Abstract
The Leishmania donovani complex, which consists of L. donovani, L. infantum-L. chagasi, and L. archibaldi, is responsible for visceral manifestations of leishmaniasis. Multilocus enzyme electrophoresis is the standard method for the characterization and identification of strains of Leishmania. For L. infantum, the predominance of zymodeme MON-1 significantly reduces the discriminative power of this approach. In the present study, we developed 17 independent polymorphic microsatellite markers for the typing of strains of L. infantum, with the main emphasis on zymodeme MON-1. The discriminative powers of 11 markers selected from among these markers were tested by using a panel of 63 isolates of the L. donovani complex. Unique multilocus genotypes were observed for the strains analyzed, with only three exceptions. Model-based and distance-based analyses of the data set showed comparable results. It was possible to discriminate between L. donovani sensu stricto, a non-MON-1 group of L. infantum isolates, and a MON-1 group of L. infantum isolates. Within MON-1, three clusters with geographical correlations became apparent. The frequency of heterozygosity in the alleles analyzed varied extremely between the different groups of isolates. The main clusters described are not consistent with species definitions based on isoenzyme analysis but confirm the results of former PCR-based investigations.
Leishmania is a genus of protozoan flagellates that cause a broad spectrum of diseases, ranging from self-limiting localized cutaneous lesions to visceral leishmaniasis with fatal spontaneous evolution (2). The majority of visceral manifestations are caused by parasites of the Leishmania donovani complex (26), which consists of L. donovani Ross, 1903; L. infantum Nicolle, 1908-L. chagasi Cunha and Chagas, 1937; and L. archibaldi Castellani and Chalmers, 1919.
Currently, multilocus enzyme electrophoresis (MLEE) is the generally accepted “gold standard” for the identification and classification of isolates of Leishmania. By this method, strains are divided into groups with identical enzyme patterns, called “zymodemes.” The main criticism of this approach is that genotypes are assayed indirectly, with the consequence that nucleotide substitutions may not be observed in synonymous sites or in nonsynonymous sites, if it is assumed that subsequent changes in the amino acid composition do not lead to different electrophoretic mobilities. In contrast, posttranslational modifications may change the electrophoretic mobilities, despite identical genotypes. Furthermore, the method is quite slow, laborious, and costly. Growth in vitro is inevitable, and the data sets of the few laboratories in which these analyses are performed are difficult to compare.
Unlike, e.g., L. donovani or L. tropica, which present extended genetic and enzymatic polymorphisms (27, 53), L. infantum is a relatively uniform species (46). More than 80% of the strains of L. infantum isolated thus far belong to the predominant zymodeme, MON-1 (40, 42). The alternative methods for species and/or strain discrimination include analysis with monoclonal antibodies (17); molecular karyotyping (7, 18); PCR fingerprinting-random amplified polymorphic DNA analysis (1, 19, 36, 51, 60); and PCR amplification of distinct nuclear multicopy target sequences or kinetoplast DNA, followed by an analysis of the amplificates by sequencing, fragment length polymorphism analysis, restriction fragment length polymorphism analysis (6, 29, 30, 32, 35, 44), and single-strand conformation polymorphism analysis (10). These methods are all limited in the intrinsic level of polymorphism that they can detect. These approaches are able to distinguish between strains belonging to the predominant zymodeme, MON-1, only in exceptional cases, thus making epidemiological or population genetic investigations of L. infantum almost impossible.
Recently, analysis of the length polymorphisms of microsatellite-containing regions has become an important tool for population and genetic studies for many different species (14, 58). Microsatellites are tandemly repeated stretches of short nucleotide motifs of 1 to 6 bp ubiquitously distributed in the genomes of eukaryotic organisms. They mutate at rates five to six orders of magnitude higher than that of the bulk of DNA. Microsatellite loci present high variability mainly due to allelic repeat length variation (8). The length variation of individual loci can easily be screened after amplification with primers that anneal specifically to their flanking regions. The results of these analyses are reproducible and exchangeable between laboratories. In contrast to MLEE, selection does not seem to act on polymorphisms in microsatellite length, and allelic variants are detectable because of the codominant nature of these markers. The major obstacle is the need to develop a new panel of 10 to 20 markers for nearly every species. Additionally, polymorphic repeats are not conserved between different species of Leishmania (21, 54).
Leishmania is relatively rich in microsatellites (47). Consequently, 13 markers were designed for L. major (21), 16 markers were designed for L. tropica (54), and 20 markers were designed for L. donovani (22). Two independent microsatellite loci described by Rossi et al. (47) and three genomic fragments containing several different microsatellite tracts (5) were polymorphic in L. infantum. Only two of the microsatellite sequences described by Bulle et al. (5), however, varied within strains of MON-1.
In the present study, we developed a panel of 11 independent microsatellite markers that showed polymorphisms within L. infantum. These loci were tested with different isolates of L. infantum from various geographic sites, with the main emphasis being on zymodeme MON-1 as well as other representatives of the L. donovani complex.
MATERIALS AND METHODS
Parasite culture and DNA preparation.
The sources, clinical manifestations, geographical origins, and zymodemes of the strains are listed in Table 1. Parasite promastigotes were maintained in Novy-Nicolle-McNeal medium and cultured in RPMI 1640 medium supplemented with 15% fetal calf serum. After the promastigotes were harvested, they were washed in buffered saline (52). DNA was extracted by the phenol-chloroform method as described previously (33), suspended in TE (Tris-EDTA) buffer (50), and stored at 4°C.
TABLE 1.
