Abstract
The accuracy and usefulness of laboratory-developed real-time PCR procedures using a Light Cycler instrument (Roche Diagnostics) for detecting and quantifying human immunodeficiency virus type 1 (HIV-1) RNA and DNA as well as herpes simplex virus type 1 (HSV-1)/HSV-2 DNA in cervicovaginal secretions from women coinfected with HIV and HSV were evaluated. For HIV-1, the use of the NEC152 and NEC131 primer set and the NEC-LTR probe in the long terminal repeat gene allowed us to detect accurately the majority of HIV-1 subtypes of group M circulating in sub-Saharan Africa, including subtypes A, B, C, D, and G as well as circulating recombinant forms 02 and 11. The detection threshold of real-time PCR for HIV in cervicovaginal lavage samples was 5 copies per assay for both RNA and DNA; the intra- and interassay coefficients of variation of CT values were 1.30% and 0.69% (HIV-1 RNA) and 1.84% and 0.67% (HIV-1 DNA), respectively. Real-time PCR for HSV using primers and probe targeting the HSV DNA polymerase gene allowed both detection and quantification of HSV DNA and also differentiation between HSV-1 and HSV-2 genotypes. The detection threshold of real-time PCR for HSV was 5 copies per assay; the intra- and interassay coefficients of variation of CT values were 0.96% and 1.49%, respectively. Both manual and automated silica-based procedures were appropriate for combined extraction of HIV and HSV genomes from female genital secretions. Taken together, these findings indicate that real-time PCR may be used as a unique nucleic acid amplification procedure to detect and quantify HIV and HSV genomes in cervicovaginal secretions and thus to assess at reduced costs the genital shedding of both viruses in women included in intervention studies.
Herpes simplex virus type 2 (HSV-2) infection is the primary cause of genital herpes, increasingly regarded as the most common cause of genital ulcer disease worldwide (10, 49). In developing countries, the major public health importance of HSV-2 relates to its potential role in facilitating human immunodeficiency virus (HIV) transmission. HSV-2 seems to be highly prevalent in most regions with severe HIV epidemics (49). Genital ulcer disease is known to enhance the infectiousness of HIV-positive subjects and the susceptibility of HIV-negative subjects, and clinical research has shown effects of herpes on genital HIV shedding (3, 26, 32, 42). Accumulating evidence suggests that HSV-2 may play a key role in HIV transmission in some parts of Africa (48). The reciprocal effect of HIV immune suppression on the exacerbation of HSV-2 symptoms implies that there may be a positive feedback loop, with HIV enhancing HSV-2 expression which in turn may enhance HIV transmission (2, 42). A workshop convened in London in 2001 by the World Health Organization, UNAIDS, and the London School of Hygiene and Tropical Medicine reviewed the evidence for these important interactions between the two viruses and recommended that intervention trials be conducted to test the impact of HSV control on HIV transmission (50). Since then, a number of randomized trials, using either episodic or suppressive antiviral therapy or other strategies (e.g., microbicides, vaccines, circumcision, or behavioral methods) to control genital herpes, which have measured the impact on HIV acquisition (among HIV-seronegative individuals) or HIV transmission (from HIV-seropositive individuals or within serodiscordant couples) have been initiated in various parts of the developing world (50). In particular, many trials have had HIV and/or HSV-2 genital shedding as major trial endpoints (50). It is believed that the presence of HIV and HSV in the genital fluids will be an important indirect marker of sexual transmissibility of the viruses, and reduction in the frequency and quantity of these viruses should translate into some reduction in sexual transmission (7). The ability to assess the viral burden of both HIV-1 and HSV-2 in genital secretions is essential in understanding the pathophysiology of interactions between both viruses and the effect of HSV-2 replication on the infectiousness of HIV and in evaluating the effect of antiherpes therapy on genital HIV shedding (10). Most studies evaluate genital viral loads of both viruses in HIV-1- and HSV-2-coinfected women (50). Inclusion of females instead of males offers the possibility to do relatively easy genital sampling (37).
Given the importance of these biological outcomes for the concerned trials and the high international expectations surrounding these trials, it would be timely to agree on standardization of some of the laboratory procedures in order to best interpret, compare, and perhaps even pool in meta-analysis the results of these trials. The availability of standardized assays for the quantification of HIV-1 RNA and HSV-2 DNA that are sensitive, specific, and reproducible therefore constitutes a prerequisite for intervention studies.
Little information is available about the accuracy of different nucleic acid or signal amplification techniques in quantifying HIV-1 RNA and DNA and HSV-2 DNA in the genital tract of women coinfected with HIV-1 and HSV-2 (9, 20, 32, 41). Real-time PCR-based assays may allow us both to simplify the detection and quantification of HIV and HSV genomes in genital fluids and to decrease the cost of laboratory evaluation, which is very important in intervention studies generally including a huge number of samples. Some studies have evaluated the ability of the real-time one-step reverse transcriptase PCR (RT-PCR) to assess HIV-1 and HIV-2 RNA load in plasma (11, 38, 40) and HIV-1 DNA proviral load in peripheral blood mononuclear cells (12, 19, 22, 47). To our knowledge, the use of real-time PCR assays has not yet been validated for the quantification of HIV-1 RNA and proviral DNA in the female genital tract. The main goal of the present study was to evaluate the analytical performance characteristics of real-time PCR to quantify HIV-1 RNA, HIV-1 proviral DNA, and HSV-2 DNA in the female genital secretions in order to propose a useful and accurate tool to assess the genital shedding of both viruses in women included in intervention studies.
MATERIALS AND METHODS
Clinical specimens.
Cervicovaginal secretions (CVS) were obtained from 103 HIV-1-seropositive adult women (including 98 HSV-2-seropositive women) and 166 HIV-seronegative adult women (including 106 HSV-2-seropositive women) suffering from genital ulcer disease who were enrolled in sexually transmitted disease and family planning clinics in Ghana (n = 188) and the Central African Republic (n = 81). Ethical approval was given by the London School of Hygiene and Tropical Medicine, United Kingdom, and by national health authorities from Ghana and the Central African Republic. Written informed consent was obtained from all participants. Genital secretions were collected outside menstruation successively by two different procedures. Briefly, following the introduction of a speculum, a standardized 60-second lavage was performed with 3 ml of 1 M phosphate-buffered saline (pH 7.2), as previously described (4). Second, a sterile dry swab was applied gently on the cervical os. All samples were processed within 1 h of collection by centrifuging the cervicovaginal lavage fluid at 1,000 × g for 15 min followed by freezing the cell-free supernatant and cell pellet separately at −80°C. All of the swab samples were rapidly stored at −80°C until use. Blood-contaminated samples were excluded after the detection of hemoglobin (Hb) by using Multistix 8 SG strips (Bayer Diagnostics, Puteaux, France). To avoid possible misclassification of shedding that could result from the presence of seminal plasma in the vagina, cervicovaginal lavage samples were further evaluated for Y chromosome traces, as described previously (8), with the slight modification of using a real-time PCR assay on a Light Cycler instrument. Cervicovaginal lavage samples positive for Y chromosome traces were excluded from further analyses. Finally, 81 Hb- and Y chromosome-negative CVS from HIV-positive women and 149 CVS from HIV-negative women were used in the study.
In addition, Hb- and Y chromosome-negative CVS from HIV-seronegative women were tested for HSV-2 DNA by real-time PCR in the DNA polymerase gene, as described below. HSV-2 DNA PCR-negative CVS were then pooled to be used for further HIV-1 RNA, HIV-1 DNA, and HSV-2 DNA quantification assays.
Nucleic acid purifications.
Nucleic acids were extracted from a 0.2-ml volume of either a cervicovaginal lavage cell pellet suspension or cell-free supernatant. DNA from cervicovaginal cell pellets and both RNA and DNA from cervicovaginal lavage supernatants were purified with a DNA extraction protocol from cells (QiaAmp DNA; QIAGEN, Courtaboeuf, France) and an RNA and DNA extraction protocol on a silica column system (High Pure viral nucleic acid kit; Roche Diagnostics, Meylan, France), respectively, according to the manufacturers' recommendations. The total nucleic acids were eluted in 100 μl of RNase- and DNase-free water. To compare manual to automated nucleic acid extraction procedures, DNA from CVS cell pellet and RNA and DNA from CVS supernatant samples were automatically purified on a MagNA Pure LC instrument (software version 2.1) with a MagNA Pure LC DNA I isolation kit and a MagNA Pure LC total nucleic acid isolation kit (Roche Diagnostics), respectively, according to the manufacturer's instructions. Elution was performed with 100 μl of elution buffer. All purification runs included one previously tested negative control sample. For the PeliCheck HSV-1/2 DNA-02 reference panel (see below), nucleic acids were extracted from 1 ml of sample with a MagNA Pure LC total nucleic acid isolation large-volume kit (Roche Diagnostics) and eluted in 50 μl.
