Abstract
Cryptosporidium sp. isolates from a waterborne outbreak of diarrhea in France were analyzed by PCR-restriction fragment length polymorphism analysis and sequencing of the Cpgp40/15 locus. Ninety-one percent of the isolates were Cryptosporidium hominis type Ib. The results of this study and those of studies of other outbreaks suggest that the type Ib allele is the predominant allele associated with waterborne cryptosporidiosis.
Cryptosporidium spp. are enteric apicomplexan protozoa responsible for a number of waterborne outbreaks of diarrheal disease worldwide (5, 14). The most notorious outbreak occurred in 1993, in Milwaukee, Wisconsin, when over 400,000 people contracted cryptosporidiosis from contaminated water (7).
Previous investigations have identified two major species that infect humans. Cryptosporidium hominis, previously referred to as genotype I (8), is anthroponotic and is almost exclusively confined to human infections (10). In contrast, C. parvum (previously referred to as genotype II) is zoonotic and is able to infect humans and animals. These genotypic distinctions have been confirmed at several loci through restriction fragment length polymorphism (RFLP) and sequence analysis (10). Overall, C. hominis appears to be responsible for more waterborne outbreaks of cryptosporidiosis than C. parvum (12).
Recently, we and others identified a highly polymorphic Cryptosporidium gene, Cpgp40/15 (2, 11, 13, 15). The degree of sequence variation at this locus is far greater than that of any other Cryptosporidium gene described thus far. Since highly polymorphic loci are particularly useful for subgenotyping, analysis of sequence polymorphism at this locus has facilitated the identification of a number of allelic subgroups, particularly among C. hominis isolates (1, 6, 9, 13, 16). It has been shown that the Cpgp40/15 allelic subgroup Ib had a major role in the Milwaukee outbreak (17). Although subgenotypic data from outbreaks in other parts of the world are scarce, two studies from Ireland also found an elevated rate of infection attributed to isolates with the Cpgp40/15 type Ib allele (4).
We previously reported on a waterborne outbreak of cryptosporidiosis in Dracy le Fort in eastern France (3). Briefly, on 20 September 2001, clustered cases of gastroenteritis occurred in the county of Dracy le Fort. Complaints about the quality of the public water supply had been reported during the previous week. Public health authorities declared an outbreak; and a series of three investigations followed: (i) analysis of fecal samples from symptomatic patients, (ii) analysis of water samples collected from the public water supply, and (iii) epidemiologic investigations based on telephone surveys of physicians from the cities served by the same public water system and all households in the city of Dracy le Fort (3).
Analysis of fecal samples showed that the outbreak was polymicrobial. Altogether, 61.2% of the samples were positive for Cryptosporidium DNA by PCR for the Hsp70 and the 18S rRNA genes. Genotyping of the isolates by direct sequencing of the PCR products showed that all isolates were genotype 1 (3). Water samples collected on 20 and 21 September confirmed a bacterial contamination of the public network. Of 56 water samples tested, 15 were positive for Cryptosporidium sp. oocysts, with the maximum oocyst concentration reaching 0.19/liter on 1 October. Samples collected after 3 October were negative for Cryptosporidium spp.
The telephone survey of physicians showed that the outbreak was almost exclusively limited to Dracy le Fort. A retrospective telephone survey reached 387 households; 291 were included in the analysis. It showed that the attack rate increased in children aged 0 to 9 years (P < 0.05) and with the amount of water consumed before 20 September (P < 0.001) (Table 1). The present study was conducted to determine the subgenotypes of isolates from the Dracy le Fort outbreak by PCR-RFLP and sequencing of the Cpgp40/15 locus. Samples from 25 symptomatic patients that tested positive for Cryptosporidium spp. were analyzed to determine which allelic subgroups were associated with the outbreak.
TABLE 1.
Gastroenteritis attack rates determined from a retrospective telephone survey in the city of Dracy le Fort
| Population | Attack rate (%) |
|---|---|
| Overall population, definite casesa | 50.8 |
| Overall population, probable casesb | 11 |
| Children aged 0 to 9 yr | 71.4c |
| Tap water consumption prior to 20 September | |
| One to three glasses/day | 56.5c |
| More than seven glasses/day | 67c |
A definite case is defined as three or more liquid stools per day or vomiting.
A probable case is defined as one or two liquid stools per day or abdominal pain.
Definite cases.
PCR-RFLP analysis of Cpgp40/15.