Strains of the L. donovani complex analyzed in this study
| Speciesa | WHO codeb | Origin | Zymodeme (MON)c | Clinical patternd |
|---|---|---|---|---|
| L. infantum (L. chagasi) | MHOM/TN/1980/IPT1Te | Tunisia | 1 | VL |
| MHOM/FR/1978/LEM75 | France | 1 | VL | |
| MHOM/FR/1995/LPN114e | France | 1 | VL | |
| MHOM/ES/1993/PM1e | Spain, Majorca | 1 | VL-HIV+ | |
| MHOM/FR/1997/LSL29 | France | 1 | CL | |
| MHOM/ES/1986/BCN16 | Spain | 1 | CL | |
| MHOM/PT/2000/IMT260 | Portugal | 1 | CL | |
| MHOM/ES/2001/LLM983 | Spain | 1 | VL-HIV+ | |
| MHOM/ES/2001/LLM1049 | Spain, Majorca | 1 | VL-HIV+ | |
| MHOM/ES/2002/LLM1109 | Spain, Majorca | 1 | VL | |
| MCAN/ES/2001/LLM1038 | Spain, Majorca | 1 | CanL | |
| MCAN/ES/2002/LLM1203 | Spain, Ibiza | 1 | CanL | |
| MCAN/ES/2002/LLM1139 | Spain, Ibiza | 1 | CanL | |
| MCAN/ES/2002/LLM1141 | Spain, Ibiza | 1 | CanL | |
| MCAN/PT/1989/IMT162 | Portugal | 1 | CanL | |
| MHOM/PT/2002/IMT288 | Portugal | 1 | VL | |
| MHOM/PT/2003/IMT337 | Portugal | 1 | CL | |
| MCAN/PT/2003/IMT300 | Portugal | 1 | CanL | |
| MCAN/PT/2003/IMT327 | Portugal | 1 | CanL | |
| MCAN/PT/2003/IMT338 | Portugal | 1 | CanL | |
| MCAN/PT/2003/IMT339 | Portugal | 1 | CanL | |
| MCAN/PT/1994/IMT193 | Portugal | 1 | CanL | |
| MHOM/GR/2001/GH1 | Greece | 1 | VL | |
| MHOM/GR/2001/GH2 | Greece | 1 | VL | |
| MHOM/GR/2001/GH3 | Greece, Crete | 1 | VL | |
| MHOM/GR/2001/GH5 | Greece, Crete | 1 | VL | |
| MHOM/GR/2001/GH8 | Greece | 1 | VL | |
| MHOM/GR/2001/GH9 | Greece | 1 | VL | |
| MCAN/GR/2001/GD7 | Greece, Crete | 1 | CanL | |
| MHOM/CN/1978/D2e | China | NDf | VL | |
| MHOM/CN/1954/Peking | China | ND | VL | |
| MCAN/TR/1996/EP16 | Turkey | ND | CanL | |
| MHOM/TR/1994/EP3 | Turkey | ND | ND | |
| MCAN/IL/1994/LRC-L639 | Israel | ND | CanL | |
| MCAN/IL/1996/LRC-L685 | Israel | ND | CanL | |
| MHOM/BR/1974/PP75T | Brazil | 1 | VL | |
| MHOM/PA/1979/WR317 | Panama | ND | CL | |
| MHOM/BR/1985/M9702 | Brazil | ND | VL | |
| MHOM/BR/19??/Edmael | Brazil | ND | ND | |
| MHOM/CR/199?/LVCR | Costa Rica | ND | ND | |
| MCAN/ES/1986/LEM935e | Spain | 77 | CanL | |
| MCAN/FR/1987/RM1e | France | 108 | CanL | |
| MHOM/FR/1962/LRC-L47 | France | ND | VL | |
| MHOM/ES/1987/Lombardi | Spain | 24 | CL | |
| MHOM/ES/1988/LLM175e | Spain | 198 | VL-HIV+ | |
| MHOM/FR/1996/LEM3249 | France | 29 | CL | |
| MHOM/ES/1991/LEM2298 | Spain | 183 | VL-HIV+ | |
| MHOM/FR/1980/LEM189 | France | 11 | CL | |
| MHOM/ES/1992/LLM373e | Spain | 199 | VL-HIV+ | |
| MHOM/IT/1994/ISS1036 | Italy | 228 | VL | |
| MHOM/MT/1985/BUCK | Malta | 78 | CL | |
| MHOM/SD/1962/3S | Sudan | 81 | VL | |
| MHOM/SD/1997/LEM3472e | Sudan | 267 | PKDL | |
| L. donovani | MHOM/IN/1980/DD8Te | India | 2 | VL |
| MHOM/IN/2000/DEVI | India | 2 | VL | |
| MHOM/IN/1996/THAK35 | India | 2 | VL | |
| MHOM/ET/1967/HU3 | Ethiopia | 18 | VL | |
| MHOM/ET/2000/HUSSEN | Ethiopia | 31 | VL | |
| MHOM/SD/1982/GILANI | Sudan | 30 | VL | |
| MCAN/SD/2000/LEM3946 | Sudan | 274 | VL | |
| L. archibaldi | MHOM/ET/1972/GEBRE1e | Ethiopia | 82 | VL |
| MHOM/SD/1997/LEM3429 | Sudan | 257 | VL | |
| MHOM/SD/97/LEM3463 | Sudan | 258 | VL |
Species as defined by MLEE (40).
The World Health Organization (WHO) code is as follows: host (MHOM, Homo sapiens; MCAN, Canis familiaris)/country/year of isolation/name of strain. T, World Health Organization reference strain.
Zymodemes are indicated as typed by the reference laboratory, Montepellier (MON).
VL, visceral leishmaniasis; CL, cutaneous leishmaniasis; PKDL, post-kala-azar-dermal leishmaniasis; CanL, leishmaniasis manifestation of infected dog; VL-HIV+: Leishmania-human immunodeficiency virus coinfection.
The strain is part of the set of 11 isolates for initial marker testing.
ND, not determined.
PCR amplification assays.
Unless mentioned otherwise, PCRs were performed in a volume of 50 μl containing 200 nM of each deoxynucleoside triphosphate, 1 U AmpliTaq DNA polymerase (Roche), and 10 mM Tris-HCl buffer (pH 8.3) containing 50 mM KCl and 1.5 mM MgCl2. The concentrations of the primers and the templates are indicated below. After an initial denaturation step of 10 min at 95°C, samples were processed through 35 cycles consisting of 30 s at 95°C, 30 s at the annealing temperature (TA) indicated in Table 2, and 1 min at 72°C, followed by a terminal elongation step of 10 min at 72°C.
TABLE 2.
Microsatellite markers developed in this study
| Markera | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) | TA (°C) | Polymorphisms within MON-1b | Chromosomec |
|---|---|---|---|---|---|
| Li22-35 | CTTGATGTTCGGGTTAGCAAGT | ATGCACACCAAAAATCATGTG | 52 | p | ND |
| Li23-41 | GATCGGAGGTGACAGCGT | CCTTTAACTGCCAGTGCG | 52 | p | 25 |
| Li41-56 | TTGCTTCATGATAACAACTTGG | CCTGTTGGTGTGAGTTCGTG | 50 | p | 36 |
| Li45-24 | GCGCCTACAGGCATAAAGGA | CTGGCGCATCAACGGTGT | 54 | p | 16 |
| Li46-67 | TCTTCTTTCGTTAGCTGAGTGC | CTGTATCACCCATGAGGGGC | 50 | m | 31 |
| Li71-5/2 | GCACGGTCGGCATTTGTA | GATAAACGAGATGGCCGC | 56 | p | 35 |
| Li71-7 | GCTGCAGCAGATGAGAAGG | GTGAGAAGGCAGGGATTCAA | 50 | p | ND |
| Li71-33 | CTCCTTTCACACCGCCTCT | GAGAGAAGACGAGCCGAAGT | 50 | p | 31 |
| Lm2TG | AAAAAGCGAGGAATGAAAGAA | TCCCTCCCCTCTACAACCTT | 53 | p | 1 |
| Lm4TA | TTTGCCACACACATACACTTAG | GTAGACGACATCGCGAGCAC | 54 | p | 1 |
| TubCA | GGCGTGGTTGCTAAACTGAT | GCCTGCGCACACAGAGAC | 58 | p | 34 |
| Li21-34 | GAGAAAGCAAGACACGAGATGA | GAGGCGTTTTCCTTCTGGTAG | 52 | m | 1 |
| Li71-19 | CAAACCGGTGTTCTGCTTTT | CGGACATTGGCAACACACT | 52 | m | ND |
| Li71-42 | CCAAGCACCACTCAAGCATA | TCTTGTCCTCTTCGGTGCTT | 54 | m | 32 |
| Li72-14 | AGAGTGTCTGCGCGTGAGTA | AAGAAAAGAAGGTGCAGCGA | 54 | m | ND |
| Li72-17/2 | CTGTTTTACGCCCAACCATC | CTTTTGGGATCGCAGGTTT | 48 | m | 35 |
| Li72-20 | GATCCCTTCGGATTACTGC | CTGCTAGCGAGGGGATAGG | 50 | p | 31 |
All markers showed polymorphisms within the L. donovani complex.
p, polymorphic within MON-1; m, not polymorphic within MON-1.