HIV-1 RNA panels.
Three HIV-1 RNA panels were prepared with viral stock sources diluted in pooled cell-free CVS or plasma from HIV-seronegative control women whose vaginal secretions were negative for HSV-2 DNA. The first panel consisted of the PeliCheck HIV-1 RNA-02 reference panel (prepared by the Central Laboratory of the Blood Transfusion Service, Amsterdam, The Netherlands, for the international Viral Quality Control [VQC] Programme), containing 12 different samples with various viral load levels and HIV subtypes; this panel was used as viral stock to assess the specificity and the sensitivity of the real-time RT-PCR assay for different HIV-1 subtypes. The second panel was obtained by spiking pooled CVS or plasma with serial dilutions of HIV-1 RNA subtypes A and C reference stocks (at 109 copies/ml) obtained from the VQC Programme. The third panel consisted of serial dilutions (5 × 107, 5 × 106, 5 × 105, 5 × 104, 5 × 103, 5 × 102, 2.5 × 102, 1 × 102, and 5 × 101 copies/ml) of culture supernatant of the HIV-1 subtype A strain.
HIV-1 DNA provirus panels.
The HIV-1 DNA panel was obtained by spiking pooled cell-free CVS from HIV-seronegative control women whose vaginal secretions were negative for HSV-2 DNA with successive dilutions of lymphoblastoid T-cell line 8E5 (NIH; ATCC 8993) containing a single proviral genome of HIV-1 LAV per cell, with the following HIV-1 DNA predicted values: 100,000, 10,000, 1,000, 100, 50, 10, 5, and 1 copy for 106 cells of cervicovaginal pellet. The number of cells was determined by cell numeration under a microscope with confirmatory real-time PCR quantification of the albumin gene, as described previously (28).
HIV-1 subtyping.
HIV-1 subtyping was determined by phylogenetic analyses of env gene sequences encoding the V3 loop gene (V3) and, for undetermined genotypes only, of reverse transcriptase pol gene sequences, as previously described (21). V3 and RT sequences were obtained from plasma HIV-1 RNA as previously described, with slight modifications concerning the choice of primer sets (31). In brief, HIV-1 virions in 1.0 ml of plasma were pelleted by centrifugation at 30,000 × g for 60 min. The pellets were then resuspended in 200 μl of RNase-free sterile distilled water, and RNA was recovered by using a Total Nucleic Acid kit on the MagNA Pure system (Roche Diagnostics). RNA was further reverse transcribed to cDNA. For RT sequences, two RT fragments (RT1, codons 6 to 152, and RT2, codons 157 to 252) were amplified using the previously published forward internal primers A and B (27) and the reverse internal primers 5′-GGTGATCCTTTCCATCC-3′ for RT1 and 5′-TCATTGGACAGTCCAGCT-3′ for RT2. For V3 sequences, an RT-nested PCR amplification was carried out using the following primers, as previously described (46): E00, ES8B, E20, and E11. The final PCR products were purified, direct sequencing of amplicons was performed using an automatic sequencer, and sequences were aligned with the software Sequencher (Gene Codes Corporation, Ann Arbor, Michigan). The HIV-1 subtype was assessed by the BLAST program at www.ncbi.nlm.nih.gov. Sequences yielding ambiguous results were further compared to reference sequences by phylogenetic analysis using the parsimony method, as described previously (24).
Cloning HSV DNA and plasmid construction.
A 324-bp PCR product encompassing nucleotides 2660 to 2963 of the UL30 gene of HSV-2 strain G (ATCC VR-734) was generated with the primers HSVpol-1 (5′-CGGAATTCCGTCATCTCACGGGGACAC-3′) and HSVpol-2 (5′-CGGGATCCCGACGGTATCGTCGTAAA-3′). This amplicon was further digested with BamHI and EcoRI and inserted and cloned into the pcDNA3.1/HisC plasmid (Invitrogen, Cergy Pontoise, France). The resulting pcDNA3.1/HisC-ul30hsv plasmid was then purified with a QIAGEN plasmid Maxi kit (QIAGEN) and was sequenced by the dideoxynucleotide chain termination method (ABI Prism dRhodamine terminator cycle sequencing ready mix; Applied Biosystems, Foster City, CA), according to the manufacturer's recommendations. The DNA concentration was assessed by spectrophotometry at 260 nm and was calculated as the average of three measurements. The plasmid was used in all experiments as a standard to quantify HSV DNA genital load.
HSV DNA panels.
Two HSV-1 and HSV-2 DNA panels were prepared with viral stock sources diluted in pooled cell-free CVS from HIV-seronegative control women whose vaginal secretions were negative for HSV-1 and HSV-2 DNA. The PeliCheck HSV-1/2 DNA-02 reference panel (VQC Programme) was composed of serial 10-fold dilutions of HSV-1 and HSV-2 standards. The panel was used as HSV stocks with the following predicted HSV DNA quantities (expressed as nucleic acid technology-detectable units by limiting dilution [NDU]/ml) and type (1 or 2): 5, 50, 500, 5,000, and 50,000 NDU/ml for HSV-1 and 2.5, 25, 250, 2,500, and 25,000 NDU/ml for HSV-2. Concentrations were expressed in NDU by probit analysis (the DNA input in PCR assays at the overall 63% detection endpoint is equivalent to 1 NDU). One NDU is expected to be close to 1 HSV DNA genome. The second panel was obtained by serial 10-fold dilution of the pcDNA3.1/HisC-ul30hsv plasmid.
Detection and quantification of HIV-1 RNA and DNA in genital secretions.
HIV-1 RNA was detected and quantified in the acellular fraction of genital secretions by real-time RT-PCR, and HIV-1 DNA was detected and quantified in the cell pellet of genital secretions by real-time PCR. HIV-1 RNA and DNA quantification used the forward NEC152 (5′-GCCTCAATAAAGCTTGCCTTGA-3′) and reverse NEC131 (5′-GGCGCCACTGCTAGAGATTTT-3′) primers and the dually labeled NEC-LTR probe (5′-6-carboxyfluorescein-AAGTAGTGTGTGCCCGTCTGTTRTKTGACT-6-carboxytetramethylrhodamine-3′) in the long terminal repeat gene, as previously published (39). Primers and probes were synthesized by Eurogentec SA (Seraing, Belgium).
HIV-1 RNA quantification was carried out by real-time RT-PCR with a Light Cycler RNA amplification kit HybProbe specifically adapted for one-step RT-PCR in glass capillaries using a Light Cycler instrument (Roche Diagnostics). The master mix contained 1× RNA master hybridization buffer, including the Tth DNA polymerase and deoxynucleoside triphosphate mix (containing dUTP instead of dTTP), 2.5 mM Mn(OAC)2, and 0.3 μM concentrations of each primer and probe. Cycling conditions were as follows: initial reverse transcription at 61°C for 30 min, denaturation at 95°C for 30 s, and 45 cycles of denaturation at 95°C for 1 s, annealing at 55°C for 15 s, and elongation at 65°C for 1 min with a ramp of 5°C/s (with fluorescence acquisition at the end of each elongation stage). The positive control consisted of culture supernatant of the HIV-1 subtype A strain. As a control for cross-contamination, a sample of distilled water was subjected to the RNA extraction procedure, and the resulting extract was amplified in duplicate. A standard graph of the CT values was obtained from serial dilutions (5 × 107 to 50 copies/ml) of the HIV-1 subtype A reference strain. The CT values from unknown samples were plotted on the standard curves, and the number of HIV-1 RNA copies per milliliter was calculated. Finally, HIV-1 RNA loads in cervicovaginal lavage samples were expressed as HIV-1 RNA copies per milliliter of cervicovaginal lavage sample.
