of a 5% CO2-enriched atmosphere. The organism is catalase positive and contains mycolic acids with short chain lengths (C26 to C36).
Acid is produced from glucose and maltose but not from ribose, xylose, mannitol, lactose, sucrose, or glycogen. Nitrate is not reduced. It displays pyrazinamidase and alkaline phosphatase activities but not pyrrolidonyl arylamidase, beta-glucuronidase, beta-galactosidase, alpha-glucosidase, beta-N-acetyl-glucosaminidase, urease, or esterase activity. It hydrolyzes hippurate but does not hydrolyze tyrosine, gelatin, or esculin. The results of both the CAMP test and the reverse CAMP test are negative. The organism was isolated from the blood of a patient with endocarditis. The type strain, ISS-5309, has been deposited in ATCC as strain ATCC BAA-1141 and in CCUG as strain CCUG 51321.
Acknowledgments
We thank P. Cammarano (University “La Sapienza,” Rome, Italy) for advice with phylogenetic analysis and G. Mandarino and M. Pataracchia (Istituto Superiore di Sanità, Rome, Italy) for secretarial and technical assistance, respectively.
This study was supported by the Italian Ministry of Heath Project (1%); grant 0AD-F (to C.V.H.) and by ISS grant 1024/RI (to C.V.H.).
REFERENCES
- 1.Collins, M. D., T. Pirouz, W. Goodfellow, and D. E. Minnikin. 1977. Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100:221-230. [DOI] [PubMed] [Google Scholar]
- 2.Collins, M. D., M. Goodfellow, and D. E. Minnikin. 1982. A survey of the structures of mycolic acids in Corynebacterium and related taxa. J. Gen. Microbiol. 128:129-149. [DOI] [PubMed] [Google Scholar]
- 3.Collins, M. D., M. Goodfellow, and D. E. Minnikin. 1982. Fatty acid composition of some mycolic acid-containing coryneform bacteria. J. Gen. Microbiol. 128:2503-2509. [DOI] [PubMed] [Google Scholar]
- 4.De Briel, D., F. Couderc, P. Riegel, F. Jehl, and R. Minck. 1992. High-performance liquid chromatography of corynomycolic acids as a tool in identification of Corynebacterium species and related organisms. J. Clin. Microbiol. 30:1407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duga, S., A. Gobbi, R. Asselta, L. Crippa, M. L. Tenchini, T. Simonic, and E. Scanziani. 1998. Analysis of the 16S rRNA gene sequence of the coryneform bacterium associated with hyperkeratotic dermatitis of athymic nude mice and development of a PCR-based detection assay. Mol. Cell Probes 12:191-199. [DOI] [PubMed] [Google Scholar]
- 6.Funke, G., A. von Graevenitz, J. E. Clarridge, and K. A. Bernard. 1997. Clinical microbiology of coryneform bacteria. Clin. Microbiol. Rev. 10:125-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funke, G., P. A. Lawson, and M. D. Collins. 1997. Corynebacterium mucifaciens sp. nov., an unusual species from human clinical material. Int. J. Syst. Bacteriol. 47:952-957. [DOI] [PubMed] [Google Scholar]
- 8.Funke, G., and K. A. Bernard. 2003. Coryneform gram-positive rods. p. 319-345. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 9.Hall, V., M. D. Collins, R. A. Hutson, P. A. Lawson, E. Falsen, and B. I. Duerden. 2003. Corynebacterium atypicum sp. nov., from a human clinical source, does not contain corynomycolic acids. Int. J. Syst. Evol. Microbiol. 53:1065-1068. [DOI] [PubMed] [Google Scholar]
- 10.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 11.Keddie, R. M., and G. L. Cure. 1977. The cell wall composition and distribution of free mycolic acids in named strains of coryneform bacteria and in isolates from various natural sources. J. Appl. Bacteriol. 42:229-252. [DOI] [PubMed] [Google Scholar]
- 12.Khamis, A., D. Raoult, and B. La Scola. 2004. rpoB gene sequencing for identification of Corynebacterium species. J. Clin. Microbiol. 43:3925-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed., approved standard M7-A5, vol. 20, no. 2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Pascual, C., P. A. Lawson, J. A. E. Farrow, M. Navarro Gimenez, and M. D. Collins. 1995. Phylogenetic analysis of the genus Corynebacterium based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 45:724-728. [DOI] [PubMed] [Google Scholar]
- 15.Riegel, P., D. De Briel, G. Prevost, F. Jehl, and H. Monteil. 1994. Genomic diversity among Corynebacterium jeikeium strains and comparison with biochemical characteristics and antimicrobial susceptibilities. J. Clin. Microbiol. 32:1860-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruimy, R., P. Riegel, P. Boiron, H. Monteil, and R. Christen. 1995. Phylogeny of the genus Corynebacterium deduced from analyses of small-subunit ribosomal DNA sequences. Int. J. Syst. Bacteriol. 45:740-746. [DOI] [PubMed] [Google Scholar]
- 17.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic nite: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 19.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. J. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and L. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 20.Yassin, A. F., U. Steiner, and W. Ludwig. 2002. Corynebacterium appendicis sp. nov. Int. J. Syst. Evol. Microbiol. 52:1165-1169. [DOI] [PubMed] [Google Scholar]
- 21.Yassin, A. F., R. M. Kroppenstedt, and W. Ludwig. 2003. Corynebacterium glaucum sp. nov. Int. J. Syst. Evol. Microbiol. 53:705-709. [DOI] [PubMed] [Google Scholar]