Based on the corresponding sequences of L. major found in the database. ND, not determined.
Sequencing.
Direct cycle sequencing of the PCR products was performed with an automatic sequencer (ABI Prism 377; Applied Biosystems) and a Taq DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems). PCR was carried out as recommended by the manufacturer. Afterward, the PCR samples were resolved on a 6% polyacrylamide gel attached to the sequencer.
For sequencing of the cloned fragments, primers SEQ_fw24 (5′-ACG ACG TTG TAA AAC GAC GGC CAG-3′) and SEQ_rev23 (5′-TTC ACA CAG GAA ACA GCT ATG AC-3′) (Martin Meixner, personal communication) were used. The α-tubulin intergenic region was sequenced with primers Tub1 (5′-AGG AGG AGG CGC GTA GCA T-3′) and Tub2 (5′-ACG CAT GGT TGA CAA GGA AG-3′) (Michal Shapira, personal communication) for several different strains. Additional primers, primer pair Tub3f (5′-TCC GAG AAC GGA GTC TAC A-3′) and Tub3r (5′-AAT AAA ATC ACG TTG ATG CG-3′) and primer pair Tub4f (5′-GAC TTT ATT GGT GTA GGC CC-3′) and Tub4r (ACA CGA ACA ACA AGA ACA GG-3′), both with TAs of 56°C, were developed based on the consensus sequences. The Tub3 and Tub4 fragments were sequenced separately in both directions.
Design of microsatellite markers.
The construction of an enriched genomic library was based on the protocols of Bloor et al. (3) and Jamjoom et al. (22). Ten micrograms of DNA extracted from a MON-1 strain from Spain (MHOM/ES/93/PM1) was digested with Sau3A (New England BioLabs). The fragments were ligated to specific adaptors consisting of “Oligo A” (5′-GGC CAG AGA CCC CAA GCT TCG-3′) and “Oligo B” (5′ PO4-GAT CCG AAG CTT GGG GTC TCT GGC C-3′) (45). Fragments between 400 and 1,000 bp were excised from the agarose gel, extracted, and concentrated by using YM-50 spin columns (Millipore). The fractionated DNA was denatured and hybridized at a temperature of 55°C to (GT)10 3′-biotinylated oligonucleotides bound to M-280 streptavidin-coated magnetic beads (Dynal, Norway) (23). After incubation at 72°C for 2 h, unbound DNA and excess oligonucleotides were eluted following differential stringency washes. For amplification of the immobilized fragments, PCR was conducted by using a suspension of 40 μg magnetic beads with the enriched fragments attached, 3.5 mM MgCl2, and 30 pmol of “Oligo A.” The terminal elongation step was extended to 30 min. The amplified fragments were ligated in a pDrive vector system (QIAGEN, The Netherlands) and transformed into competent Escherichia coli, according to the manufacturer's instructions. Cells were plated out on Luria-Bertani agar plates containing isopropyl-β-d-thiogalactopyranoside and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and incubated for 18 h at 37°C. For screening of the library, colony PCRs were conducted by using a TA of 52°C, 10 pmol of standard primers M13F and M13R, oligonucleotide (AC)10, and several cells of the white colonies as the template. Microsatellite-containing fragments produced double bands in the subsequent gel electrophoresis (3). For sequencing, positive colonies were amplified again by using 10 pmol of primers SEQ_fw24 and SEQ_rev23 (see above) at a TA of 55°C.
By using Primer3 software (48), PCR primers between 18 bp and 22 bp in length were designed against sequences flanking the detected microsatellites. Primers were deduced from sequences 5 to 25 nucleotides upstream and downstream of the repeat, with three exceptions due to unsuitable sequence patterns. A BLAST search was conducted for all markers to find corresponding sequences of Leishmania in order to determine the chromosome on which the amplified region was localized. One fragment with an insufficient flanking region was supplemented with a published sequence of L. major for primer design (template sequence of Li21-34R from strain MHOM/IL/80/Friedlin).
Primers Lm2TGf and Lm2TGr and primers Lm4TAf and Lm4TAr (Table 2) were designed by using the sequences published for two anonymous fragments, Lm2 and Lm4 (5). Lm2 (∼900 bp) contains three microsatellite tracts, the most polymorphic of which is a poly(TG) repeat located in the middle of the fragment. Primers Lm2TGf and Lm2TGr anneal to the flanking sequences of this microsatellite. Primers Lm4TAf and Lm4TAr were designed to amplify the variable poly(TA) repeat close to the 3′ end of fragment Lm4 (∼560 bp).
Amplification of the α-tubulin intergenic region by primers Tub1 and Tub2 (see above) at a TA of 65°C yielded a PCR product of approximately 900 bp. Primers TubCAf and TubCAr (Table 2) were designed to amplify the most polymorphic microsatellite of the α-tubulin intergenic region.
Analysis of microsatellite variation.
Amplification reactions were performed by using 40 ng DNA as a template and 10 pmol of each primer. All PCR assays were optimized with regard to the annealing temperature and by using the DNA of the originally cloned strain as the template. Polyacrylamide gels (12%; Roth, Germany) 350 by 450 by 0.8 mm were prepared by following the manufacturer's instructions. Separation was carried out in a vertical electrophoresis system for 18 h at 10 W. Gels were fixed in 1% nitric acid for 10 min, stained in 0.2% silver nitrate for 25 min, and developed by the use of 0.28 M sodium carbonate with 250 μl 37% formalin added to each liter of solution. The gels were fixed with 10% acetic acid and blotted on filter paper. The dried gels were scanned and evaluated by using Bionumerics version 2.5 software (Applied Maths BVBA, Belgium).
Screening of the length variations of the amplified markers was also performed by gel electrophoresis in 4% MetaPhor agarose gels (BioWhittaker Molecular Applications), prepared by following the manufacturer's instructions. Separation took 3.5 h at 6 V/cm of interelectrode distance in a standard horizontal electrophoresis system.
In addition, PCR was performed with fluorescence-conjugated forward primers (Proligo, France), and the amplified products were analyzed in an automated capillary sequencer (CEQ 8000; Beckman Coulter) by using the fragment analysis tool.
Analysis of population structure.
The multilocus genotype data, consisting of the repeat numbers determined by polyacrylamide gel electrophoresis (PAGE) or automated fragment analysis, were processed by using STRUCTURE software (43). This application can be used to infer a population structure by a model-based clustering method independent of a particular mutation model. The complete set of individuals is divided into K subpopulations, with K ranging from 1 to 10. Isolates are assigned probabilistically to clusters or, in the case of admixed genotypes, jointly to more than one cluster, with the membership coefficients of all subpopulations adding up to 1 by use of a Bayesian algorithm. A series of runs (a burn-in period of 20,000 iterations and a run of 200,000 iterations) was performed for each value of K between 1 and 10.
