Abstract
Mucormycosis is an emerging infection associated with a high mortality rate. Identification of the causative agents remains difficult and time-consuming by standard mycological procedures. In this study, internal transcribed spacer (ITS) sequencing was validated as a reliable technique for identification of Zygomycetes to the species level. Furthermore, species identification directly from infected tissues was evaluated in experimentally infected mice. Fifty-four Zygomycetes strains belonging to 16 species, including the most common pathogenic species of Rhizopus spp., Absidia spp., Mucor spp., and Rhizomucor spp., were used to assess intra- and interspecies variability. Ribosomal DNA including the complete ITS1-5.8S-ITS2 region was amplified with fungal universal primers, sequenced, and compared. Overall, for a given species, sequence similarities between isolates were >98%. In contrast, ITS sequences were very different between species, allowing an accurate identification of Zygomycetes to the species level in most cases. Six species (Rhizopus oryzae, Rhizopus microsporus, Rhizomucor pusillus, Mucor circinelloides, and Mucor indicus) were also used to induce disseminated mucormycosis in mice and to demonstrate that DNA extraction, amplification of fungal DNA, sequencing, and molecular identification were possible directly from frozen tissues.
Mucormycosis is a severe disease associated with a high mortality rate occurring mostly in immunocompromised patients such as those with diabetes mellitus or with neutropenia (8, 20, 23). Recent studies have shown an increasing incidence of mucormycosis, particularly in allogeneic bone marrow/stem cell-transplanted patients (17, 21). Moreover, Zygomycetes are resistant to many antifungal drugs, including new agents such as caspofungin (9) and voriconazole (6). One of the consequences of voriconazole-intrinsic resistance of these fungi is the occurrence of breakthrough mucormycosis in patients receiving voriconazole for prophylaxis or for treatment of other fungal infections such as aspergillosis (14, 16, 22, 27).
Diagnosis of mucormycosis remains difficult. First, cultures of infected tissues are often negative, and no serological tests for the diagnosis are routinely available. Then, diagnosis depends mainly on histopathology that evidences broad, rarely septate hyphae with right-angled branching. However, it is not possible to identify the different genera and species belonging to the Zygomycetes by histopathology. Second, when cultures are positive, identification to the species level is time-consuming and may require the expertise of a reference laboratory (26, 32). Culture is currently the major method used to identify the organisms responsible for mucormycosis. The main pathogens belong to the genera Rhizopus, Absidia, Mucor, and Rhizomucor. Other species such as Apophysomyces elegans, Cunninghamella bertholletiae, Cokeromyces recurvatus, Saksenaea vasiformis, and Syncephalastrum racemosum are also causative agents of mucormycosis (8, 23). The development of a method to reliably identify Zygomycetes species would be of major interest and would provide a tool for a better understanding of the epidemiology of mucormycosis. Moreover, Zygomycetes are a heterogeneous group for which antifungal susceptibility to both polyenes and azoles is variable (5, 6). Therefore, an accurate identification of the infecting species could help to guide therapy. Because of the difficulties associated with a microbiological diagnosis, new diagnostic tools are needed for species identification either from cultures or directly from infected tissues.
Molecular identification has been evaluated for several groups of medically important fungi (15). Different molecular targets have been used, including conserved ribosomal DNA genes and the more variable internal transcribed spacer (ITS) regions between those genes which allow identification to the species level. However, few data are available for the molecular identification of Zygomycetes (30). Each species is often represented by few isolates in data banks. A large number of isolates have to be tested to assess the intra- and interspecies variability.
Therefore, the first aim of the present study was to validate ITS sequencing as a reliable technique for identification of Zygomycetes to the species level. For this purpose, a large number of isolates were analyzed. The second aim of the study was to evaluate the possibility of species identification directly from infected tissues. We therefore developed animal models of mucormycosis to get infected tissues and assess this possibility.
MATERIALS AND METHODS
Molecular identification of Zygomycetes isolates from fungal cultures.
The 54 isolates, including 14 type strains, used in this study are presented in Table 1. They were obtained from the Centraalbureau voor Schimmelcultures (CBS) collection, from the Pasteur Institute Collection of Fungi (IP), and from the collection of the National Reference Center for Mycoses and Antifungals (CNRMA). None of the strains were epidemiologically linked. They comprised 17 Rhizopus oryzae isolates, 6 Absidia corymbifera isolates, 7 Rhizomucor pusillus isolates, 6 Rhizopus microsporus isolates, 5 Mucor circinelloides isolates, 2 Mucor indicus isolates, and 1 isolate each for the other genera and species (n = 11). For all clinical strains, phenotypic identification was done using standard mycological procedures (7). All strains were stored as spore suspensions at −20°C in 40% glycerol until used.
TABLE 1.