For HIV-1 DNA quantification, the master mix contained 1× Fast-Start Taq DNA polymerase reaction buffer (Roche Diagnostics) including deoxynucleoside triphosphate mix, 3 mM MgCl2, and 0.3 μM concentrations of each primer and probe. Cycling conditions were as follows: initial denaturation/Fast-Start Taq DNA polymerase activation at 95°C for 10 min and then 45 cycles of denaturation at 95°C for 10 s and annealing and extension at 60°C for 30 s with a ramp of 5°C/s. The positive control consisted of DNA purified from lymphoblastoid T-cell line 8E5. As a control for cross-contamination, a procedure similar to the one described above was used for HIV-1 RNA quantification. A standard graph of the CT values was obtained from serial dilutions (105 to 1 copy per assay) of the 8E5 cells. The level of albumin DNA copies in the CVS cell pellet was used as an endogenous reference to normalize the variations in cell number in cervicovaginal lavage or DNA extraction samples. The CT values from unknown samples were plotted on the 8E5 standard curves, and the number of HIV-1 proviral DNA copies per assay was calculated. The normalized value of cell-associated HIV-1 DNA load corresponding to the ratio (HIV-1 copy number/albumin copy number) × 2 × 106 was finally expressed as the number of HIV-1 DNA copies per 106 cells of the cervicovaginal cell pellet.
Plasma HIV-1 RNA load quantification by branched DNA assay.
To validate the ability of laboratory-developed real-time RT-PCR to accurately quantify the most frequent HIV-1 subtypes circulating in Africa, including A, C, G, and recombinant forms (CRF02 and CRF11), which are highly prevalent in Central Africa and West Africa (17, 36), plasma samples from HIV-1-seropositve women were tested in parallel by the commercial Quantiplex HIV RNA 3.0 assay (Bayer Diagnostics). This assay has been shown to have high sensitivity to detect and quantify accurately the majority of HIV-1 subtypes of group M (14, 39, 45). The Quantiplex HIV RNA 3.0 assay was carried out with 200 μl of plasma sample, according to the manufacturer's recommendations.
Detection and quantification of HSV in genital secretions.
HSV DNA was detected and quantified in the acellular fraction of genital secretions by real-time PCR. The real-time PCR for HSV used the HSVpolF (5′-GCTCGAGTGCGAAAAAACGTTC-3′) and HSVpolR (5′-TGCGGTTGATAAACGCGCAGT-3′) primer set, allowing amplification of a 140-bp product, as previously published (6). A pair of fluorescently labeled probes, HSV-2 FLU (5′-GCGCACCAGATCCACGCCCTTGATGAGC-FLUOR-3′) and HSV-2 LCR (5′-LC-Red 640-CTTGCCCCCGCAGATGACGCC-phos-3′), was used for real-time quantification and melting temperature differentiation of HSV-1 and HSV-2, as described previously (6). In order to check the sensitivity of the assay to detect HSV-1, we compared the results of HSV-1 DNA detection by using an HSV-1 ligase chain reaction (LCR) probe (5′-LC-Red 640-CTTACCCCCGTAGATGACGCC-phos-3′), which perfectly matched with the HSV-1 DNA polymerase sequence in 10-fold serial dilutions of HSV-1 DNA panel number 5 (HSV-1 at 50,000 NDU/ml) from the PeliCheck HSV-1/2 DNA-02 reference panel and in 10-fold serial dilutions of culture supernatant from an HSV-1 laboratory strain propagated in Vero cells. The LC PCR master mix contained 1× Fast-Start Taq DNA polymerase reaction buffer (Roche Diagnostics), 3 mM MgCl2, 0.5 μM concentrations of each primer, 0.2 μM HSV-2 FLU, and 0.4 μM HSV-2 LCR. Cycling conditions were as follows: initial denaturation and Fast-Start Taq DNA polymerase activation at 95°C for 10 min, 45 cycles of denaturation at 95°C for 5 s, annealing at 58°C for 10 s (with fluorescence acquisition at the end of each annealing stage), and extension at 72°C for 12 s. After amplification was complete, melting curve analysis was performed as follows: denaturation at 95°C for 20 s and annealing at 45°C for 30 s followed by a gradual increase in temperature (transition rate of 0.1°C/s) to 85°C with continuous fluorescence acquisition.
The positive control consisted of the 324-bp pcDNA3.1/HisC-ul30hsv plasmid containing targeted nucleotides 2660 to 2963 of the UL30 gene, as described above.
As a control for cross-contamination, a sample of distilled water was also subjected to the DNA extraction procedure, and the resulting extract was amplified in duplicate. A standard graph of the CT values was obtained from serial dilutions (5 × 108, 5 × 107, 5 × 106, 5 × 105, 5 × 104, 5 × 103, 5 × 102, 2.5 × 102, 1 × 102, and 5 × 101 copies/ml) of the plasmid. The CT values from unknown samples were plotted on the standard curves, and the number of HSV genomes per milliliter was calculated. Each sample was analyzed in duplicate.
Differentiation between HSV-1 and HSV-2 by melting curve analysis.
The melting temperature of the HSV-2 LCR probe was used to determine whether the amplified DNA sequence was related to HSV-1 or HSV-2, as described previously (6). The HSV-2 LCR probe sequence is exactly complementary to HSV-2 DNA, and it has a melting peak at 58°C when hybridized to HSV-1 DNA and a peak at 71°C with a shoulder at 66°C when hybridized to HSV-2 DNA.
HIV-1 and HSV-2 detection and quantification in pools of cervicovaginal secretions spiked with various levels of HIV-1 and HSV-2.
Pooled cell-free CVS from HIV-seronegative women whose vaginal secretions were negative for HSV-2 DNA were spiked with large (250,000 copies/ml) or small (2,500 copies/ml) amounts of HIV-1 RNA from culture supernatant of the HIV-1 subtype A strain and with large (100,000 copies/ml) or small (1,000 copies/ml) amounts of HSV-2 DNA from culture supernatant of HSV-2 VR-734 (ATCC). HIV-1 RNA and HSV-2 DNA were further detected and quantified by real-time RT-PCR or real-time PCR, as described above.
Statistical analysis.
Standards were tested in duplicate in 10 separate runs to determine the threshold, reproducibility, and linearity of the real-time PCR assays. Quantitative HIV RNA and DNA and HSV-2 DNA values were log10 transformed for analyses. Linear regression analysis on log10-transformed values was performed to compare expected values and those obtained by real-time PCR-based quantifications. Comparison of the laboratory-developed real-time RT-PCR assay with the commercial branched DNA assay by the mean differences (bias) was calculated by the Bland-Altman plot analysis (5). A comparison of both extraction procedures was done with a paired-samples Student t test. Genital viral HIV-1 RNA loads between HSV-2 shedders and HSV-2 nonshedders were compared using the Student t test. Correlation analysis between viral loads was assessed by using Pearson's correlation coefficient. Statistical analyses were carried out using the software StatView SE 1 (Abacus Concepts, Inc., Berkeley, Calif.).
RESULTS
HIV-1 RNA quantification by real-time PCR.
Tenfold serial dilutions of culture supernatant of the HIV-1 subtype A strain from 107 to 50 copies/ml (106 to 1 copy per assay) were used to generate a standard curve. Each dilution was detected in triplicate. One copy per assay of HIV-1 subtype A RNA was inconsistently detected (not shown), as expected from a Poisson distribution. The point of 5 copies per assay was detected in 100% of runs and was thus considered to be the lowest limit of quantification corresponding to 250 copies/ml. The linearity and dynamic range of at least 6 log10 decades were confirmed by the strong linear relationship between CT values and the log10 of the starting copy number up to 107 copies (r2 > 0.99). To determine the intra-assay reproducibility, six HIV-1 RNA dilutions (10 to 105 copies per assay) from HIV-1 RNA subtype A culture supernatant were carried out simultaneously 10 times. The intra-assay coefficients of variation (CV) for the CT values ranged from 0.8 to 2.2%. The corresponding standard deviation of log10 copies/assay ranged from 0.01 for log10 5 to 0.15 for log10 1 (Table 1). These values are in the range of 0.1 to 0.2 log10 of intra-assay variations usually admitted with the Virology Quality Assessment Program. The interassay reproducibility determinations in 10 different runs gave similar results, with CV for the CT values ranging from 1.26 to 2.62%.
TABLE 1.