Alternatively, pairwise distance matrices based on the genetic distance (DSA) and the microsatellite distance (Ddm) were calculated. DSA is based on the infinite allele model. For individual pairwise comparison, DSA is estimated by 1 − PSA (where PSA is the proportion of shared alleles) (4). Ddm was developed for microsatellite application; it follows the single-step mutation model and is based on the average squared difference in allele size (15, 16). Distances matrices (100 bootstrap replicates) were calculated by using MICROSAT software (34). Phylograms based on the algorithm of the unweighted pair group method with arithmetic averages (UPGMA) (55) and consensus trees (majority rule, semistrict) were constructed by using PHYLIP 3.5c (11) and PAUP version 4.0b8 software (56).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been submitted to the EMBL database with the accession numbers AM050044 to AM050057.
RESULTS
Altogether, 1,154 clones of the (CA)n-enriched genomic library were screened, and 104 of these clones contained microsatellite structures. Redundancies and poor sequences reduced the number of suitable sequences to 28. For primer design, attention was paid to avoiding long flanking regions in which additional mutational events could interfere with the length differences in the microsatellite tracts. In addition, fragment length analysis in polyacrylamide and MetaPhor agarose gels is easier to perform with shorter PCR products.
Initially, 28 microsatellite markers were tested with 11 strains of the L. donovani complex (Table 1). Fourteen of these produced an amplificate of the expected size and were polymorphic within the L. donovani complex (Table 2). Of these, eight markers which presented polymorphisms within Mediterranean strains of L. infantum were selected for further analyses. Three microsatellite markers based on Lm2 and Lm4 sequences and the α-tubulin intergenic region completed the panel.
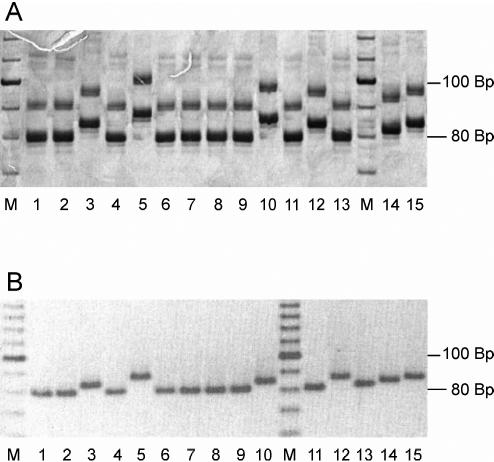
For fragment analysis, three different methods were used. PAGE (Fig. 1A), MetaPhor agarose gel electrophoresis (Fig. 1B), and fragment analysis with an automated capillary sequencer (data not shown) were all found to be suitable for the detection of fragment length variations and yielded reproducible and comparable results. While PAGE analysis is a time-consuming method, automated fragment analysis can be performed more quickly, and the ability to process three different markers in one lane makes this approach suitable for high-throughput analysis. Separation on MetaPhor agarose gels has proven to be a quick and easy method for the screening of large numbers of samples for small length polymorphisms. Differences of a single dinucleotide repeat can be reliably determined between fragments run in neighboring lanes (Fig. 1B).
FIG. 1.
Fragment length analysis by PAGE (A) and MetaPhor agarose gel electrophoresis (B) of PCR products amplified with marker TubCA. Lanes (repeat numbers are in parentheses): 1, PP75 (n = 9); 2, WR317 (n = 9); 3, Lombardi (n = 11); 4, LEM75 (n = 9); 5, LPN114 (n = 13); 6, PM1 (n = 9); 7, LSL29 (n = 9); 8, BCN16 (n = 9); 9, IMT260 (n = 9); 10, LRC-L47 (n = 12); 11, RM1 (n = 9); 12, LEM3249 (n = 11); 13, LEM2298 (n = 9); 14, BUCK (n = 10); 15, LEM189 (n = 11); M, 10-bp ladder.
The set of 11 microsatellite markers was tested with 51 strains of L. infantum from various areas of endemicity in the Mediterranean Basin and South America, with the main emphasis on strains of zymodeme MON-1. For comparison, 12 East African and Indian strains representing different taxonomic groups of the L. donovani complex were included (Table 1). Previous genotypic analysis has not confirmed the existence of three different species, namely, L. donovani, L. infantum, and L. archibaldi, in Sudan and Ethiopia, as identified by MLEE (9, 20, 25, 30, 32, 44, 60). In this study, the term “L. infantum” is therefore applied only to strains from the Mediterranean Basin, Latin America (synonymous with L. chagasi [31]), or China, while all isolates from East Africa and India are considered “L. donovani.”
With the exception of marker Li71-5/2, which did not amplify five MON-1 isolates from Greece (isolates GH2, GH3, GH5, GH8, and GD7), one or two PCR products of the expected size were obtained for all strains and markers tested. The lengths of the amplified fragments, and thus the number of repeats, were always compared to the lengths of the fragments from cloned strain PM1.
All markers used here with the exception of Li46-67 showed polymorphisms within strains of zymodeme MON-1; Li46-67 was, however, polymorphic for strains of other L. infantum zymodemes (Table 3). Three markers (Lm2, Li46-67, and Li71-7) showed species-specific alleles for either L. donovani or L. infantum.
TABLE 3.
Microsatellite markers tested with all strains
| Marker | Repeat arraya | No. of allelesb
|
||
|---|---|---|---|---|
| All strains | L. infantum | MON-1 | ||
| Li22-35 | (CA) 8-39 | 18 | 17 | 8 |
| Li23-41 | (GT) 6-32 | 14 | 9 | 4 |
| Li41-56 | (AC) 7-13 | 7 | 6 | 2 |
| Li45-24 | (CA) 7-20 | 10 | 9 | 4 |
| Li46-67 | (CA) 6-9 | 4 | 4 | 1 |
| Li71-5/2 | (CA) 7-16 | 5 | 5 | 4 |
| Li71-7 | (AC) 8-13 | 6 | 3 | 2 |
| Li71-33 | (TG) 8-27 | 11 | 9 | 3 |
| Lm2TG | (TG) 9-28 | 14 | 13 | 6 |
| Lm4TA | (TA) 6-16 | 9 | 7 | 7 |
| TubCA | (CA) 9-13 | 5 | 5 | 2 |
Repeat array indicates the motif and the number of repeats, with the shortest and the longest alleles observed indicated.
Number of different alleles observed for all strains investigated, within L. infantum, and within MON-1.
The analysis of microsatellite repeat variation revealed double bands at several of the loci analyzed. The appearance of three or more alleles per locus was never observed. Altogether, 9.7% of the 11 loci tested with 63 strains were apparently heterozygous for their microsatellite repeat numbers. The number of double bands per strain varied between zero and eight, depending on which main cluster the strain was a part of. While isolates of MON-1 presented almost no double bands or one at the most, all isolates from Sudan and Ethiopia had double bands for at least two loci. The non-MON-1 cluster from France, Spain, Italy, and Latin America showed a comparable frequency of double bands (25% of all loci analyzed). Remarkably, for many loci the same double band patterns were present in two or more strains within a particular cluster.
Based on the analysis of variation at 11 microsatellite loci, multilocus microsatellite types were established for the 63 strains studied. Among all isolates tested, only three multilocus microsatellite types were represented twice (the pairs GH8 and GD7, M9702 and LEM75, and DEVI and THAK35 each produced identical patterns). A correlation of the microsatellite patterns and the clinical manifestation caused by a particular strain or the host from which it was isolated could not be observed.