Strains used and accession numbers of ITS1-5.8S-ITS2 sequences
| Isolate | Strain numbera | Origin | Accession no. |
|---|---|---|---|
| Absidia corymbifera | IP 1129.75 | Environment, outdoor air, Morocco | DQ118982 |
| Absidia corymbifera | IP 1279.81 | Unknown | DQ118980 |
| Absidia corymbifera | IP 1280.81 | Unknown | DQ118985 |
| Absidia corymbifera | CNRMA 03.611 | Human, bronchia, France | DQ118983 |
| Absidia corymbifera | CNRMA 03.697 | Human, bone, France | DQ118984 |
| Absidia corymbifera | CNRMA 04.732 | Human, lung, France | DQ118981 |
| Cokeromyces recurvatus | CBS 158.50T | Animal, feces, USA | DQ118986 |
| Mucor circinelloides f. circinelloides | CBS 195.68NT | Environment, air, The Netherlands | DQ118991 |
| Mucor circinelloides | IP 1873.89 | Human, feces, France | DQ118989 |
| Mucor circinelloides | CNRMA 03.154 | Human, skin, France | DQ118987 |
| Mucor circinelloides | CNRMA 03.371 | Human, muscle, France | DQ118988 |
| Mucor circinelloides | CNRMA 04.805 | Human, muscle, France | DQ118990 |
| Mucor hiemalis f. hiemalis | CBS 201.65NT | United States | DQ118992 |
| Mucor indicus | CBS 226.29T | Switzerland | DQ118994 |
| Mucor indicus | CNRMA 03.894 | Human, stomach, Germany | DQ118993 |
| Mucor racemosus f. racemosus | CBS 260.68T | Switzerland | DQ118996 |
| Mucor ramosissimus | CBS 135.65NT | Human, nose, Uruguay | DQ118997 |
| Mucor rouxii | CBS 416.77 | Environment, fermenting rice | DQ118998 |
| Rhizomucor miehei | CBS 182.67T | Environment, P. argentatum, United States | DQ118995 |
| Rhizmomucor pusillus | CBS 354.68NT | Environment, cornmeal, The Netherlands | DQ119005 |
| Rhizmomucor pusillus | ATCC 36606 | Animal, brain | DQ119001 |
| Rhizmomucor pusillus | IP 1127.75 | Unknown | DQ119002 |
| Rhizmomucor pusillus | IP 1956.90 | Human, bronchia, France | DQ119003 |
| Rhizmomucor pusillus | CNRMA 03.1205 | Human, lung, France | DQ119000 |
| Rhizmomucor pusillus | CNRMA 04.210 | Human, bone, France | DQ118999 |
| Rhizmomucor pusillus | CNRMA 04.503 | Human, sputum, France | DQ119004 |
| Rhizomucor variabilis | CBS 103.93T | Human, wrist and hand, China | DQ119006 |
| Rhizomucor variabilis var. regularior | CBS 384.95T | Human, skin, China | DQ119007 |
| Rhizopus azygosporus | CBS 357.93T | Environment, tempeh, Indonesia | DQ119008 |
| Rhizopus microsporus var. chinensis | CBS 631.82T | Environment, bread, China | DQ119009 |
| Rhizopus microsporus var. microsporus | IP 1124.75 | Unknown | DQ119010 |
| Rhizopus microsporus var. oligosporus | CBS 339.62 | Environment, tempeh, Indonesia | DQ119011 |
| Rhizopus microsporus var. rhizopodiformis | IP 676.72 | Human, skin, France | DQ119014 |
| Rhizopus microsporus var. rhizopodiformis | IP 1123.75 | Unknown | DQ119013 |
| Rhizopus microsporus var. rhizopodiformis | CNRMA 04.1469 | Unknown | DQ119012 |
| Rhizopus oryzae | CBS 112.07T | Human, lung, The Netherlands | DQ119031 |
| Rhizopus oryzae | IP 4.77 | Human, brain | DQ119024 |
| Rhizopus oryzae | IP 1443.83 | Unknown | DQ119023 |
| Rhizopus oryzae | CNRMA 03.253 | Human, lung, France | DQ119029 |
| Rhizopus oryzae | CNRMA 03.375 | Human, sinus, France | DQ119021 |
| Rhizopus oryzae | CNRMA 03.395 | Human, skin, France | DQ119020 |
| Rhizopus oryzae | CNRMA 03.410 | Human, sputum, France | DQ119018 |
| Rhizopus oryzae | CNRMA 03.411 | Human, sputum, France | DQ119028 |
| Rhizopus oryzae | CNRMA 03.412 | Human, sputum, France | DQ119019 |
| Rhizopus oryzae | CNRMA 03.413 | France | DQ119026 |
| Rhizopus oryzae | CNRMA 03.909 | Human, sinus, France | DQ119022 |
| Rhizopus oryzae | CNRMA 03.918 | Human, lung, France | DQ119025 |
| Rhizopus oryzae | CNRMA 04.48 | Human, skin, France | DQ119030 |
| Rhizopus oryzae | CNRMA 04.160 | Human, sputum, France | DQ119027 |
| Rhizopus oryzae | CNRMA 04.785 | Human, rhino-cerebral, France | DQ119032 |
| Rhizopus oryzae | CNRMA 04.1253 | Human, BAL,b France | DQ119033 |
| Rhizopus oryzae | CNRMA 04.1468 | Human, sinus | DQ119017 |
| Rhizopus schipperae | CBS 138.95T | Human, bronchial wash, United States | DQ119015 |
| Syncephalastrum racemosum | CNRMA 03.414 | Human, skin, France | DQ119016 |
ATCC, American Type Culture Collection, Manassas, Va.; CBS, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; CNRMA, National Reference Center for Mycoses and Antifungals, Institut Pasteur, Paris, France; IP, Pasteur Institute Collection of Fungi, Institut Pasteur, Paris, France. Superscript letters: T, type strain; NT, neotype strain.
BAL, bronchoalveolar lavage.
(i) Culture.
Strains were subcultured from frozen stocks on Sabouraud-chloramphenicol agar slants for 7 days at 20, 28, or 35°C, depending on the optimum growth temperature for each species (7). From the cultures, spore suspensions were prepared in 0.9% NaCl. Twenty milliliters of RPMI 1640 (Sigma-Aldrich, Saint Quentin Fallavier, France) with l-glutamine but without sodium bicarbonate buffered to pH 7 with 0.165 M morpholinepropanesulfonic acid (Sigma) was inoculated with the spore suspension, and the mycelium was grown for 48 to 72 h at 30°C with agitation. Before use, the mycelium was washed once with 20 ml 0.9% NaCl.
(ii) DNA extraction.
Complete genomic DNA was extracted from approximately 50 mg of mycelium grown in RPMI 1640 according to the CTAB (hexacetyltrimethylammonium bromide; Sigma) protocol described by Voigt et al. (30) with modifications. Briefly, the mycelium was homogenized in a 50-ml tube containing 1 ml CTAB extraction buffer (100 mM Tris-HCl [pH 8.4], 1.4 M NaCl, 25 mM EDTA, 2% CTAB), 3 glass beads (0.5 cm) (Sigma), and 500 mg of 425- to 600-μm-diameter glass beads (Sigma). The suspension was vigorously vortexed and put into liquid nitrogen for 1 min followed by immersion at 37°C for 1 min. After vortexing again for 1 min, 700 μl of the suspension was transferred into a 2-ml microcentrifuge tube. An equal volume of chloroform was added to the mixture, vortexed for 5 s, and spun for 10 min at 14,000 × g. Five hundred microliters of the upper phase was transferred to a new 2-ml microcentrifuge tube, and DNA was precipitated by the addition of an equal volume of 2-propanol. DNA was pelleted at 14,000 × g for 1 min. After supernatant was discarded, the pellet was washed with 100% ethanol and resuspended in 200 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Genomic DNA was stored at −20°C.
(iii) PCR, primers, and sequencing.