Repeatability assay resultsa
| log10 copies | HIV-1 RNA
|
HIV-1 DNA
|
HSV-2 DNA
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| CV (%) of CT values | SD of CT values | SD of log10 copies | CV (%) of CT values | SD of CT values | SD of log10 copies | CV (%) of CT values | SD of CT values | SD of log10 copies | |
| 5 | 1.20 | 0.25 | 0.014 | 0.12 | 0.02 | 0.003 | 0.29 | 0.06 | 0.005 |
| 4 | 0.78 | 0.21 | 0.011 | 0.33 | 0.07 | 0.008 | 0.31 | 0.07 | 0.006 |
| 3 | 1.33 | 0.41 | 0.031 | 0.51 | 0.16 | 0.012 | 0.35 | 0.09 | 0.014 |
| 2 | 1.00 | 0.36 | 0.099 | 1.34 | 0.46 | 0.057 | 1.00 | 0.21 | 0.112 |
| 1 | 2.18 | 0.82 | 0.158 | 1.17 | 0.45 | 0.160 | 2.87 | 0.82 | 0.503 |
Repeatability precision is determined by the CV of CT values and standard deviation (SD) of the values obtained by 10 simultaneous determinations of serial dilutions from HIV-1 RNA culture supernatant, HIV-1 proviral DNA from 8E5 cells, and HSV-2 DNA from pcDNA3.1/HisC-ul30hsv plasmid.
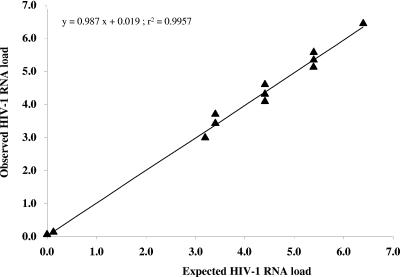
Table 2 depicts the raw data of HIV-1 RNA estimations from CVS by real-time RT-PCR with the LC assay tested from the PeliCheck HIV-1 RNA-02 panel. The mean Δlog10 (defined as the log10 of the observed RNA level minus the log10 of the expected RNA level) was −0.06 with the LC assay. All Δlog10 values are below 0.25 log10 units. The linear regression analysis of observed LC values versus expected values showed a slope of 0.98 and an r2 value of 0.99 (P < 0.001) (Fig. 1).
TABLE 2.
Quantification of HIV-1 RNA from the PeliCheck HIV-1 RNA-02 reference panel (VQC Programme) by HIV-1 RNA real-time RT-PCR using a Light Cycler apparatus
| Reference panel no. | Genotype | Expected result (log10 copies/ml) | Light Cycler assay observed result (log10 copies/ml) | Δlog10a |
|---|---|---|---|---|
| 1 | C | 3.20 | 3.10 | 0.10 |
| 2 | B | 6.40 | 6.49 | 0.09 |
| 3 | NA | 0.00 | 0.00 | 0.00 |
| 4 | B | 3.40 | 3.47 | 0.07 |
| 5 | D | 3.40 | 3.62 | 0.22 |
| 6 | C | 4.40 | 4.34 | 0.06 |
| 7 | B | 5.40 | 5.19 | −0.21 |
| 8 | NA | 0.00 | 0.00 | 0.00 |
| 9 | D | 4.40 | 4.44 | 0.04 |
| 10 | C | 5.40 | 5.42 | 0.02 |
| 11 | B | 4.40 | 4.17 | −0.23 |
| 12 | D | 5.40 | 5.22 | −0.18 |
Δlog10 is the difference between expected log10 values and observed log10 values obtained with a real-time RT-PCR assay. NA, not attributable.
FIG. 1.
Scatter plot of observed versus expected HIV-1 RNA levels, expressed in log10 copies, estimated by HIV-1 RNA Light Cycler assay in HIV-1-negative CVS spiked with a known amount of HIV-1 subtype B, C, and D from an HIV-1 RNA PeliCheck-02 reference panel.
We next evaluated the accuracy of the real-time PCR assay in genital fluid and in plasma for HIV subtypes A and C by spiking pooled CVS and plasma from HIV-negative women with serial dilutions of HIV-1 RNA subtype A from the PeliCheck HIV-1 RNA subtype A and subtype C. Tables 3 and 4 depict the raw data of expected and observed values of HIV-1 RNA log10 copies/ml. The mean Δlog10 values (defined as the log10 of the expected RNA level minus the log10 of the observed RNA level obtained with the LC assay) were −0.06 and 0.11, respectively, for CVS- and plasma-spiked samples for subtype A and −0.17 and −0.04 for subtype C.
TABLE 3.
Quantification of a known amount of HIV-1 RNA subtype A in pooled CVS lavage fluids and plasma samples from HIV-negative women spiked with a known amount of HIV-1 subtype A reference control by HIV-1 RNA real-time RT-PCR using a Light Cycler apparatusa
| Expected result (log10 copies)b | Cervicovaginal secretion
|
Plasma
|
||
|---|---|---|---|---|
| Observed result (log10 copies) | Δlog10 | Observed result (log10 copies) | Δlog10 | |
| 6.00 | 5.96 | 0.04 | 5.77 | 0.23 |
| 5.00 | 5.15 | −0.15 | 4.80 | 0.20 |
| 4.00 | 4.21 | −0.21 | 3.91 | 0.09 |
| 3.00 | 3.15 | −0.15 | 2.83 | 0.17 |
| 2.70 | 2.82 | −0.12 | 2.70 | 0.00 |
| 2.40 | 2.45 | −0.05 | 2.36 | 0.04 |
| 2.10 | 2.20 | −0.10 | 2.04 | 0.06 |
| 1.80 | 1.53 | 0.27 | 1.70 | 0.10 |
| Mean | −0.06 | 0.11 | ||
Δlog10 is the difference between expected log10 values and observed log10 values obtained with a real-time RT-PCR assay.
HIV-1 RNA subtype A at 109 copies/ml was obtained from the Viral Diagnostic Quality Control Programme, Amsterdam, The Netherlands.
TABLE 4.
Quantification of a known amount of HIV-1 RNA subtype C in pooled CVS lavage fluids and plasma samples from HIV-negative women spiked with a known amount of HIV-1 subtype C reference control by HIV-1 RNA real-time RT-PCR using a Light Cycler apparatusa
| Expected result (log10 copies)b | Cervicovaginal secretion
|
Plasma
|
||
|---|---|---|---|---|
| Observed result (log10 copies) | Δlog10 | Observed result (log10 copies) | Δlog10 | |
| 6.00 | 5.90 | 0.10 | 5.73 | 0.27 |
| 5.00 | 4.95 | 0.05 | 5.02 | −0.02 |
| 4.00 | 4.27 | −0.27 | 3.99 | 0.01 |
| 3.00 | 3.21 | −0.21 | 3.02 | −0.02 |
| 2.70 | 2.92 | −0.22 | 2.67 | 0.03 |
| 2.40 | 2.62 | −0.22 | 2.71 | −0.31 |
| 2.10 | 2.37 | −0.28 | 2.05 | 0.04 |
| 1.80 | 2.13 | −0.33 | 2.10 | −0.31 |
| Mean | −0.17 | −0.04 | ||
Δlog10 is the difference between expected log10 values and observed log10 values obtained with a real-time RT-PCR assay.
HIV-1 RNA subtype C at 109 copies/ml was obtained from the Viral Diagnostic Quality Control Programme, Amsterdam, The Netherlands.
In order to check possible detection of HIV-1 DNA in the nucleic acids extracted from cell-free supernatant, we carried out HIV-1 DNA detection by real-time PCR assay in 52 HIV-1 RNA-positive cervicovaginal lavage (CVL) supernatant samples. HIV-1 DNA was detected in only 11 samples (21%). The proportion of HIV-1 RNA load in the total HIV-1 RNA load and HIV-1 DNA load in the 11 HIV-1 RNA- and DNA-positive samples was estimated as the ratio (RNA load − DNA load)/RNA load = 99.9%, clearly indicating that the load of HIV-1 DNA when present in the supernatant CVL did not influence significantly the level of HIV-1 RNA load.
We determined the HIV-1 RNA load in CVS supernatant and in plasma from 81 HIV-1-seropositive women. HIV-1 RNA was detected in 68% of cervicovaginal lavage fluids, with a mean viral load of 3.73 log10 copies/ml and values ranging from 2.40 to 5.54 log10 copies/ml. Plasma was found to be positive for HIV-1 RNA in 84% of women, with a mean viral load of 5.15 log10 copies/ml, values ranging from 2.39 to 6.72 log10 copies/ml, and values higher than 500,000 copies/ml (5.69 log10) for 22 samples. HIV-1 RNA genital and plasma loads were significantly correlated (r = 0.50, P < 0.0001).
Comparison of plasma HIV-1 RNA loads with branched DNA assay.