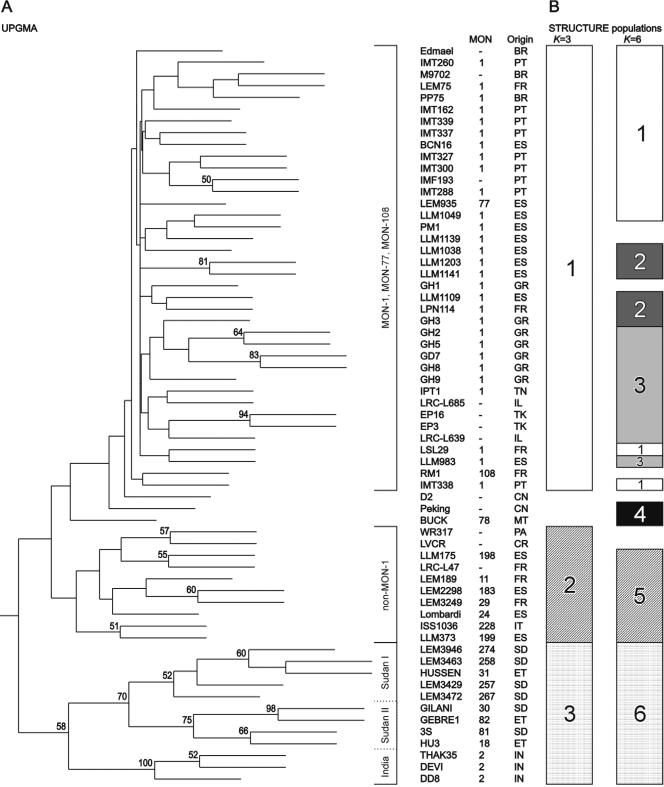
The genetic distance between the 63 strains of the L. donovani complex was calculated by using the coefficients DSA, and Ddm, both of which resulted in the same tree topologies. The semistrict UPGMA phenograms, based on the distance matrices obtained, produced three major clusters corresponding to MON-1 strains of L. infantum, non-MON-1 strains of L. infantum, and East African and Indian strains of L. donovani (Fig. 2A). Nine isolates not classified by MLEE as well as two strains assigned to MON-77 and MON-108, respectively, were grouped together with the 29 MON-1 strains. The L. donovani cluster was further subdivided into three stable groups. Two of these, Sudan I and Sudan II, contained all strains from Sudan or Ethiopia. The third contained three MON-2 strains from India. The basal nodes separating the major MON-1, non-MON-1, and L. donovani clusters were, however, not supported by significant bootstrap values. Bootstrap values >50 were obtained only for nodes between subgroups inside the major clusters.
FIG. 2.
(A) Bootstrap analysis of all isolates analyzed (DSA, UPGMA). Bootstrap values >50 are shown above the branches. (B) Population inference by model-based analysis. Populations are as inferred by STRUCTURE for K equal to 3 and K equal to 6. Strains that could not be clearly assigned to a population because of admixture were excluded. For the strain code, see Table 1.
The Bayesian model-based algorithm implemented in the program STRUCTURE attempts to identify genetically distinct subgroups on the basis of allelic frequencies (43). The analysis of all 63 isolates produced the same major groups at K equal to 3, MON-1, non-MON-1, and L. donovani, as seen in the UPGMA tree (Fig. 2). Again, one large cluster represented all MON-1 strains together with several isolates of an unknown zymodeme, and the two strains were assigned to MON-77 and MON-108, respectively. At K equal to 6, the MON-1 group became further subdivided. The clusters comprising the strains of L. donovani from East Africa and India and non-MON-1 L. infantum remained stable. The only exceptions were two strains of L. chagasi, strains WR317 and LVCR, which showed a mixed assignment at K equal to 6. Strain BUCK from Malta, as well as the two isolates from China, showed admixture assignment at K equal to <6 and were interpreted as separate populations at K equal to 6.
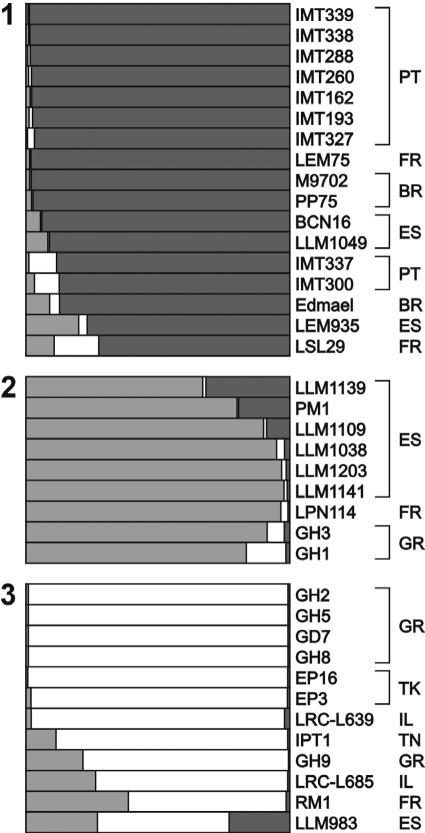
Subsequently, for the 38 isolates assigned to the MON-1 cluster at K equal to 3, a second STUCTURE analysis was carried out. For these strains, an optimal number of three populations (K = 3) was defined, leading to three clearly separated subgroups (Fig. 3). The first group consists predominantly of strains from Portugal, the second consists of Spanish isolates, whereas the third group is mainly composed of strains from the eastern part of the Mediterranean Basin.
FIG. 3.
Model-based analysis of the MON-1 corresponding cluster (K = 3). The lengths of the bars within boxes represent joint assignment to defined populations. The origin is the code defined by World Health Organization nomenclature (Table 1). PT, Portugal; FR, France; BR, Great Britain; ES, Spain; GR, Greece; TK, Turkey; IL, Ireland; TN, Tunisia.
DISCUSSION
An important impetus for this study was the de novo isolation of hypervariable microsatellite loci to compose a set of markers usable for future epidemiological and population genetic studies with closely related strains of the L. donovani complex. Altogether, 17 polymorphic markers were developed, and 11 of these were analyzed with 63 different strains in order to test their discriminative power within the L. donovani complex, particularly within L. infantum MON-1. One of the markers tested was monomorphic within MON-1, and individual multilocus microsatellite patterns were observed for each isolate analyzed, with only three exceptions. Only amplification of kinetoplast DNA followed by restriction fragment length polymorphism analysis with a set of restriction enzymes can lead to a similarly high discriminative power (35); however, the results of microsatellite analysis are much easier to compare between laboratories or to store in databases.
By using a model-based clustering method, it was possible to delimit a group of strains of L. infantum which corresponds mainly to the phenotype MON-1. Furthermore, three definite subgroups were observed in this, until now, highly monomorphic group.
The microsatellite analysis has proven to be a powerful tool for population genetic investigations, as well as epidemiological investigations, of Leishmania species. These short sequence repeats are highly polymorphic, codominant, and dispersed throughout the parasite genome. It has been shown that microsatellite loci of the family Trypanosomatidae are stable under laboratory conditions and can be detected directly in biological samples containing low amounts of parasitic DNA (5, 28; our unpublished data).