The Cpgp40/15 gene was amplified by nested PCR with primer set 5′-ATGCAAAAATACGTGGACTGGG-3′ and 5′-TCGCACGAAAGATTTCCATTG-3′ in the primary PCR and primer set 5′-TTACTCTCCGTTATAGTCTCCGCTG-3′ and 5′-CGAATAAGGCTGCAAAGATTGC-3′ (2) in the secondary PCR. The PCR conditions were 95°C for 15 min (hot start) and 40 cycles of 94°C for 40 s, 55°C for 50 s, and 72°C for 1 min 30 s; this was followed by a final extension for 10 min at 72°C. The PCR products were digested with AluI and separately with RsaI at 37°C for 1 h. Since sequence variation may occur within Cpgp40/15 allelic subgroups (6, 13), digestion with two different enzymes was performed to ensure the accuracy of the PCR-RFLP method for subgenotyping. Fragments were resolved on 2% agarose gels and were visualized by ethidium bromide staining.
Cpgp40/15 sequence analysis.
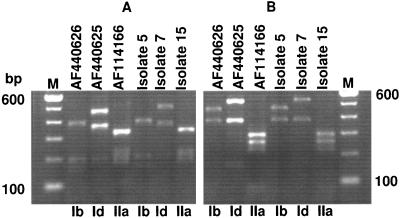
The Cpgp40/15 gene was amplified by nested PCR by using the conditions mentioned above and Proofstart Taq polymerase (QIAGEN, Inc., Valencia, Calif.). The secondary PCR products were purified by using a QIAquick kit (QIAGEN) and were sequenced by the dye-terminator method at the Tufts University Core Facility with a Perkin-Elmer ABI 377 sequencer. The nucleotide sequences from this study were compared with each other and with sequences deposited in GenBank by using the Clustal W alignment algorithm in the MegAlign 6.2 program from the Lasergene suite 6.0 of DNASTAR Inc. (Madison, Wis.). Three Cpgp40/15 allelic subgroups were found in the Dracy le Fort outbreak. Of 23 isolates, 21 were type Ib by PCR-RFLP (products were not amplified from two of the original samples). Isolate 7 belonged to the type Id subgroup, while isolate 15, previously identified as genotype I, was found to be type IIa at this locus (Fig. 1). These results were confirmed through sequence analysis. A complete or a partial nucleotide sequence could be obtained from the PCR products of all 21 isolates. The phylogenetic tree in Fig. 2 shows the relationship of the Cpgp40/15 nucleotide sequences of isolates from this study to each other and to those of previously identified representative subgroup Id, Ib, and IIa isolates deposited in GenBank. The sequences of the isolates of type Ib, which was the predominant subtype identified in this study, were almost identical to each other, suggesting that the outbreak may have occurred from a common source.
FIG. 1.
PCR-RFLP analysis of the Cpgp40/15 locus from representative isolates. (A) Secondary PCR amplicons digested with RsaI; (B) secondary PCR amplicons digested with AluI. The first three lanes of both panels show the RFLP profiles for known subgenotype Ib, Id, and IIa Cpgp40/15 sequences previously deposited in GenBank (GenBank accession numbers AF440626, AF440625, and AF114166, respectively), while the last three lanes in each panel show the PCR-RFLP profiles from isolates 5 (a representative type Ib isolate), 7, and 15, respectively. Lane M, markers. Following RsaI digestion, the expected sizes of the PCR products, based on the known sequences of isolates with GenBank accession numbers AF440626 (type Ib), AF440625 (type Id), and AF114166 (type IIa), were 387, 200, 195, 159, and 35 bp for the Ib subtype; 483, 367, 200, and 30 bp for the Id subtype; and 329, 207, 195, 177, and 35 bp for the IIa subtype. Following AluI digestion, the expected sizes of the PCR products were 457, 380, 85, and 56 bp for the Ib subtype; 520, 385, and 175 bp for the Id subtype; and 294, 258, 214, 81, 60, and 36 bp for the IIa subtype.
FIG. 2.
Genetic relationship among isolates from the outbreak. The Cpgp40/15 nucleotide sequences from the type Ib, Id, and IIa isolates from the outbreak were aligned with each other and with known Cpgp40/15 subgenotype sequences previously deposited in GenBank (GenBank accession numbers AF440626, AF440625, and AF114166, respectively) by using the Clustal W algorithm in the MegAlign 6.2 program from the Lasergene suite 6.0 of DNASTAR Inc. A phylogenetic tree was constructed from the alignment by using the neighbor-joining algorithm in the MegAlign 6.2 program. The nucleotide substitutions are in hundreds.
Despite the diversity of other infectious agents contaminating the water supply in Dracy le Fort, it is surprising that only one Cryptosporidium subgroup, subgroup Ib, should predominate among the cases of cryptosporidiosis examined. Furthermore, this homogeneity in Cryptosporidium sp. infection does not appear to be unique to the outbreak in question. Cpgp40/15 type Ib isolates have been implicated as the major source of illness in at least three other waterborne outbreaks, although an explanation for this is unknown (4, 17). If other outbreaks fit this emerging pattern, further research will be needed to determine whether Cpgp40/15 allele Ib represents an elevated threat to public health.
Nucleotide sequence accession numbers.