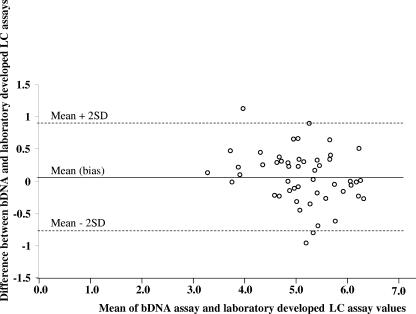
Twenty-seven plasma samples from women living in the Central African Republic (from patients 110 to 148) and 23 from women living in Ghana (from patients 152 to 178) were quantified with both procedures. The results of viral load determination by both assays and HIV subtyping show that HIV-1 RNA quantifications gave similar results by both methods for all subtypes (P = 0.4), which include 5 subtype A (patients 124, 135, 136, 138, and 167), 1 subtype C (121), 2 subtype F (120 and 133), 1 subtype G (118), 1 subtype H (139), 20 subtype CRF02 (110, 112, 117, 122, 134, 137, 141, 153, 154, 155, 157, 163, 164, 170, 172, 173, 174, 176, 177, and 178), 3 subtype CRF04 (126, 132, and 162), 5 subtype CRF06 (152, 160, 161, 166, and 165), 4 subtype CRF11 (129, 140, 143, and 148), 3 CRF13 (131, 128, and 171), and 5 undetermined (145, 146, 147, 158, 159) samples. The differences in log10 copies/ml results versus the mean assay results were homogeneously distributed. Thus, the Bland-Altman analysis demonstrated a mean difference (bias) between the branched DNA assay and the real-time RT-PCR assay of +0.07 log10 copies/ml (Fig. 2). For four samples, the quantification by laboratory-developed real-time RT-PCR assay gave higher values (Δlog10 > 0.5) than those observed with branched DNA (124, 128, 129, and 163), while the latter gave higher viral loads (Δlog10 > 0.5) for five samples (138, 139, 160, 176, and 177). Most of the largest differences occurred for recombinant forms of HIV.
FIG. 2.
Differences between the mean loads measured by the branched DNA assay and real-time RT-PCR by using the Bland-Altman plot analysis. The mean difference (bias) is indicated by a solid line, and the 95% confidence interval (±2 standard deviations) is represented by dotted lines. The mean difference ± standard deviation was 0.07 ± 0.41 log10 copies/ml.
HIV-1 DNA quantification by real-time PCR.
The standard curve of HIV-1 DNA was generated by successive dilutions of a known number of 8E5 cells. The number of cells was confirmed by evaluating the number of albumin gene copies by real-time PCR. Tenfold serial dilutions of the 8E5 cells from 105 to 5 copies per assay were used to generate a standard curve. Each dilution was amplified in triplicate. One copy of HIV-1 subtype A DNA was inconsistently detected (not shown). The point corresponding to 5 copies per assay that could always be detected was considered to be the lowest limit of quantification. The linearity and the dynamic range of at least 5 log10 were confirmed by the strong linear relationship between CT values and the log10 of the starting copy number up to 105 copies (r2 > 0.99).
To determine intra-assay reproducibility, six HIV-1 DNA dilutions (10 to 105 copies per assay) from 8E5 cell lines were carried out simultaneously 10 times. The intra-assay CV for the CT values ranged from 0.1 to 1.2%. The corresponding standard deviation of log10 copies/assay ranged from 0.002 for log10 5 to 0.15 for log10 1 (Table 1). These values are in the range of 0.1 to 0.2 log10 of intra-assay variations usually admitted with the Virology Quality Assessment Program. The interassay reproducibility determinations in 10 different runs gave similar results, with CV for the CT values ranging from 0.14 to 1.41%.
We determined the HIV-1 DNA proviral load in CVS cell pellets from 73 HIV-1-seropositive women. No cells could be detected in two samples. In the 71 remaining samples, 34 cell pellets with numbers of cells ranging from 10,000 to 5 × 106 were found to be negative for HIV-1 DNA. The mean HIV-1 DNA proviral load in the 37 positive samples was 1,501 copies/106 cells, with values ranging from 16 to 10,629 copies/106 cells.
HSV-2 DNA quantification by real-time PCR.
Tenfold serial dilutions of the pcDNA3.1/HisC-ul30hsv plasmid from 5 × 108 to 50 copies/ml were used to generate a standard curve. Each dilution was detected in triplicate. One and 2 copies per assay of the pcDNA3.1/HisC-ul30hsv plasmid were inconsistently detected (not shown), while 5 copies per assay could be detected in 100% of runs. This point was considered to be the lowest limit of quantification. The linearity and dynamic range of at least 7 log10 were confirmed by the strong linear relationship between CT values and the log10 of the starting copy number up to 107 copies per assay (r2 > 0.99).
To determine intra-assay reproducibility, six HSV-2 DNA dilutions (10 to 105 copies per assay) from pcDNA3.1/HisC-ul30hsv were carried out simultaneously 10 times. The intra-assay CV for the CT values ranged from 0.3 for log10 5 to 2.9% for log10 1 (Table 1). The interassay reproducibility determinations in 10 different runs gave similar results, with CV for the CT values ranging from 0.65 to 2.42%.
Despite the presence of two mismatches between the HSV-2 LCR probe and the sequence of the HSV-1 DNA polymerase, real-time PCR assays carried out with either HSV-1 LCR or HSV-2 LCR gave similar results. The CT values were not significantly different between the two probes in both 10-fold serial dilutions of HSV-1 DNA panel number 5 (HSV-1 at 50,000 NDU/ml) from the PeliCheck HSV-1/2 DNA-02 reference panel (mean ΔCT, 0.14; maximum ΔCT, 0.46) and HSV-1 laboratory strain culture supernatant (mean ΔCT, 0.18; maximum ΔCT, 0.68), indicating that HSV DNA real-time PCR assay using the HSV-2 LCR probe was similarly reliable for accurate detection and quantification of HSV-1 DNA and HSV-2 DNA.
Table 5 depicts the raw data of HSV DNA level and typing of PeliCheck HSV-1/2 DNA determined by real-time PCR quantitation and melting curve analysis, respectively. All positive as well as negative results expected from the PeliCheck HSV-1/2 DNA panel assay were confirmed by real-time PCR assay, except one with low viral burden at the threshold value (for patient 6). Sample 11 was positive after 40 cycles, which does not allow us to give an accurate viral load. HSV genotypes determined by melting curve analysis were all identical to the PeliCheck HSV types. The linear regression analysis of LC observed log10 values versus PeliCheck expected log10 values (with 6 and 11 excluded) showed a slope of 0.97 and an r2 value of 0.99 (P < 0.001).
TABLE 5.
Quantification and typing of HSV-1 and HSV-2 DNA from the PeliCheck HSV-1/2 DNA panel by HSV DNA real-time PCR using a Light Cycler apparatus
| Reference panel no. | PeliCheck HSV-1/2 DNA panel typea | Expected result (NDU/ml) | Light Cycler assay observed result (copies/ml) | Melting point |
|---|---|---|---|---|
| 1 | 2 | 250 | 507 | 71°C |
| 2 | NA | Negative | 0 | NA |
| 3 | 1 | 50 | 159 | 58°C |
| 4 | 2 | 25,000 | 39,475 | 71°C |
| 5 | 1 | 50,000 | 77,450 | 58°C |
| 6 | 1 | 5 | Negative | NA |
| 7 | NA | Negative | 0 | NA |
| 8 | 2 | 25 | 31 | 71°C |
| 9 | 1 | 500 | 713 | 58°C |
| 10 | 2 | 2,500 | 3,731 | 71°C |
| 11 | 2 | 2.5 | Positive <25 | 71°C |
| 12 | 1 | 5,000 | 6,890 | 58°C |
NA, not attributable.
Among 269 women suffering from genital ulceration whose cervicovaginal lavage samples were tested for HSV DNA, 103 (38%) were found to be positive for HSV-2 DNA, with viral loads ranging from 100 to 2.7 × 108 copies/ml, and 1 was positive for HSV-1 DNA. For 223 samples, cervicovaginal secretions were collected by both washing and endocervical swabbing. When cervicovaginal secretions were collected by washing, HSV-2 DNA was detected in 98 (44%) cervicovaginal lavage samples, whereas HSV-2 DNA was detected in only 57 (26%) endocervical swabbing samples (P < 0.01). All but one of the HSV-2 DNA-containing cervicovaginal secretions collected by endocervical swabbing were also positive when collected by cervicovaginal washing.
HIV-1 and HSV-2 in pools of cervicovaginal secretions spiked with various levels of HIV-1 and HSV-2.