Ribosomal DNA including the complete ITS1-5.8S-ITS2 region was amplified with the fungal universal primers V9D (5′-TTAAGTCCCTGCCCTTTGTA-3′) and LS266 (5′-GCATTCCCAAACAACTCGACTC-3′) (10). Amplification mixtures (100 μl) contained 5 μl of the extracted genomic DNA, 2.5 μl of 20 μM concentrations of each primer 10 μl of 2.5 mM [each] dATP, dTTP, dGTP, dCTP [Roche Diagnostics GmbH, Mannheim, Germany]), 10 μl of 25 mM MgCl2, 6.25 U of AmpliTaq polymerase (Roche), and 10 μl of 10× PCR buffer (Roche). Amplification of the PCR products was done in a Bio-Rad iCycler thermocycler (Hercules, CA) with the following cycling parameters: initial denaturizing step of 10 min at 94°C, 30 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 58°C, and elongation for 30 s at 72°C, with a final extension for 10 min at 72°C. PCR products were purified on P100 gel fine (Bio-Rad), and both strands were sequenced once by the BigDye terminator cycle sequencing ready reaction kit, version 3.1 (Applied Biosystems, Foster City, CA), with the primer set V9D and LS266. Reaction products were analyzed on an ABI Prism 3700 automated DNA analyzer (Applied Biosystems).
(iv) Sequence analysis.
Sequences were manually corrected with Chromas version 2.24 (Technelysium, Helensvale, Queensland, Australia) and analyzed with BioEdit sequence alignment editor (Isis Therapeutics, Carlsbad, CA). Multiple-sequence alignments were performed with ClustalW. Sequences for all 54 strains analyzed in the present study have been deposited in GenBank (see “Nucleotide sequence accession numbers” below).
Molecular identification of Zygomycetes using tissues from experimentally infected mice. (i) Organisms.
Six isolates, mostly of clinical origin, were used for in vivo studies: A. corymbifera (CNRMA 03.697), M. circinelloides (CNRMA 03.154), M. indicus (CNRMA 03.894), R. pusillus (CNRMA 04.210), R. microsporus var. rhizopodiformis (IP 1123.75), and R. oryzae (CNRMA 03.918).
(ii) Mice.
Female OF-1 outbred mice (Charles River Laboratories, L'Arbresle, France), 7 weeks old, weighing 22 to 24 g, were used for the experiments. Mice were maintained in a room at 21°C with a 12-h dark-light cycle. Animal studies were performed according to the recommendations of the European Community (Directive 86/609/EEC, 24 November 1986) and were approved by the ethical committee of the Institut Pasteur. Mice were given food and water ad libitum.
(iii) Infection.
Inocula were prepared from cultures of the strains on potato-dextrose agar slants for 7 days at 28 or 35°C to obtain sufficient sporulation. Spores were harvested by washing the agar surface with sterile 0.9% NaCl containing 0.05% Tween 80. Suspensions of spores were filtered through a nylon filter (pore size, 11 μm), counted in a hemacytometer, and adjusted to the desired concentration. Viability determination was performed by plating 10-fold dilutions prepared in 0.9% NaCl with 0.05% Tween 80. Plates were incubated at 35°C, and CFU were counted after 16 h. Mice were infected with 104, 105, or 106 spores per animal in 100-μl volumes given intravenously into a lateral tail vein. Each group contained 3 to 6 mice. Mice were not immunocompromised. Mice were sacrificed 3 to 4 days postinfection, and brains and kidneys were removed aseptically and stored at −20°C until used.
(iv) DNA extraction from tissues and assessment of infection.
Tissues were homogenized in a tissue grinder in 2 ml 0.9% NaCl. As it has been shown that stilbene derivatives (e.g., calcofluor) are useful to demonstrate the presence of Zygomycetes in clinical samples (23, 24), the homogenized tissues were examined for the presence of hyphae under a epifluorescence microscope (filters: excitation, 340 to 380 nm; emission, 425 nm; Leitz, Wetzlar, Germany) after staining with 0.1% calcofluor white (Sigma) in water containing 1% KOH. Fifty microliters of the homogenized tissues were placed on microscope slides, allowed to dry at room temperature, and stained with 20 μl calcofluor white. The volume corresponding to 50 mg of tissue was transferred to a 2-ml microcentrifuge tube and spun for 2 min at 14,000 × g. The supernatant was discarded, and the pellet was transferred to a 50-ml tube. Complete genomic DNA was extracted, and PCR was performed as described above. Standard precautions to prevent cross-contamination of samples were taken (19). In each set of experiments, brain and kidney from uninfected mice were used as negative controls.
Nucleotide sequence accession numbers.
Sequences for all 54 strains analyzed in the present study have been deposited in GenBank under accession numbers DQ118980 to DQ119033 (Table 1).
RESULTS
Sequences of ITS and 5.8S regions.
The length of the ITS1 region varied from 132 to 269 bp, while the ITS2 region was from 173 to 257 bp in length. Interestingly, the sequence corresponding to the fungal universal primer ITS2/ITS3 differed by 1 to 2 bp for all studied Zygomycetes strains compared to the conserved sequence found in other fungi.
Sequence similarity within species.
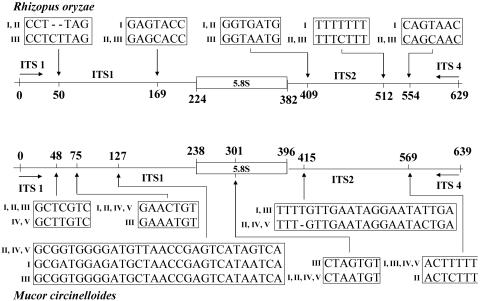
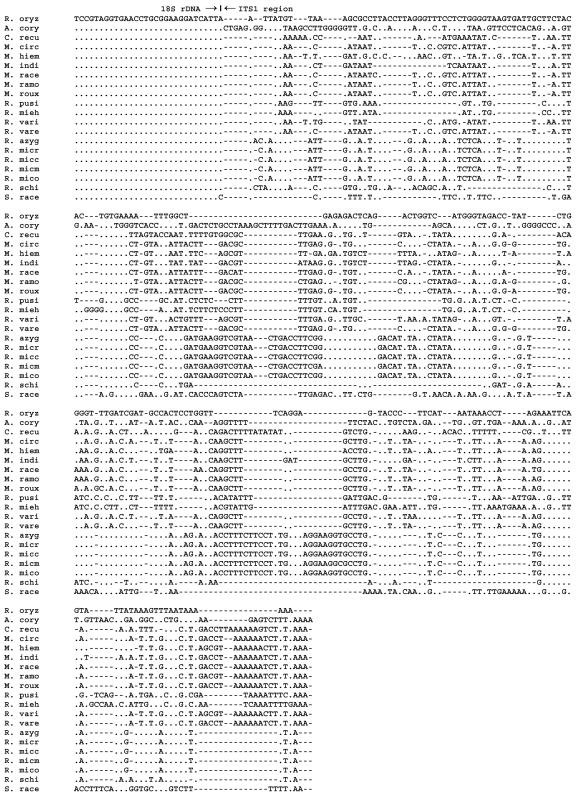
Sequences of the whole ITS1-5.8S-ITS2 region of A. corymbifera, M. circinelloides, R. pusillus, R. microsporus, and R. oryzae were examined for intraspecies similarities (Table 2; Fig. 1). Overall, for a given species, sequence similarities between isolates were >98%. Within the 6 A. corymbifera isolates, 0- to 4-bp differences were found, with overall similarities from 99.5 to 100%. Sequences of the 7 isolates of R. pusillus were 100% identical. Within the 17 R. oryzae isolates, differences of 0 to 6 bp were found, with overall similarities from 98.9 to 100%. Three types were recognized with sequence variations at 5 different positions in both the ITS1 and ITS2 regions (Fig. 1). For the 6 strains (including 4 varieties) of the R. microsporus group, similarities ranged from 98.9 to 100% (0- to 8-bp differences). The 5 M. circinelloides isolates showed the lowest homogeneity within the examined regions, with 1- to 8-bp differences and overall similarities from 98.7 to 99.8%. The sequence for each isolate of M. circinelloides was unique (Fig. 1).