The Cpgp40/15 sequences from isolates in this study have been deposited in GenBank under accession numbers DQ184508, AY677199, AY677200, and AY702619 to AY702638.
Acknowledgments
This work was supported by National Institutes of Health grant AI05786 (to H.D.W.) and center grant DK34928-18 to the GRASP Digestive Diseases Center at Tufts—New England Medical Center.
We thank Anne Kane and the Intestinal Microbiology Core of the GRASP Center for reagents and Anne Kane and Roberta O'Connor for careful review of the manuscript.
REFERENCES
- 1.Alves, M., L. Xiao, I. Sulaiman, A. A. Lal, O. Matos, and F. Antunes. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 41:2744-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cevallos, A. M., X. Zhang, M. K. Waldor, S. Jaison, X. Zhou, S. Tzipori, M. R. Neutra, and H. D. Ward. 2000. Molecular cloning and expression of a gene encoding Cryptosporidium parvum glycoproteins gp40 and gp15. Infect. Immun. 68:4108-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalle, F., P. Roz, G. Dautin, M. Di-Palma, E. Kohli, C. Sire-Bidault, M. G. Fleischmann, A. Gallay, S. Carbonel, F. Bon, C. Tillier, P. Beaudeau, and A. Bonnin. 2003. Molecular characterization of isolates of waterborne Cryptosporidium spp. collected during an outbreak of gastroenteritis in South Burgundy, France. J. Clin. Microbiol. 41:2690-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glaberman, S., J. E. Moore, C. J. Lowery, R. M. Chalmers, I. Sulaiman, K. Elwin, P. J. Rooney, B. C. Millar, J. S. Dooley, A. A. Lal, and L. Xiao. 2002. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8:631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leav, B. A., M. Mackay, and H. D. Ward. 2003. Cryptosporidium species: new insights and old challenges. Clin. Infect. Dis. 36:903-908. [DOI] [PubMed] [Google Scholar]
- 6.Leav, B. A., M. R. Mackay, A. Anyanwu, R. M. O'Connor, A. M. Cevallos, G. Kindra, N. C. Rollins, M. L. Bennish, R. G. Nelson, and H. D. Ward. 2002. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect. Immun. 70:3881-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacKenzie, W. R., N. J. Hoxie, M. E. Proctor, M. S. Gradus, K. A. Blair, D. E. Peterson, J. J. Kazmierczak, D. G. Addiss, K. R. Fox, J. B. Rose, et al. 1994. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 331:161-167. [DOI] [PubMed] [Google Scholar]
- 8.Morgan-Ryan, U. M., A. Fall, L. A. Ward, N. Hijjawi, I. Sulaiman, R. Fayer, R. C. Thompson, M. Olson, A. Lal, and L. Xiao. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433-440. [DOI] [PubMed] [Google Scholar]
- 9.Peng, M. M., S. R. Meshnick, N. A. Cunliffe, B. D. Thindwa, C. A. Hart, R. L. Broadhead, and L. Xiao. 2003. Molecular epidemiology of cryptosporidiosis in children in Malawi. J. Eukaryot. Microbiol. 50(Suppl.):557-559. [DOI] [PubMed] [Google Scholar]
- 10.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. Mac Kenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Priest, J. W., J. P. Kwon, M. J. Arrowood, and P. J. Lammie. 2000. Cloning of the immunodominant 17-kDa antigen from Cryptosporidium parvum. Mol. Biochem. Parasitol. 106:261-271. [DOI] [PubMed] [Google Scholar]
- 12.Rose, J. B., D. E. Huffman, and A. Gennaccaro. 2002. Risk and control of waterborne cryptosporidiosis. FEMS Microbiol. Rev. 26:113-123. [DOI] [PubMed] [Google Scholar]
- 13.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzipori, S., and H. Ward. 2002. Cryptosporidiosis: biology, pathogenesis and disease. Microbes Infect. 4:1047-1058. [DOI] [PubMed] [Google Scholar]
- 15.Winter, G., A. A. Gooley, K. L. Williams, and M. B. Slade. 2000. Characterization of a major sporozoite surface glycoprotein of Cryptosporidum parvum. Funct. Integr. Genomics 1:207-217. [DOI] [PubMed] [Google Scholar]
- 16.Wu, Z., I. Nagano, T. Boonmars, T. Nakada, and Y. Takahashi. 2003. Intraspecies polymorphism of Cryptosporidium parvum revealed by PCR-restriction fragment length polymorphism (RFLP) and RFLP-single-strand conformational polymorphism analyses. Appl. Environ. Microbiol. 69:4720-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou, L., A. Singh, J. Jiang, and L. Xiao. 2003. Molecular surveillance of Cryptosporidium spp. in raw wastewater in Milwaukee: implications for understanding outbreak occurrence and transmission dynamics. J. Clin. Microbiol. 41:5254-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]