In order to check that CVS infection by both viruses does not modify the specificity and sensitivity of quantification of each virus, HIV-1- and HSV-2-negative CVS were spiked with a known amount of HIV-1 subtype A and with a known amount of HSV-2. The results depicted in Table 6 show that quantifications of HIV-1 RNA and HSV-2 DNA were not changed by the presence of both viruses at high or low levels.
TABLE 6.
HIV-1 RNA and HSV-2 DNA quantifications by real-time RT-PCR or real-time PCR in four pools of HIV- and HSV-negative cervicovaginal lavage samples spiked with large (250,000 copies/ml) or small (2,500 copies/ml) amounts of HIV-1 RNA and with large (100,000 copies/ml) or small (1,000 copies/ml) amounts of HSV-2 DNA
| Pool | Amount of HIV-1 RNA | Amount of HSV-2 DNA | HIV-1 RNAa
|
HSV-2 DNAa
|
||
|---|---|---|---|---|---|---|
| Expected result | Observed result | Expected result | Observed result | |||
| Pool 1 | Large | Large | 5.40 | 5.39 | 5.00 | 5.11 |
| Pool 2 | Small | Large | 3.70 | 3.89 | 5.00 | 5.00 |
| Pool 3 | Large | Small | 5.40 | 5.39 | 3.00 | 3.10 |
| Pool 4 | Small | Small | 3.70 | 3.59 | 3.00 | 2.96 |
Observed results are expressed as log10 copies/ml and are the mean values obtained for each pool tested in triplicate.
HIV-1 and HSV-2 detection in cervicovaginal secretions from women coinfected with HIV-1 and HSV-2.
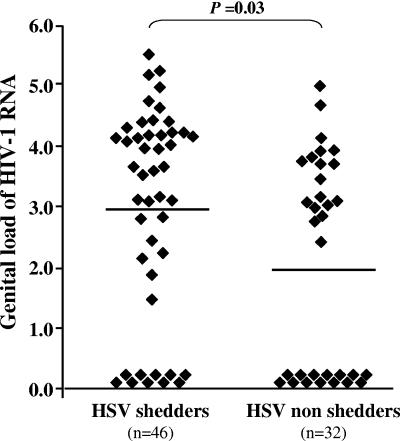
Among the 78 semen-free cervicovaginal lavage samples collected from women coinfected with HIV-1 and HSV-2, 46 were positive for HSV-2 DNA, with a mean viral load of 3.77 log10 HSV-2 DNA copies/ml (range, 1.70 to 7.10). Women with HSV-2 DNA genital shedding were more likely to shed HIV-1 RNA than were HSV-2 DNA-nonshedding women (30/46 [78%] versus 18/32 [56%]; P = 0.04). In addition, the mean HIV-1 RNA genital tract load was significantly higher in HSV-2 DNA-shedding women than in HSV-2 DNA-nonshedding women (2.98 log10 copies/ml versus 2.04 log10 copies/ml, respectively; P = 0.03) (Fig. 3). In contrast, there was no difference between HSV-2 DNA shedders and nonshedders in HIV-1 RNA plasma load (4.63 log10 copies/ml versus 4.61 log10 copies/ml, respectively) or in HIV-1 DNA cell-associated genital load (1.41 log10 copies/106 cells versus 1.36 log10 copies/106 cells, respectively).
FIG. 3.
Genital HIV-1 RNA shedding in HSV-2 DNA-shedding and -nonshedding women coinfected with HSV-2 and HIV-1. Genital HIV-1 RNA loads from HSV-2 DNA-shedding and HSV-2-nonshedding women, expressed in log10 copies/ml, are represented by triangles. Means of log10 copies/ml HIV-1 RNA load are indicated in each group by a plain bar. HIV-1 non-RNA-shedding women were given a log-transformed value of zero.
Comparison between manual and automated nucleic acid extraction procedures.
Finally, manual versus automated extraction procedures of nucleic acid followed by real-time RT-PCR detection/quantification of HIV-1 RNA or real-time PCR detection/quantification of HSV-2 DNA and HIV-1 DNA were compared in a series of 40 cervicovaginal samples from HIV-1- and HSV-2-coinfected women. No significant differences (e.g., >0.5 log10) in resulting HIV-1 RNA or HSV-2 DNA loads from CVS supernatants and HIV-1 DNA load from CVS cell pellets were observed between the extraction procedures (not shown).
DISCUSSION
In the present study, we demonstrate the accuracy and usefulness of laboratory-developed real-time PCR procedures for detecting and quantifying HIV-1 RNA and DNA as well as HSV DNA in cervicovaginal secretions from women coinfected with HIV and HSV.
Several molecular procedures have previously been proposed to detect and quantify HIV or HSV genomes in female genital secretions, including laboratory-developed quantitative competitive RT-PCR assay (18) and various commercially available assays (13, 25, 43) for HIV-1 RNA; laboratory-developed competitive PCR (44) and modified commercial PCR amplification assay (1, 33) for HIV-1 DNA; and finally, competitive PCR (20), real-time PCR assay (41), and PCR detection followed by DNA enzyme immunoassay hybridization (32) for HSV DNA. All of these procedures do not allow a common technique for the detection and quantification of both HIV and HSV genomes, as needed, for example, in the assessment of HIV and HSV genital shedding in intervention studies. Furthermore, only a few of them have been previously evaluated according to reference panels of HIV-1 or HSV-1/2 strains. Finally, the use of commercially available kits remains expensive, especially when large series of clinical samples may be tested.
We herein proposed to use real-time PCR assays to detect and quantify both HIV and HSV genomes in female genital fluids. Real-time PCR-based assays combine a wide dynamic range of quantification of 7 to 8 logarithmic decades, a high technical sensitivity, a high precision, the absence of carryover contamination, lower analytical turnaround times, and substantially decreased costs (29, 39).
HIV-1 RNA was quantified by one-step real-time RT-PCR assay on a Light Cycler instrument using the primer sets and exonuclease probe in the long terminal repeat gene, previously validated for HIV quantitation in plasma by the quantification working group of the French National Agency for AIDS Research (39). We checked the accuracy of this real-time PCR assay, which was able to detect similarly HIV-1 subtypes A, B, C, D, and G as well as circulating recombinant forms 02 (AG) and 11 (which are the most prevalent HIV-1 subtypes circulating in Africa) and which showed a high correlation with expected values from international controls and those obtained with branched DNA assay. HIV-1 proviral DNA was measured by using the same set of primers and probe as those used for HIV RNA quantification. To express HIV-1 proviral load in cervicovaginal lavage samples, it was necessary to normalize HIV DNA quantification by the number of cells extracted in the cervicovaginal secretion cell pellet by the additional quantification of the albumin gene, due to the variation of the number of genital cells recovered in cervicovaginal lavage samples. The threshold of sensitivity and the intra- and interassay variations we observed for HIV-1 RNA detection and quantification by real-time RT-PCR and for HIV-1 DNA detection by real-time PCR were similar to those previously reported by Rouet and colleagues (39).
The quantification of HSV genomes by real-time PCR in genital fluid has been previously reported in a few studies (3, 16, 34, 35, 41), but no published procedure has been validated yet. We have herein validated a laboratory-developed one-step real-time PCR assay to quantify accurately HSV DNA in cervicovaginal fluid by using as standard a plasmid containing HSV DNA polymerase target. Our results showed a high correlation with VQC values (r2 > 0.99) and a slope close to 1. The use of hybridization probes allowed perfect typing of HSV DNA by the analysis of the melting curve at the end of the PCR. This genotyping is presumably not required for studies set in developing countries, where HSV-2 is the almost exclusive etiology of herpes genital ulceration, but it may be useful for studies taking place in industrialized countries, where HSV-1 is an increasing cause of genital ulceration (30). The threshold of sensitivity and the intra- and interassay variations we observed for HSV DNA detection and quantification by real-time PCR were of similar order to those observed for HIV-1 RNA and HIV-1 DNA detection and quantification by real-time PCR. In addition, we checked that spiking pools of cervicovaginal secretions with both viruses does not modify the ability to quantify each virus in mixed samples. These observations provide the rationale for the use of real-time PCR procedures to quantify with similar accuracy levels both HIV and HSV in clinical samples.
In the present series of women with genital ulceration, nearly half of the cervicovaginal secretion lavage samples found to be positive for HSV-2 DNA were negative when endocervical swabbing samples were used to detect HSV-2 DNA. Although the cervicovaginal lavage washing carried out prior to the endocervical swabbing may have removed some cervical secretions, these observations are in keeping with previous data on asymptomatic genital herpes shedding showing that the cervicovaginal washing procedure may be considered more sensitive than the endocervicovaginal swabbing procedure for detecting HSV-2 DNA in genital fluids (37).