TABLE 2.
Number of nucleotide differences in ITS1, 5.8S, and ITS2 regions within a single species
| Species and isolatea | No. of bp differences compared to reference strain
|
% Similarityb | ||
|---|---|---|---|---|
| ITS1 | 5.8S | ITS2 | ||
| Rhizopus oryzae | ||||
| CBS 112.07T | ||||
| IP 4.77 | 0 | 0 | 0 | 100 |
| IP 1443.83 | 1 | 0 | 0 | 99.8 |
| CNRMA 03.253 | 3 | 0 | 3 | 98.9 |
| CNRMA 03.375 | 1 | 0 | 0 | 99.8 |
| CNRMA 03.395 | 1 | 0 | 0 | 99.8 |
| CNRMA 03.410 | 0 | 0 | 0 | 100 |
| CNRMA 03.411 | 0 | 0 | 0 | 100 |
| CNRMA 03.412 | 0 | 0 | 0 | 100 |
| CNRMA 03.413 | 0 | 0 | 0 | 100 |
| CNRMA 03.909 | 1 | 0 | 0 | 99.8 |
| CNRMA 03.918 | 1 | 0 | 0 | 99.8 |
| CNRMA 04.48 | 0 | 0 | 0 | 100 |
| CNRMA 04.160 | 3 | 0 | 3 | 98.9 |
| CNRMA 04.785 | 1 | 0 | 0 | 99.8 |
| CNRMA 04.1253 | 1 | 0 | 0 | 99.8 |
| CNRMA 04.1468 | 0 | 0 | 0 | 100 |
| Rhizopus microsporus | ||||
| CBS 631.82T | ||||
| CBS 339.62 | 0 | 0 | 0 | 100 |
| IP 676.72 | 0 | 0 | 0 | 100 |
| IP 1123.75 | 0 | 0 | 0 | 100 |
| IP 1124.75 | 2 | 0 | 6 | 98.9 |
| CNRMA 04.1469 | 0 | 0 | 0 | 100 |
| Absidia corymbifera | ||||
| IP 1129.75 | ||||
| IP 1279.81 | 0 | 0 | 0 | 100 |
| IP 1280.81 | 4 | 0 | 0 | 99.5 |
| CNRMA 03.611 | 0 | 0 | 0 | 100 |
| CNRMA 03.697 | 0 | 0 | 0 | 100 |
| CNRMA 04.732 | 0 | 0 | 0 | 100 |
| Mucor circinelloides | ||||
| CBS 195.68NT | ||||
| IP 1873.89 | 6 | 0 | 2 | 98.7 |
| CNRMA 03.154 | 2 | 0 | 1 | 99.5 |
| CNRMA 03.371 | 1 | 0 | 0 | 99.8 |
| CNRMA 04.805 | 5 | 1 | 2 | 98.7 |
| Rhizomucor pusillus | ||||
| CBS 354.68NT | ||||
| ATCC 36606 | 0 | 0 | 0 | 100 |
| IP 1127.75 | 0 | 0 | 0 | 100 |
| IP 1959.90 | 0 | 0 | 0 | 100 |
| CNRMA 03.1205 | 0 | 0 | 0 | 100 |
| CNRMA 04.210 | 0 | 0 | 0 | 100 |
| CNRMA 04.503 | 0 | 0 | 0 | 100 |
ATCC, American Type Culture Collection, Manassas, Va.; CBS, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; CNRMA, National Reference Center for Mycoses and Antifungals, Institut Pasteur, Paris, France; IP, Pasteur Institute Collection of Fungi, Institut Pasteur, Paris, France. Superscript letters: T, type strain; NT, neotype strain. CBS 631.82 is the type strain of R. microsporus var. chinensis, and CBS 195.68 is the type strain of M. circinelloides f. circinelloides.
Percent similarity value reflects similarity across all three rRNA gene regions (ITS1, 5.8S, and ITS2).
FIG. 1.
Intraspecies sequence variability within the ITS1-5.8S-ITS2 region for strains of R. oryzae and M. circinelloides. Among the 17 isolates of R. oryzae, 3 types were distinguished. Five types were found for the 5 isolates of M. circinelloides. Different types are represented by Roman numerals. Numbers represent positions on the sequence alignment (position 0 corresponds to the 5′ end of the universal fungal primer ITS 1).
Sequence similarity between species and varieties.