A high number of independent microsatellite markers developed by us and other authors (5, 22, 47) are now available for multilocus microsatellite typing (MLMT) (12) of the L. donovani complex. In order to make high-throughput typing feasible, optimization of fragment analysis is most important. We have shown that PAGE, MetaPhor gel electrophoresis, and automated capillary sequencing all produced comparable and reproducible results. Automated fragment analysis can be performed more quickly, particularly because three different samples can be run in one lane. Theoretically, sequencing can also be used to determine the number of repeats. This is indispensable for the analysis of large fragments containing more than one microsatellite. This method is expensive, however, and sequences containing small tandem repeats are difficult to process. For the typing of large numbers of isolates, we recommend screening for polymorphisms in the different loci by using MetaPhor agarose gel electrophoresis and subsequently determining the repeat numbers in samples of different sizes by using fragment analysis in an automated capillary sequencer. This will reduce the number of necessary fragment analyses considerably and thus reduce the costs of microsatellite typing.
Unique multilocus microsatellite patterns have been observed for 34 of the 38 strains of the MON-1 cluster. MLMT thus offers the possibility to track down strains of this predominant zymodeme of L. infantum and to investigate its population dynamics. To infer a DNA-based population structure within L. infantum, a model-based analysis was carried out. This clustering method proved to be superior to distance-based approaches for the processing of data sets with low variability, like those presented by L. infantum MON-1. A separate analysis of the MON-1 cluster revealed three geographical subgroups. The results are promising, but many more isolates must be processed to confirm the spatial subdivision of the MON-1 group.
In general, the three main clusters identified by all phenetic methods were in good agreement with the results of previous studies that used different PCR-based methods (27, 32, 39) and were strongly associated with the zymodeme types (41). The MON-1 cluster identified in this study also contained numerous isolates not typed by MLEE, as well as two strains assigned to MON-77 and MON-108. The last two zymodemes are, however, closely related to MON-1 (41). The two strains from China were assigned to the MON-1 cluster by distance-based analyses but held an intermediate position between that cluster and that of L. donovani obtained by model-based processing.
Distance-based analysis yielded essentially the same clusters, but bootstrap support of >50% was observed only between single strains or subclusters within the main groups and was never observed for basal nodes discriminating the major clusters (Fig. 2A). This is in agreement with the observation that microsatellite data become less informative for distantly related taxa (4). DSA follows the infinite allele mutation model, in which every new mutation is assumed to give rise to a new allele, while Ddm is based on the single-step mutation model, in which alleles can mutate only by the gain or the loss of one repeat unit. Goldstein et al. (16) showed that although an approach based on the single-step mutation model more accurately reflects the relationships of closely related isolates, DSA becomes more reliable as the genetic distance grows. Consequently, in our study DSA produced higher bootstrap values for major groups in the tree (Fig. 2A), while Ddm was more suitable for the detection of significant relationships within the MON-1 group (data not shown).
The significance of microsatellite typing is influenced by the number of loci tested (49). Microsatellites tend to gain repeat units, which is counteracted by constraints on allele sizes (57). This will result in homoplasy caused by alleles that are identical in size but not by descent. When fewer microsatellite loci are analyzed, the differences estimated between isolates depend more upon the markers selected (24). Consequently, whether the analysis of more loci will improve the discriminatory power of MLMT significantly must always be examined. On the other hand, genetic distances between distantly related isolates are underestimated in microsatellite analysis (13), a phenomenon which increases with the degree of polymorphism of the markers used (our unpublished data). By consideration of this phenomenon, for the analysis of population structures within the entire L. donovani complex, it could be useful to complete this panel with markers of low discriminatory power.
Heterozygosity has often been reported in studies of microsatellite variation in other trypanosomatid protozoa (37, 47, 54). The occurrence of double bands that vary in their microsatellite repeat numbers can be explained either by heterozygosity at individual loci or by the possibility that the locus analyzed is part of an isogene sequence (59) or a spacer region between such tandem-like repeated genes. Because the strains tested were not cloned, mixtures of strains cannot be excluded. Remarkably, in this study the frequency of “heterozygous” microsatellite loci was significantly higher in the Sudan-Ethiopia group and the non-MON-1 cluster than in MON-1 strains. The fact that all strains of the L. donovani complex showed patterns of only one or two microsatellite bands at all loci is most consistent with the strains being diploid, although our results do not preclude the possibility that some strains might be aneuploid or even polyploid. Heterozygosity may, however, be underestimated by conventional microsatellite assays because of mutation events that do not lead to length variations and the possible existence of null alleles (38).
Multilocus microsatellite typing with the markers developed in this study has great potential for use in epidemiological and population genetic studies of strains within L. infantum MON-1 in order to investigate the structure and dynamics of the corresponding natural foci. It will also help to answer specific clinical questions, such as the role of parasite persistence after subclinical infection as well as endogenous and/or exogenous reinfection associated with immunosuppression and vector-independent transmission, such as congenital transmission and transmission via blood transfusion or shared syringes.
Acknowledgments
This work was supported by a grant (grant QLK2-CT-2001-01810) from the European Union to G.S.
We thank Carmen Chicharro (Instituto de Salud Carlos III, Madrid, Spain), Lenea Campino (Instituto de Higiene e Medicina Tropical, Universidade Nova de Lisboa, Lisbon, Portugal), Maria Antoniou (Faculty of Medicine, University of Crete, Greece), and Isabel Mauricio (London School of Hygiene and Tropical Medicine, London, United Kingdom) for providing strains and DNA; Renate Rebenstorff for parasite cultivation; and Carola Schweynoch for technical assistance.
REFERENCES
- 1.Andresen, K., M. E. Ibrahim, T. G. Theander, and A. Kharazmi. 1996. Random amplified polymorphic DNA for the differentiation of Leishmania donovani isolates from Sudan. Trans. R. Soc. Trop. Med. Hyg. 90:204-205. [DOI] [PubMed] [Google Scholar]
- 2.Ashford, R. W. 2000. The leishmaniases as emerging and reemerging zoonoses. Int. J. Parasitol. 30:1269-1281. [DOI] [PubMed] [Google Scholar]
- 3.Bloor, P. A., F. S. Barker, P. C. Watts, H. A. Noyes, and S. J. Kemp. 2001. Microsatellite libraries by enrichment, v 1, September. [Online.] http://www.liv.ac.uk/∼kempsj/genomics.html. Accessed January 2002.
- 4.Bowcock, A. M., A. Ruíz-Linares, J. Tomfohrde, E. K. Minch, J. R. Kidd, and L. L. Cavalli-Sforza. 1994. High resolution human evolutionary trees with polymorphic microsatellites. Nature 368:455-457. [DOI] [PubMed] [Google Scholar]
- 5.Bulle, B., L. Millon, J.-M. Bart, M. Gállego, F. Gambarelli, M. Portús, L. Schnur, C. L. Jaffe, S. Fernandez-Barredo, J. M. Alunda, and R. Piarroux. 2002. Practical approach for typing strains of Leishmania infantum by microsatellite analysis. J. Clin. Microbiol. 40:3391-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cupolillo, E., G. Grimaldi, Jr., H. Momen, and S. M. Beverley. 1995. Intergenic region typing (IRT): a rapid molecular approach to the characterization and evolution of Leishmania. Mol. Biochem. Parasitol. 73:145-155. [DOI] [PubMed] [Google Scholar]
- 7.Dujardin, J. C., J. P. Dujardin, M. Tibayrenc, G. Timperman, S. De Doncker, D. Jacquet, J. Arevalo, A. Llanos-Cuentas, H. Guerra, and H. Bermudez. 1995. Karyotype plasticity in neotropical Leishmania: an index for measuring genomic distance among L. (V.) peruviana and L. (V.) braziliensis populations. Parasitology 110:21-30. [DOI] [PubMed] [Google Scholar]
- 8.Ellegren, H. 2000. Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet. 16:551-558. [DOI] [PubMed] [Google Scholar]
- 9.El Tai, N. O., M. El Fari, I. Mauricio, M. A. Miles, L. Oskam, S. H. El Safi, W. Presber, and G. Schönian. 2001. Leishmania donovani: intraspecific polymorphisms of Sudanese isolates revealed by PCR-based analyses and DNA sequencing. Exp. Parasitol. 97:35-44. [DOI] [PubMed] [Google Scholar]
- 10.El Tai, N. O., O. F. Osman, M. El Fari, W. Presber, and G. Schönian. 2000. Genetic heterogeneity of ribosomal internal transcribed spacer (ITS) in clinical samples of Leishmania donovani spotted on filter papers as revealed by single-strand conformation polymorphisms (SSCP) and sequencing. Trans. R. Soc. Trop. Med. Hyg. 94:575-579. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1993. PHYLIP (phylogeny inference package) v 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle.