In African women coinfected with HIV-1 and HSV-2 harboring genital secretions positive for HSV-2 DNA, the genital replication of HSV-2 was associated with a 10-fold increase of HIV-1 RNA genital shedding. These findings extend previous observations reporting on the frequent detection of HIV-1 RNA within the herpetic genital ulcer lesions in men (42) or women (26). These features are also in agreement with a previous report showing that HSV-2 DNA genital shedding was associated with higher genital shedding of HIV-1 RNA in asymptomatic HSV-2 DNA-shedding HIV-1-infected women (32) and with the results from a recent randomized clinical trial of vitamin A supplementation demonstrating that HSV shedding was associated with significantly higher vaginal and cervical HIV-1 shedding (3). The levels of HSV-2 DNA in cervicovaginal secretions were not correlated with genital HIV-1 proviral DNA, as previously reported for genital secretions among asymptomatic HSV-2 DNA-shedding women (33). Our data also confirm that HSV-2 DNA shedding did not lead to an increase in the HIV-1 RNA plasma viral load, confirming that HSV-2 genital replication does not influence HIV-1 systemic production (32). Taken together, these results provide the basis of increased genital HIV-1 infectivity during HSV-2 genital replication in women.
The analytical turnaround times were below 2 hours for each of the three assays. The use of a Light Cycler instrument allowed quantification of 29 samples per run. HIV-1 RNA, HIV-1 proviral DNA, and albumin DNA may also be quantified by a TaqMan instrument in 84 samples in a single run for large series, while HSV quantification followed by genotyping using melting curve analysis requires the use of a Light Cycler instrument. The use of real-time PCR assays dramatically decreases the cost of quantification. For each real-time PCR on a Light Cycler instrument, the cost per reaction was calculated at 6.9 euros, while the price of HIV-1 RNA quantification by branched DNA assay in our institution is roughly 38 euros. Such cost savings should be considered in the design of international clinical studies. In addition to the costs associated with viral load testing, the dynamic ranges of real-time PCR-based assays may provide additional potential cost savings. The branched DNA assay has a dynamic range of 50 to 500,000 copies/ml (15), while roughly 25% of plasma samples in our series had a higher viral load. The Light Cycler assay, with a dynamic range up to 107 copies/ml, avoids retesting diluted samples.
One major concern for a transition from commercial kits to real-time PCR is the need for an efficient automated method for the extraction of nucleic acid. Extraction procedures for nucleic acids contained in the female genital tract are also critical to guarantee accurate and reproducible results. Since a silica-based extraction procedure is likely one of the most effective to extract both DNA and RNA at the same time in corporeal fluids (23), an automated method could be useful to process a great number of samples with increased reliability. We analyzed the performance of a MagNA Pure system for the purification of both HSV DNA and HIV RNA in CVL supernatant, HIV RNA in plasma, and DNA from cervicovaginal lavage cell pellet. Both manual and automated procedures were found to be appropriate for combined extraction of HIV and HSV genomes from genital secretions. For large series of nucleic acid purifications, an automated method dramatically decreases technical time. For 32 samples, processing by manual extraction required 110 to 120 min of hands-on time, compared to only 25 to 30 min with a MagNA Pure system. The MagNA Pure cost per test was 3.38 euros compared to 2.50 euros for manual extraction by High Pure viral nucleic acid kit; however, the automated method had a better overall cost consideration by reducing technical time. Finally, the cost of HIV quantification by real-time PCR, including automated extraction and real-time PCR assay, remains 5 times cheaper than a commercial assay.
Our results indicate that real-time PCR assays, in combination with automated nucleic acid extraction, are efficient, reliable, and economic tools for HIV and HSV detection and quantification in plasma and female genital fluid and should be considered for clinical trials. The use of qualitative and quantitative quality control should also be systematically carried out in order to be able to compare results between studies.
Acknowledgments
This work was supported by the Agence Nationale de Recherches sur le SIDA et les Hépatites (ANRS 12-12) and the Institut National de la Santé et de la Recherche Médicale (INSERM), France. H.B. was supported by the Agence Nationale de Recherches sur le SIDA et les Hépatites, France.
REFERENCES
- 1.Andreoletti, L., N. Chomont, G. Gresenguet, M. Matta, J. de Dieu Longo, M. P. Carreno, A. Si-Mohamed, J. Legoff, M. D. Kazatchkine, and L. Belec. 2003. Independent levels of cell-free and cell-associated human immunodeficiency virus-1 in genital-tract secretions of clinically asymptomatic, treatment-naive African women. J. Infect. Dis. 188:549-554. [DOI] [PubMed] [Google Scholar]
- 2.Augenbraun, M., J. Feldman, K. Chirgwin, J. Zenilman, L. Clarke, J. DeHovitz, S. Landesman, and H. Minkoff. 1995. Increased genital shedding of herpes simplex virus type 2 in HIV-seropositive women. Ann. Intern. Med. 123:845-847. [DOI] [PubMed] [Google Scholar]
- 3.Baeten, J. M., R. S. McClelland, L. Corey, J. Overbaugh, L. Lavreys, B. A. Richardson, A. Wald, K. Mandaliya, J. J. Bwayo, and J. K. Kreiss. 2004. Vitamin A supplementation and genital shedding of herpes simplex virus among HIV-1-infected women: a randomized clinical trial. J. Infect. Dis. 189:1466-1471. [DOI] [PubMed] [Google Scholar]
- 4.Belec, L., D. Meillet, M. Levy, A. Georges, C. Tevi-Benissan, and J. Pillot. 1995. Dilution assessment of cervicovaginal secretions obtained by vaginal washing for immunological assays. Clin. Diagn. Lab. Immunol. 2:57-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1:307-310. [PubMed] [Google Scholar]
- 6.Burrows, J., A. Nitsche, B. Bayly, E. Walker, G. Higgins, and T. Kok. 2002. Detection and subtyping of Herpes simplex virus in clinical samples by LightCycler PCR, enzyme immunoassay and cell culture. BMC Microbiol. 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celum, C. L. 2004. The interaction between herpes simplex virus and human immunodeficiency virus. Herpes 11(Suppl. 1):36A-45A. [PubMed] [Google Scholar]
- 8.Chomont, N., G. Gresenguet, M. Levy, H. Hocini, P. Becquart, M. Matta, J. Tranchot-Diallo, L. Andreoletti, M. P. Carreno, M. D. Kazatchkine, and L. Belec. 2001. Detection of Y chromosome DNA as evidence of semen in cervicovaginal secretions of sexually active women. Clin. Diagn. Lab. Immunol. 8:955-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coombs, R. W., P. S. Reichelderfer, and A. L. Landay. 2003. Recent observations on HIV type-1 infection in the genital tract of men and women. AIDS 17:455-480. [DOI] [PubMed] [Google Scholar]
- 10.Corey, L., and H. H. Handsfield. 2000. Genital herpes and public health: addressing a global problem. JAMA 283:791-794. [DOI] [PubMed] [Google Scholar]