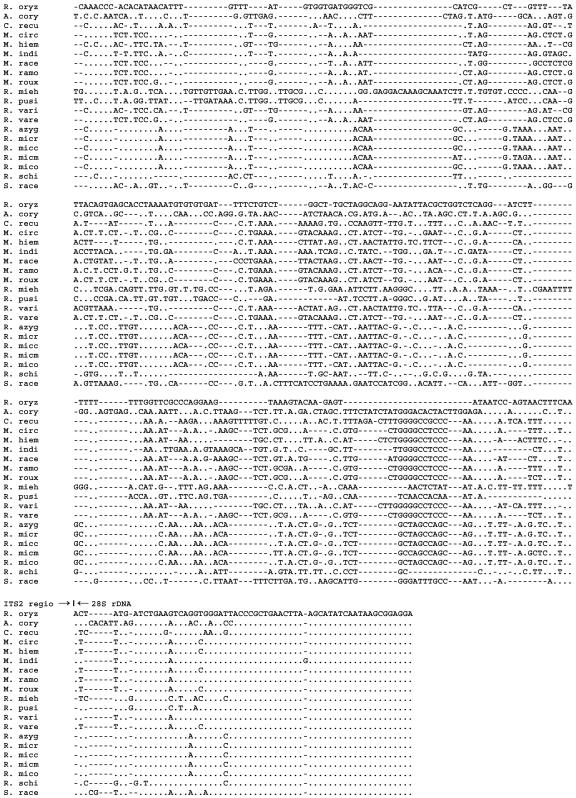
Figures 2 and 3 show the alignment of 16 Zygomycetes species, and the similarity matrix of the examined species is shown in Table 3. Between the four Rhizopus species, sequence similarities from 61.5 to 100% were found. All varieties of R. microsporus showed a strong homology when compared to each other, varying from 98.9 to 100%. Rhizopus azygosporus was also very homologous to the R. microsporus varieties (98.9 to 100%). In contrast, when R. oryzae was compared with the R. microsporus group, the percentage of similarity was about 70%. Rhizopus schipperae showed a low level of sequence similarity with both the R. microsporus group (61%) and with R. oryzae (66%). Except for other Rhizopus species, all Rhizopus species demonstrated less than 58% sequence similarity with other Zygomycetes. Sequences of A. corymbifera were very different from those of all other species, with similarities of less than 49%. Within the genus Rhizomucor, sequence similarities ranged from 46.1 to 81.1%. Interestingly, Rhizomucor variabilis var. regularior showed a higher homology with sequences of some Mucor species (with up to 99.3% of similarity) than with Rhizomucor variabilis. When Rhizomucor spp. were compared to the other species, percentages of homology were between 41.7 and 71.6%. Between the six Mucor species, sequence similarities from 75.2 to 98.5% were found. Except for Rhizomucor species, Mucor species demonstrated sequence similarities of less than 73% with all other Zygomycetes.
FIG. 2.
Alignment of Zygomycetes ITS1 sequences, including the 3′ end of 18S rRNA gene: R. oryz, R. oryzae CBS 112.07T; A cory, A. corymbifera IP 1129.75; C. recu, C. recurvatus CBS 158.50T; M. circ, M. circinelloides CBS 195.68NT; M. hiem, M. hiemalis CBS 201.64NT; M. indi, M. indicus CBS 226.29T; M. race, M. racemosus CBS 260.68 T; M. ramosissimus CBS 135.65NT; M. roux, M. rouxii CBS 416.77; R. mieh, R. miehei CBS 182.67T; R. pusi, R. pusillus CBS 354.68NT; R. vari, R. variabilis CBS 103.93T; R. vare, R. variabilis var. regularior CBS 384.95T; R. azyg, R. azygosporus CBS 357.93T; R. micr, R. microsporus var. rhizopodiformis IP 676.72; R. micc, R. microsporus var. chinensis CBS 631.82T; R. micm, R. microsporus var. microsporus IP 1124.75; R. mico, R. microsporus var. oligosporus CBS 339.62, R. schi, R. schippereae CBS 138.95T; S. race, S. racemosum CNRMA 03.414. T, type strain; NT, neotype strain. Alignment was made with the type strain of the species or with a reference strain from an international collection when no type strain was available, except for S. racemosum.
FIG. 3.
Alignment of Zygomycetes ITS2 sequences, including the 5′ end of 28S rRNA gene. Strain identification is the same as indicated in the legend to Fig. 2. Alignment was made with the type strain of the species or with a reference strain from an international collection when no type strain was available, except for S. racemosum.
TABLE 3.
Matrix of ITS1-5.8S-ITS2 similarities of twenty different genera, species, and varieties of Zygomycetesa
| Species | % Similarity
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. corymbifera | C. recurvatus | M. circinelloides | M. hiemalis | M. indicus | M. racemosus | M. ramosissimus | M. rouxii | R. pusillus | R. miehei | R. variabilis | R. variabilis var. regularior | R. azygosporus | R. microsporus var. chinensis | R. microsporus var. microsporus | R. microsporus var. oligosporus | R. microsporus var. rhizopodiformis | R. oryzae | R. schipperae | |
| C. recurvatus | 46.0 | ||||||||||||||||||
| M. circinelloides | 48.6 | 72.2 | |||||||||||||||||
| M. hiemalis | 46.8 | 69.3 | 79.3 | ||||||||||||||||
| M. indicus | 47.8 | 70.3 | 79.7 | 75.2 | |||||||||||||||
| M. racemosus | 48.3 | 70.9 | 91.8 | 79.3 | 78.9 | ||||||||||||||
| M. ramosissimus | 48.9 | 71.2 | 95.7 | 79.2 | 79.0 | 91.0 | |||||||||||||
| M. rouxii | 48.0 | 71.8 | 98.5 | 79.3 | 79.1 | 91.8 | 95.9 | ||||||||||||
| R. pusillus | 44.2 | 48.0 | 47.3 | 46.8 | 48.7 | 48.0 | 48.5 | 47.2 | |||||||||||
| R. miehei | 44.1 | 45.8 | 46.5 | 45.1 | 47.5 | 45.8 | 46.7 | 46.1 | 75.8 | ||||||||||
| R. variabilis | 47.4 | 71.3 | 81.0 | 85.3 | 78.2 | 80.4 | 80.7 | 80.8 | 48.3 | 47.4 | |||||||||
| R. variabilis var. regularior | 48.1 | 71.6 | 98.9 | 79.5 | 79.4 | 91.6 | 95.9 | 99.3 | 47.2 | 46.2 | 81.1 | ||||||||
| R. azygosporus | 46.5 | 53.5 | 55.6 | 54.1 | 56.4 | 56.4 | 55.0 | 55.4 | 43.0 | 41.7 | 56.1 | 55.5 | |||||||
| R. microsporus var. chinensis | 46.5 | 53.5 | 55.6 | 54.1 | 56.4 | 56.4 | 55.0 | 55.4 | 43.0 | 41.7 | 56.1 | 55.5 | 100 | ||||||
| R. microsporus var. microsporus | 46.2 | 53.3 | 55.1 | 54.4 | 56.1 | 56.1 | 54.6 | 54.8 | 43.0 | 41.7 | 56.2 | 55.0 | 98.8 | 98.8 | |||||
| R. microsporus var. oligosporus | 46.5 | 53.5 | 55.6 | 54.1 | 56.4 | 56.4 | 55.0 | 55.4 | 43.0 | 41.7 | 56.1 | 55.5 | 100 | 100 | 98.8 | ||||
| R. microsporus var. rhizopodiformis | 46.5 | 53.5 | 55.6 | 54.1 | 56.4 | 56.4 | 55.0 | 55.4 | 43.0 | 41.7 | 56.1 | 55.5 | 100 | 100 | 98.8 | 100 | |||
| R. oryzae | 46.7 | 54.2 | 56.8 | 55.3 | 57.6 | 57.7 | 56.6 | 56.5 | 45.2 | 44.9 | 56.9 | 56.7 | 69.6 | 69.6 | 69.3 | 69.6 | 69.6 | ||
| R. schipperae | 42.9 | 53.1 | 54.5 | 54.3 | 56.3 | 55.0 | 54.9 | 54.1 | 45.2 | 44.2 | 54.5 | 54.3 | 61.5 | 61.5 | 61.5 | 61.5 | 61.5 | 66.3 | |
| S. racemosum | 46.2 | 49.5 | 50.4 | 50.0 | 50.6 | 50.7 | 51.1 | 50.3 | 58.6 | 56.2 | 50.0 | 50.6 | 45.8 | 45.8 | 45.2 | 45.8 | 45.8 | 48.3 | 47.5 |
In vivo infection, amplification, and sequencing.