- 12.Fisher, M. C., D. Aanensen, S. de Hoog, and N. Vanittanakom. 2004. Multilocus microsatellite typing system for Penicillium marneffei reveals spatially structured populations. J. Clin. Microbiol. 42:5065-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher, M. C., G. Koenig, T. J. White, and J. W. Taylor. 2000. A test for concordance between the multilocus genealogies of genes and microsatellites in the pathogenic fungus Coccidioides immitis. Mol. Biol. Evol. 17:1164-1174. [DOI] [PubMed] [Google Scholar]
- 14.FitzSimmons, N. N., C. Moritz, and S. S. Moore. 1995. Conservation and dynamics of microsatellite loci over 300 million years of marine turtle evolution. Mol. Biol. Evol. 12:432-440. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein, D. B., A. Ruíz-Linares, L. L. Cavalli-Sforza, and M. W. Feldman. 1995. An evaluation of genetic distances for use with microsatellite loci. Genetics 139:463-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein, D. B., A. Ruíz-Linares, L. L. Cavalli-Sforza, and M. W. Feldman. 1995. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc. Natl. Acad. Sci. USA 92:6720-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimaldi, G., Jr., J. R. David, and D. McMahon-Pratt. 1987. Identification and distribution of New World Leishmania species characterized by serodeme analysis using monoclonal antibodies. Am. J. Trop. Med. Hyg. 36:270-287. [DOI] [PubMed] [Google Scholar]
- 18.Guerbouj, S., I. Guizani, N. Speybroeck, D. Le Ray, and J. C. Dujardin. 2001. Genomic polymorphism of Leishmania infantum: a relationship with clinical pleomorphism? Infect. Genet. Evol. 1:49-59. [DOI] [PubMed] [Google Scholar]
- 19.Hide, M., A. L. Bañuls, and M. Tibayrenc. 2001. Genetic heterogeneity and phylogenetic status of Leishmania (Leishmania) infantum zymodeme MON-1: epidemiological implications. Parasitology 123:425-432. [DOI] [PubMed] [Google Scholar]
- 20.Jamjoom, M. B., R. W. Ashford, P. A. Bates, M. L. Chance, S. J. Kemp, P. C. Watts, and H. A. Noyes. 2004. Leishmania donovani is the only cause of visceral leishmaniasis in East Africa; previous descriptions of L. infantum and “L. archibaldi” from this region are a consequence of convergent evolution in the isoenzyme data. Parasitology 129:1-11. [DOI] [PubMed] [Google Scholar]
- 21.Jamjoom, M. B., R. W. Ashford, P. A. Bates, S. J. Kemp, and H. A. Noyes. 2002. Polymorphic microsatellite repeats are not conserved between Leishmania donovani and Leishmania major. Mol. Ecol. Notes 2:104-106. [Google Scholar]
- 22.Jamjoom, M. B., R. W. Ashford, P. A. Bates, S. J. Kemp, and H. A. Noyes. 2002. Towards a standard battery of microsatellite markers for the analysis of the Leishmania donovani complex. Ann. Trop. Med. Parasitol. 96:265-270. [DOI] [PubMed] [Google Scholar]
- 23.Koblízková, A., J. Dolezel, and J. Macas. 1998. Subtraction with 3′ modified oligonucleotides eliminates amplification artefacts in DNA libraries enriched for microsatellites. BioTechniques 25:32-38. [DOI] [PubMed] [Google Scholar]
- 24.Koskinen, M. T., H. Hirvonen, P.-A. Landry, and C. R. Primmer. 2004. The benefits of increasing the number of microsatellites utilized in genetic population studies: an empirical perspective. Hereditas 141:61-67. [DOI] [PubMed] [Google Scholar]
- 25.Kuhls, K., I. L. Mauricio, F. Pratlong, W. Presber, and G. Schönian. 2005. Analysis of ribosomal DNA internal transcribed spacer sequences of the Leishmania donovani complex. Microbes Infect. 7:1224-1234. [DOI] [PubMed] [Google Scholar]
- 26.Lainson, R., and J. J. Shaw. 1987. Evolution, classification and geographical distribution, p. 1-120. In W. Peters and R. Kilick-Kendrick (ed.), Leishmaniases in biology and medicine, vol. 1. Academic Press, London, United Kingdom. [Google Scholar]
- 27.Lewin, S., G. Schönian, N. El Tai, L. Oskam, P. Bastien, and W. Presber. 2002. Strain typing in Leishmania donovani by using sequence-confirmed amplified region analysis. Int. J. Parasitol. 32:1267-1276. [DOI] [PubMed] [Google Scholar]
- 28.Macedo, A. M., J. R. Pimenta, R. S. de Aguiar, A. I. R. Melo, E. Chiari, B. Zingales, S. D. J. Pena, and R. P. Oliveira. 2001. Usefulness of microsatellite typing in population genetic studies of Trypanosoma cruzi. Mem. Inst. Oswaldo Cruz 96:407-413. [DOI] [PubMed] [Google Scholar]
- 29.Marfurt, J., A. Nasereddin, I. Niederwieser, C. L. Jaffe, H.-P. Beck, and I. Felger. 2003. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J. Clin. Microbiol. 41:3147-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mauricio, I. L., J. R. Stothard, and M. A. Miles. 2004. Leishmania donovani complex: genotyping with the ribosomal internal transcribed spacer and the mini-exon. Parasitology 128:263-267. [DOI] [PubMed] [Google Scholar]
- 31.Mauricio, I. L., J. R. Stothard, and M. A. Miles. 2000. The strange case of Leishmania chagasi. Parasitol. Today 16:188-189. [DOI] [PubMed] [Google Scholar]
- 32.Mauricio, I. L., M. W. Gaunt, J. R. Stothard, and M. A. Miles. 2001. Genetic typing and phylogeny of the Leishmania donovani complex by restriction analysis of PCR amplified gp63 intergenic regions. Parasitology 122:393-403. [DOI] [PubMed] [Google Scholar]
- 33.Meredith, S. E. O., E. E. Zijlstra, G. J. Schoone, C. C. M. Kroon, G. J. J. Van Eys, K. U. Schaeffer, A. M. El Hassan, and P. G. Lawyer. 1993. Development and application of the polymerase chain reaction for the detection and identification of Leishmania parasites in clinical material. Arch. Inst. Pasteur Tunis 70:419-431. [PubMed] [Google Scholar]
- 34.Minch, E., A. Ruíz-Linares, D. B. Goldstein, M. Feldman, and L. L. Cavalli-Sforza. 1995. MICROSAT, the Microsatellite Distance Program. Stanford University Press, Stanford, Calif.
- 35.Morales, M. A., I. Cruz, J. M. Rubio, C. Chicharro, C. Canavate, F. Laguna, and J. Alvar. 2002. Relapses versus reinfections in patients coinfected with Leishmania infantum and human immunodeficiency virus type 1. J. Infect. Dis. 185:1533-1537. [DOI] [PubMed] [Google Scholar]
- 36.Motadezian, H., H. Noyes, and R. Maingoon. 1996. Leishmania and Sauroleishmania: the use of random amplified polymorphic DNA for the identification of parasites from vertebrated and invertebrates. Exp. Parasitol. 83:150-154. [DOI] [PubMed] [Google Scholar]
- 37.Oliveira, R. P., N. E. Broude, A. M. Macedo, C. R. Cantor, C. L. Smith, and S. D. J. Pena. 1998. Probing the genetic population structure of Trypanosoma cruzi with polymorphic microsatellites. Proc. Natl. Acad. Sci. USA 95:3776-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortí, G., D. E. Pearse, and J. C. Avise. 1997. Phylogenetic assessment of length variation at a microsatellite locus. Proc. Natl. Acad. Sci. USA 94:10745-10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oskam, L., F. Pratlong, E. E. Zijlstra, C. C. M. Kroon, J. P. Dedet, P. A. Kager, G. Schönian, H. W. Ghalip, A. M. El-Hassan, and S. E. O. Meredith. 1998. Biochemical and molecular characterization of Leishmania parasites isolated from an endemic focus in eastern Sudan. Trans. R. Soc. Trop. Med. Hyg. 92:120-122. [DOI] [PubMed] [Google Scholar]
- 40.Pratlong, F., J. A. Rioux, P. Marty, F. Faraut-Gambarelli, J. Dereure, G. Lanotte, and J. P. Dedet. 2004. Isoenzymatic analysis of 712 strains of Leishmania infantum in the south of France and relationship of enzymatic polymorphism to clinical and epidemiological features. J. Clin. Microbiol. 42:4077-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pratlong, F., J. Dereure, B. Bucheton, S. El-Safi, A. Dessein, G. Lanotte, and J. P. Dedet. 2001. Sudan: the possible original focus of visceral leishmaniasis. Parasitology 122:599-605. [DOI] [PubMed] [Google Scholar]
- 42.Pratlong, F., J. P. Dedet, P. Marty, M. Portús, M. Deniau, J. Dereure, P. Abranches, J. Reynes, A. Martini, M. Lefebvre, and J. A. Rioux. 1995. Leishmania-human immunodeficiency virus coinfection in the Mediterranean basin: isoenzymatic characterization of 100 isolates of the Leishmania infantum complex. J. Infect. Dis. 172:323-326. [DOI] [PubMed] [Google Scholar]
- 43.Pritchard, J. K., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quispe Tintaya, K. W., X. Jing, J.-P. Dedet, S. Rijal, X. De Bolle, and J.-C. Dujardin. 2004. Antigen genes for molecular epidemiology of leishmaniasis: polymorphism of cysteine proteinase B and surface metalloprotease glycoprotein 63 in the Leishmania donovani complex. J. Infect. Dis. 189:1035-1043. [DOI] [PubMed] [Google Scholar]
- 45.Refseth, U. H., B. M. Fangan, and K. S. Jakobsen. 1997. Hybridization capture of microsatellites directly from genomic DNA. Electrophoresis 18:1519-1523. [DOI] [PubMed] [Google Scholar]
- 46.Rioux, J. A., G. Lanotte, E. Serres, F. Pratlong, P. Bastien, and J. Périère. 1990. Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann. Parasitol. Hum. Comp. 65:111-125. [DOI] [PubMed] [Google Scholar]
- 47.Rossi, V., P. Wincker, C. Ravel, C. Blaineau, M. Pagés, and P. Bastien. 1994. Structural organisation of microsatellite families in the Leishmania genome and polymorphisms at two (CA)n loci. Mol. Biochem. Parasitol. 65:271-282. [DOI] [PubMed] [Google Scholar]
- 48.Rozen, S., and H. J. Skaletsky. 1998. Primer3 on the WWW for general users and biologist programmers. Mol. Biochem. Parasitol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 49.Ruzzante, D. E. 1998. A comparison of several measures of genetic distance and population structure with microsatellite data: bias and sampling variance. Can. J. Fish. Aquat. Sci. 55:1-14. [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Schönian, G., C. Schweynoch, K. Zlateva, L. Oskam, N. Kroon, Y. Gräser, and W. Presber. 1996. Identification and determination of the relationship of species and strains within the genus Leishmania using single primers in the polymerase chain reaction. Mol. Biochem. Parasitol. 77:19-29. [DOI] [PubMed] [Google Scholar]
- 52.Schönian, G., H. Akuffo, S. Lewin, K. Maasho, S. Nylén, F. Pratlong, C. L. Eisenberger, L. Schnur, and W. Presber. 2000. Genetic variability within the species Leishmania aethiopica does not correlate with clinical variations of cutaneous leishmaniasis. Mol. Biochem. Parasitol. 106:239-248. [DOI] [PubMed] [Google Scholar]
- 53.Schönian, G., L. Schnur, M. El Fari, L. Oskam, A. A. Kolesnikov, W. Sokolowska-Köhler, and W. Presber. 2001. Genetic heterogeneity in the species Leishmania tropica revealed by different PCR-based methods. Trans. R. Soc. Trop. Med. Hyg. 95:217-224. [DOI] [PubMed] [Google Scholar]
- 54.Schwenkenbecher, J., C. Fröhlich, F. Gehre, L. Schnur, and G. Schönian. 2004. Evolution and conservation of microsatellite markers for Leishmania tropica. Infect. Genet. Evol. 4:99-105. [DOI] [PubMed] [Google Scholar]
- 55.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman & Co., San Francisco, Calif.
- 56.Swofford, D. L. 2000. PAUP. Phylogenetic analysis using parsimony (and other methods). Version 4.0b8. Sinauer Associates, Sunderland, Mass.
- 57.Taylor, J. S., J. M. H. Durkin, and F. Breden. 1999. The death of a microsatellite. Mol. Biol. Evol. 16:567-572. [DOI] [PubMed] [Google Scholar]
- 58.Tóth, G., Z. Gáspári, and J. Jurka. 2000. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10:967-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Victoir, K., and J. C. Dujardin. 2002. How to succeed in parasitic life without sex? Asking Leishmania. Trends Parasitol. 18:81-85. [DOI] [PubMed] [Google Scholar]
- 60.Zemanová, M. Jirků, I. L. Mauricio, M. A. Miles, and J. Lukeš. 2004. Genetic polymorphism within the Leishmania donovani complex: correlation with geographic origin. Am. J. Trop. Hyg. 70:613-617. [PubMed] [Google Scholar]