Overall, for the 6 Zygomycetes species that had been used in animals, an active infection, as defined by the presence of hyphae in tissues, in both brain and kidneys was demonstrated in more than 50% of mice by calcofluor white staining (Table 4). Amplification of the tissue-extracted DNA was possible for every species causing infection. Overall, a positive PCR result was obtained in more than 70% of the animals, indicating that in some instances amplification of the fungus was positive despite a negative direct examination. It is not known if positive PCR results in the presence of negative calcofluor white staining results represent increased sensitivity of PCR compared to calcofluor white staining or if it was related to the presence of ungerminated spores retained in tissues. Cross-contamination of samples causing false-positive results were prevented by adherence to standard protocols (19) and by inclusion of negative controls in each set of experiments. For all tissues with a positive direct examination, a positive PCR was obtained. Sequencing of the ITS regions allowed identification to the species level for all 6 species (Table 4).
TABLE 4.
Molecular identification of Zygomycetes from frozen tissues of mice experimentally infected with six different speciesa
| Species | Presence of hyphaeb (no. of samples positive/no. tested)
|
Positive PCRc (no. of samples positive/no. tested)
|
||
|---|---|---|---|---|
| Kidney | Brain | Kidney | Brain | |
| A. corymbifera | 5/6 | 2/6 | 5/6 | 2/6 |
| M. circinelloides | 2/6 | 2/6 | 5/6 | 4/6 |
| M. indicus | 4/4 | 4/4 | 4/4 | 4/4 |
| R. pusillus | 1/5 | 4/5 | 1/5 | 4/5 |
| R. microsporus | 4/5 | 2/5 | 5/5 | 4/5 |
| R. oryzae | 2/3 | 2/3 | 3/3 | 2/3 |
Mice were infected with 104, 105, or 106 spores per animal (1 to 2 mice per inoculum).
After staining of the cell wall of the fungi with Calcofluor White, the presence of hyphae was determined by direct examination of the homogenized tissue with an epifluorescence microscope.
All tissues containing hyphae were also positive by PCR.
DISCUSSION
Species identification of Zygomycetes from cultures remains a difficult and time-consuming task that relies on morphological and physiological examination and that requires, in some instances, an expertise restricted to reference laboratories. DNA sequence analysis to differentiate among genera within the Zygomycetes or between the Zygomycetes and other fungal genera is particularly important in cases where these organisms differ in their susceptibility to antifungal drugs. For example, species of Aspergillus may not be easily differentiated from Zygomycetes using conventional tissue staining techniques, especially when the number of fungal elements present in tissues are few or when characteristic tissue morphologies of the organism are absent. Thus, there is a need for new tools that allow an accurate and more rapid identification. In recent years, molecular tools have been shown to be useful for identification of a large panel of fungi responsible for human mycoses (15), but until now there were few molecular studies that focused on Zygomycetes (1, 3, 30). Moreover, one study (12) even showed that the use of a large-subunit ribosomal DNA sequencing kit for identification of Zygomycetes is questionable, as previously commented (11). Indeed, this study showed that less than 50% of the Zygomycetes isolates that were evaluated were correctly identified by the sequencing kit. The lack of correct identification was, in part, the result of an incomplete database supplied with the kit so that some organisms could not be identified.
The aim of the present study was to investigate the usefulness of rRNA gene sequencing for species identification of Zygomycetes from pure cultures and from tissues of experimentally infected animals. Our results showed that ITS sequences shared a high level of identity between isolates within a given species, contrasting with a low level of identity between species. This indicates that ITS sequencing is a reliable molecular tool for precise identification of Zygomycetes to the genus and species level and can be used for “DNA bar coding” of this group of fungi. DNA bar coding (13) is a DNA-based approach to routine species identification by sequencing short DNA regions. ITS regions have already been successfully used for DNA bar coding of plants (18).
Only one study assessed the intraspecies variability of ITS regions for Zygomycetes on a large panel of isolates (1). In the present study, we found very few differences in ITS regions within a given species, despite evaluation of isolates from different origins. Indeed, the studied isolates were both from clinical and environmental sources and were not epidemiologically linked. The overall similarities for the analyzed sequences of the species A. corymbifera, M. circinelloides, R. pusillus, R. microsporus, and R. oryzae were approximately 99% within each of their respective species. Isolates of R. pusillus and A. corymbifera showed almost no variability within the ITS regions. In contrast, analysis of the sequences of 17 isolates of R. oryzae, including the type strain, showed that, within these 17 isolates, 3 types could be distinguished with a maximum of sequence differences at 5 positions. One recent study analyzed the ITS sequence (650 bp) of 64 isolates of R. oryzae and showed results very similar to ours, including sequence variability at the same nucleotide positions (1). The 5 isolates of M. circinelloides analyzed in the present study showed more intraspecies variations in ITS sequences than did R. oryzae isolates. Although there was a maximum of an 8-bp difference within the whole ITS1-5.8S-ITS2 region, each isolate had a unique sequence. ITS sequences for the six isolates of Rhizopus microsporus, including four varieties, were identical except for R. microsporus var. microsporus, which showed differences at eight positions. Similar results have been obtained for other genetic sequences for R. microsporus, such as nuclear small-subunit (18S) and large-subunit (28S) rRNA gene regions (30) as well as the actin gene (31). These data indicate that the individualization of varieties among R. microsporus based on differences in morphology are not supported by sequence analysis, at least for the RNA genes.
Overall, ITS sequences were very different between species. This shows the interest of ITS sequencing for identification of Zygomycetes to the species level and will be useful for epidemiological studies, particularly to assess the relative frequency of the different species causing human diseases. In particular, Rhizopus spp. that are assumed to be responsible for more than 90% of the cases of human mucormycosis (23) showed very divergent sequences from the other Zygomycetes, with less than 58% similarity. The two most common species of Rhizopus, R. oryzae and R. microsporus, could be clearly differentiated from each other, as their sequences showed only 70% similarity. The other species of Rhizopus spp. have been rarely reported as causative agents of mucormycosis in humans (2, 25). Among these rare pathogens, R. shipperae showed a different sequence but R. azygosporus showed a sequence almost identical to that of R. microsporus. Mucor spp. are generally considered the third most common causative agent of mucormycosis after Rhizopus spp. and A. corymbifera (23), but it has also been reported that Mucor spp. ranked first in cancer patients infected with Zygomycetes (17). These discrepancies could be explained in part by changes in the nomenclature of Zygomycetes, some “Mucor” species have been reassigned to other genera (23). Another explanation is the difficulty of identification of Mucor species by standard mycological procedures. Indeed, the genus Mucor is mostly defined by negative characteristics (26). In this study, we analyzed the ITS sequences of 6 different Mucor spp.: M. circinelloides, M. hiemalis, M. indicus, M. racemosus, M. ramosissimus, and M. rouxii. Among these species, similarities of 79 to 96% were observed, allowing a good identification, except for M. circinelloides and M. rouxii, for which sequences were 99% similar. ITS sequences of Mucor spp. also showed high variability compared to the other Zygomycetes. Nevertheless, for one species, R. variabilis var. regularior, 99% of sequence similarity was found compared to M. circinelloides. These findings suggest that this newly described variety of R. variabilis (33) might in fact belong to the genus Mucor. Infections due to Rhizomucor spp. in humans are rare and mostly caused by R. pusillus, which was also identified as a major animal pathogen (23). Within Rhizomucor spp., all 3 species were well distinguished based on their ITS sequences. Among the other species that are known to be human pathogens, such as A. elegans, C. recurvatus, C. bertholletiae, S. vasiformis, and S. racemosum, we have tested isolates of C. recurvatus and S. racemosum and shown that they were easily distinguished from all other species.
Overall, results of the present study showed that sequencing of ITS regions is a reliable tool for the identification of Zygomycetes from pure cultures to the species level, including rare species and species that lack typical morphological characteristics. All the sequences obtained in the present study have been deposited in GenBank and will increase the available data for sequence identification of Zygomycetes. Molecular identification could be performed within a few days and is then much faster than standard mycological identification, which takes several weeks in instances when specialized tests are needed. Moreover, in vitro (5, 6, 28) and in vivo studies with animal models (4, 29) have shown that different genera and species of Zygomycetes exhibited variable susceptibilities to antifungal drugs, including to the new azoles such as posaconazole. Therefore, accurate and rapid identification to the genus or species level may be of interest to improve antifungal therapy.
Although molecular identification of Zygomycetes species from pure cultures is an interesting tool for in vitro diagnosis, the problem is that cultures of infected tissues in patients with mucormycosis are often negative. To identify the fungus responsible directly from the infected tissues is of the utmost importance in clinical settings. For these reasons, we set up animal models of experimental disseminated mucormycosis for the species A. corymbifera, M. circinelloides, M. indicus, R. pusillus, R. microsporus, and R. oryzae and confirmed that DNA extraction, amplification of fungal DNA, sequencing, and molecular identification are possible directly from frozen tissues. These results are of importance for early discrimination of the presence of Zygomycetes versus that of other filamentous fungi in tissues to initiate the appropriate antifungal therapy. Indeed, it is known that newly used systemic antifungals such as voriconazole and echinocandins do not exhibit any significant activity against Zygomycetes (6, 9).
In conclusion, ITS sequencing is appropriate for species identification within Zygomycetes either from cultures or from infected frozen tissues. This method should now be investigated to improve the diagnosis of mucormycosis in humans.
Acknowledgments
P.S. was supported by an educational grant from Gilead Sciences, Paris, France.
We are grateful to Anne-Sophie Delannoy and Christine Bouchier for their help in DNA sequencing. We also thank Monique Coutanson and Bernard Papierok from the Pasteur Institute Collection of Fungi for providing reference strains and Marie-Antoinette Piens from Lyon, France, and Paul Verweij from Nijmegen, The Netherlands, for sharing some clinical isolates. Other clinical isolates were studied as part as the nationwide survey of mucormycosis in France. Members of the French Mycoses Study Group who sent their isolates were as follows (in alphabetical order by city): H. Chardon (Aix en Provence), A. Tottet (Amiens), F. Le Turdu (Argenteuil), C. Duhamel (Caen), X. Kubab (Corbeil), S. Ranque, L. Collet (Marseille), O. Morin (Nantes), S. Bonacorsi, G. Buot, C. Lacroix, V. Lavarde (Paris), D. Toubas (Reims), and B. Graf (Berlin, Germany).
REFERENCES
- 1.Abe, A., T. Sone, I. N. Sujaya, K. Saito, Y. Oda, K. Asano, and F. Tomita. 2003. rDNA ITS sequence of Rhizopus oryzae: its application to classification and identification of lactic acid producers. Biosci. Biotechnol. Biochem. 67:1725-1731. [DOI] [PubMed] [Google Scholar]
- 2.Anstead, G. M., D. A. Sutton, E. H. Thompson, I. Weitzman, R. A. Otto, and S. K. Ahuja. 1999. Disseminated zygomycosis due to Rhizopus schipperae after heatstroke. J. Clin. Microbiol. 37:2656-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakrabarti, A., A. Ghosh, G. S. Prasad, J. K. David, S. Gupta, A. Das, V. Sakhuja, N. K. Panda, S. K. Singh, S. Das, and T. Chakrabarti. 2003. Apophysomyces elegans: an emerging zygomycete in India. J. Clin. Microbiol. 41:783-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Council of the European Communities. 1986. Council Directive 86/609/EEC of 24 November 1986 on the approximation of laws, regulations and administrative provisions of the Member States regarding the protection of animals used for experimental and other scientific purposes. Official Journal of the European Communities. L 358, 18/12/1986, p. 0001-0028. Council of the European Communities, Brussels, Belgium.
- 4.Dannaoui, E., J. F. Meis, D. Loebenberg, and P. E. Verweij. 2003. Activity of posaconazole in treatment of experimental disseminated zygomycosis. Antimicrob. Agents Chemother. 47:3647-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dannaoui, E., J. F. Meis, J. W. Mouton, and P. E. Verweij. 2002. In vitro susceptibilities of Zygomycota to polyenes. J. Antimicrob. Chemother. 49:741-744. [DOI] [PubMed] [Google Scholar]
- 6.Dannaoui, E., J. Meletiadis, J. W. Mouton, J. F. Meis, and P. E. Verweij. 2003. In vitro susceptibilities of zygomycetes to conventional and new antifungals. J. Antimicrob. Chemother. 51:45-52. [DOI] [PubMed] [Google Scholar]
- 7.de Hoog, G. S., and J. Guarro (ed.). 1995. Atlas of clinical fungi. Centraalbureau voor Schimmelcultures, Baarn, The Netherlands.
- 8.Dromer, F., and M. R. McGinnis. 2002. Zygomycosis, p. 297-308. In M. A. Pfaller (ed.), Clinical mycology. Churchill Livingstone, New York, N.Y.
- 9.Espinel-Ingroff, A. 1998. Comparison of in vitro activities of the new triazole SCH56592 and the echinocandins MK-0991 (L-743,872) and LY303366 against opportunistic filamentous and dimorphic fungi and yeasts. J. Clin. Microbiol. 36:2950-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerrits van den Ende, A. H. G., and G. S. de Hoog. 1999. Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana. Stud. Mycol. 43:151-162. [Google Scholar]
- 11.Greenberg, R. N., L. J. Scott, H. H. Vaughn, and J. A. Ribes. 2004. Zygomycosis (mucormycosis): emerging clinical importance and new treatments. Curr. Opin. Infect. Dis. 17:517-525. [DOI] [PubMed] [Google Scholar]
- 12.Hall, L., S. Wohlfiel, and G. D. Roberts. 2004. Experience with the MicroSeq D2 large-subunit ribosomal DNA sequencing kit for identification of filamentous fungi encountered in the clinical laboratory. J. Clin. Microbiol. 42:622-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebert, P. D. N., S. Ratnasingham, and J. R. DeWaard. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B 270:S596-S599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imhof, A., S. A. Balajee, D. N. Fredricks, J. A. Englund, and K. A. Marr. 2004. Breakthrough fungal infections in stem cell transplant recipients receiving voriconazole. Clin. Infect. Dis. 39:743-746. [DOI] [PubMed] [Google Scholar]
- 15.Iwen, P. C., S. H. Hinrichs, and M. E. Rupp. 2002. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med. Mycol. 40:87-109. [DOI] [PubMed] [Google Scholar]
- 16.Kontoyiannis, D. P., M. S. Lionakis, R. E. Lewis, G. Chamilos, M. Healy, C. Perego, A. Safdar, H. Kantarjian, R. Champlin, T. J. Walsh, and I. I. Raad. 2005. Zygomycosis in a tertiary-care cancer center in the era of Aspergillus-active antifungal therapy: a case-control observational study of 27 recent cases. J. Infect. Dis. 191:1350-1360. [DOI] [PubMed] [Google Scholar]
- 17.Kontoyiannis, D. P., V. C. Wessel, G. P. Bodey, and K. V. Rolston. 2000. Zygomycosis in the 1990s in a tertiary-care cancer center. Clin. Infect. Dis. 30:851-856. [DOI] [PubMed] [Google Scholar]
- 18.Kress, W. J., K. J. Wurdack, E. A. Zimmer, L. A. Weigt, and D. H. Janzen. 2005. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. USA 102:8369-8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 20.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology. Lea & Febiger, Philadelphia, Pa.
- 21.Marr, K. A., R. A. Carter, F. Crippa, A. Wald, and L. Corey. 2002. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 34:909-917. [DOI] [PubMed] [Google Scholar]
- 22.Marty, F. M., L. A. Cosimi, and L. R. Baden. 2004. Breakthrough zygomycosis after voriconazole treatment in recipients of hematopoietic stem-cell transplants. N. Engl. J. Med. 350:950-952. [DOI] [PubMed] [Google Scholar]
- 23.Ribes, J. A., C. L. Vanover-Sams, and D. J. Baker. 2000. Zygomycetes in human disease. Clin. Microbiol. Rev. 13:236-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruchel, R., and M. Schaffrinski. 1999. Versatile fluorescent staining of fungi in clinical specimens by using the optical brightener Blankophor. J. Clin. Microbiol. 37:2694-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schipper, M. A., M. M. Maslen, G. G. Hogg, C. W. Chow, and R. A. Samson. 1996. Human infection by Rhizopus azygosporus and the occurrence of azygospores in zygomycetes. J. Med. Vet. Mycol. 34:199-203. [PubMed] [Google Scholar]
- 26.Scholer, H. J., E. Müller, and M. A. A. Schipper. 1983. Mucorales, p. 9-59. In D. H. Howard (ed.), Fungi pathogenic for humans and animals, part A. Biology. Marcel Dekker, New York, N.Y.
- 27.Siwek, G. T., K. J. Dodgson, M. De Magalhaes-Silverman, L. A. Bartelt, S. B. Kilborn, P. L. Hoth, D. J. Diekema, and M. A. Pfaller. 2004. Invasive zygomycosis in hematopoietic stem cell transplant recipients receiving voriconazole prophylaxis. Clin. Infect. Dis. 39:584-587. [DOI] [PubMed] [Google Scholar]
- 28.Sun, Q. N., A. W. Fothergill, D. I. McCarthy, M. G. Rinaldi, and J. R. Graybill. 2002. In vitro activities of posaconazole, itraconazole, voriconazole, amphotericin B, and fluconazole against 37 clinical isolates of zygomycetes. Antimicrob. Agents Chemother. 46:1581-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, Q. N., L. K. Najvar, R. Bocanegra, D. Loebenberg, and J. R. Graybill. 2002. In vivo activity of posaconazole against Mucor spp. in an immunosuppressed-mouse model. Antimicrob. Agents Chemother. 46:2310-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voigt, K., E. Cigelnik, and K. O'Donnell. 1999. Phylogeny and PCR identification of clinically important Zygomycetes based on nuclear ribosomal-DNA sequence data. J. Clin. Microbiol. 37:3957-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voigt, K., and J. Wostemeyer. 2001. Phylogeny and origin of 82 zygomycetes from all 54 genera of the Mucorales and Mortierellales based on combined analysis of actin and translation elongation factor EF-1alpha genes. Gene 270:113-120. [DOI] [PubMed] [Google Scholar]
- 32.Weitzman, I., S. Whittier, J. C. McKitrick, and P. Della-Latta. 1995. Zygospores: the last word in identification of rare or atypical zygomycetes isolated from clinical specimens. J. Clin. Microbiol. 33:781-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng, R. Y., and G. Q. Chen. 1993. Another non-thermophilic Rhizomucor causing human primary cutaneous mucormycosis. Mycosystema 6:1-12. [Google Scholar]