- 11.Damond, F., D. Descamps, I. Farfara, J. N. Telles, S. Puyeo, P. Campa, A. Lepretre, S. Matheron, F. Brun-Vezinet, and F. Simon. 2001. Quantification of proviral load of human immunodeficiency virus type 2 subtypes A and B using real-time PCR. J. Clin. Microbiol. 39:4264-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desire, N., A. Dehee, V. Schneider, C. Jacomet, C. Goujon, P. M. Girard, W. Rozenbaum, and J. C. Nicolas. 2001. Quantification of human immunodeficiency virus type 1 proviral load by a TaqMan real-time PCR assay. J. Clin. Microbiol. 39:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeVange Panteleeff, D., S. Emery, B. A. Richardson, C. Rousseau, S. Benki, S. Bodrug, J. K. Kreiss, and J. Overbaugh. 2002. Validation of performance of the Gen-Probe human immunodeficiency virus type 1 viral load assay with genital swabs and breast milk samples. J. Clin. Microbiol. 40:3929-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elbeik, T., W. G. Alvord, R. Trichavaroj, M. de Souza, R. Dewar, A. Brown, D. Chernoff, N. L. Michael, P. Nassos, K. Hadley, and V. L. Ng. 2002. Comparative analysis of HIV-1 viral load assays on subtype quantification: Bayer Versant HIV-1 RNA 3.0 versus Roche Amplicor HIV-1 Monitor version 1.5. J. Acquir. Immune Defic. Syndr. 29:330-339. [DOI] [PubMed] [Google Scholar]
- 15.Erice, A., D. Brambilla, J. Bremer, J. B. Jackson, R. Kokka, B. Yen-Lieberman, and R. W. Coombs. 2000. Performance characteristics of the QUANTIPLEX HIV-1 RNA 3.0 assay for detection and quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 38:2837-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filen, F., A. Strand, A. Allard, J. Blomberg, and B. Herrmann. 2004. Duplex real-time polymerase chain reaction assay for detection and quantification of herpes simplex virus type 1 and herpes simplex virus type 2 in genital and cutaneous lesions. Sex. Transm. Dis. 31:331-336. [DOI] [PubMed] [Google Scholar]
- 17.Fischetti, L., O. Opare-Sem, D. Candotti, F. Sarkodie, H. Lee, and J. P. Allain. 2004. Molecular epidemiology of HIV in Ghana: dominance of CRF02_AG. J. Med. Virol. 73:158-166. [DOI] [PubMed] [Google Scholar]
- 18.Hart, C. E., J. L. Lennox, M. Pratt-Palmore, T. C. Wright, R. F. Schinazi, T. Evans-Strickfaden, T. J. Bush, C. Schnell, L. J. Conley, K. A. Clancy, and T. V. Ellerbrock. 1999. Correlation of human immunodeficiency virus type 1 RNA levels in blood and the female genital tract. J. Infect. Dis. 179:871-882. [DOI] [PubMed] [Google Scholar]
- 19.Hatzakis, A. E., G. Touloumi, N. Pantazis, C. G. Anastassopoulou, O. Katsarou, A. Karafoulidou, J. J. Goedert, and L. G. Kostrikis. 2004. Cellular HIV-1 DNA load predicts HIV-RNA rebound and the outcome of highly active antiretroviral therapy. AIDS 18:2261-2267. [DOI] [PubMed] [Google Scholar]
- 20.Hobson, A., A. Wald, N. Wright, and L. Corey. 1997. Evaluation of a quantitative competitive PCR assay for measuring herpes simplex virus DNA content in genital tract secretions. J. Clin. Microbiol. 35:548-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izopet, J., G. Salama, C. Pasquier, K. Sandres, B. Marchou, P. Massip, and J. Puel. 1998. Decay of HIV-1 DNA in patients receiving suppressive antiretroviral therapy. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:478-483. [DOI] [PubMed] [Google Scholar]
- 22.Kabamba-Mukadi, B., P. Henrivaux, J. Ruelle, N. Delferriere, M. Bodeus, and P. Goubau. 2005. Human immunodeficiency virus type 1 (HIV-1) proviral DNA load in purified CD4+ cells by LightCycler real-time PCR. BMC Infect. Dis. 5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kok, T., S. Wati, B. Bayly, D. Devonshire-Gill, and G. Higgins. 2000. Comparison of six nucleic acid extraction methods for detection of viral DNA or RNA sequences in four different non-serum specimen types. J. Clin. Virol. 16:59-63. [DOI] [PubMed] [Google Scholar]
- 24.Korber, B., B. Hahn, B. Foley, J. W. Mellors, T. Leitner, G. Myers, F. McCutchan, and C. Kuiken (ed.). 1997. HIV sequence compendium: human retroviruses and AIDS, 1997. Theoretical Biology and Biophysics Group T-10, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 25.Kovacs, A., L. S. Chan, Z. C. Chen, W. A. Meyer III, L. Muderspach, M. Young, K. Anastos, and A. M. Levine. 1999. HIV-1 RNA in plasma and genital tract secretions in women infected with HIV-1. J. Acquir. Immune Defic. Syndr. 22:124-131. [DOI] [PubMed] [Google Scholar]
- 26.Kreiss, J. K., R. Coombs, F. Plummer, K. K. Holmes, B. Nikora, W. Cameron, E. Ngugi, J. O. Ndinya Achola, and L. Corey. 1989. Isolation of human immunodeficiency virus from genital ulcers in Nairobi prostitutes. J. Infect. Dis. 160:380-384. [DOI] [PubMed] [Google Scholar]
- 27.Larder, B., and C. Boucher. 1993. PCR detection of human immunodeficiency virus drug resistance mutations, p. 527-533. In D. H. Persing (ed.), Diagnostic molecular biology: principles and applications, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 28.Laurendeau, I., M. Bahuau, N. Vodovar, C. Larramendy, M. Olivi, I. Bieche, M. Vidaud, and D. Vidaud. 1999. TaqMan PCR-based gene dosage assay for predictive testing in individuals from a cancer family with INK4 locus haploinsufficiency. Clin. Chem. 45:982-986. [PubMed] [Google Scholar]
- 29.Mackay, I. M., K. E. Arden, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malkin, J. E. 2004. Epidemiology of genital herpes simplex virus infection in developed countries. Herpes 11(Suppl. 1):2A-23A. [PubMed] [Google Scholar]
- 31.Masquelier, B., E. Race, C. Tamalet, D. Descamps, J. Izopet, C. Buffet-Janvresse, A. Ruffault, A. S. Mohammed, J. Cottalorda, A. Schmuck, V. Calvez, E. Dam, H. Fleury, and F. Brun-Vezinet. 2001. Genotypic and phenotypic resistance patterns of human immunodeficiency virus type 1 variants with insertions or deletions in the reverse transcriptase (RT): multicenter study of patients treated with RT inhibitors. Antimicrob. Agents Chemother. 45:1836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mbopi-Keou, F. X., G. Gresenguet, P. Mayaud, H. A. Weiss, R. Gopal, M. Matta, J. L. Paul, D. W. Brown, R. J. Hayes, D. C. Mabey, and L. Belec. 2000. Interactions between herpes simplex virus type 2 and human immunodeficiency virus type 1 infection in African women: opportunities for intervention. J. Infect. Dis. 182:1090-1096. [DOI] [PubMed] [Google Scholar]
- 33.Mbopi-Keou, F. X., J. Legoff, G. Gresenguet, A. Si-Mohamed, M. Matta, P. Mayaud, L. Andreoletti, J. E. Malkin, H. Weiss, D. Brown, and L. Belec. 2003. Genital shedding of herpes simplex virus-2 DNA and HIV-1 RNA and proviral DNA in HIV-1- and herpes simplex virus-2-coinfected African women. J. Acquir. Immune Defic. Syndr. 33:121-124. [DOI] [PubMed] [Google Scholar]
- 34.Mbopi-Keou, F. X., G. Gresenguet, P. Mayaud, H. A. Weiss, R. Gopal, D. W. Brown, R. J. Hayes, D. C. Mabey, and L. Belec. 1999. Genital herpes simplex virus type 2 shedding is increased in HIV-infected women in Africa. AIDS 13:536-537. [DOI] [PubMed] [Google Scholar]
- 35.McClelland, R. S., C. C. Wang, J. Overbaugh, B. A. Richardson, L. Corey, R. L. Ashley, K. Mandaliya, J. Ndinya-Achola, J. J. Bwayo, and J. K. Kreiss. 2002. Association between cervical shedding of herpes simplex virus and HIV-1. AIDS 16:2425-2430. [DOI] [PubMed] [Google Scholar]
- 36.Montavon, C., L. Vergne, A. Bourgeois, E. Mpoudi-Ngole, G. Malonga-Mouellet, C. Butel, C. Toure-Kane, E. Delaporte, and M. Peeters. 2002. Identification of a new circulating recombinant form of HIV type 1, CRF11-cpx, involving subtypes A, G, J, and CRF01-AE, in Central Africa. AIDS Res. Hum. Retrovir. 18:231-236. [DOI] [PubMed] [Google Scholar]
- 37.Ndjoyi-Mbiguino, A., F. Ozouaki, J. Legoff, F. X. Mbopi-Keou, A. Si-Mohamed, I. N. Onas, E. Avoune, and L. Belec. 2003. Comparison of washing and swabbing procedures for collecting genital fluids to assess cervicovaginal shedding of herpes simplex virus type 2 DNA. J. Clin. Microbiol. 41:2662-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer, S., A. P. Wiegand, F. Maldarelli, H. Bazmi, J. M. Mican, M. Polis, R. L. Dewar, A. Planta, S. Liu, J. A. Metcalf, J. W. Mellors, and J. M. Coffin. 2003. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 41:4531-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouet, F., D. K. Ekouevi, M. L. Chaix, M. Burgard, A. Inwoley, T. D. Tony, C. Danel, X. Anglaret, V. Leroy, P. Msellati, F. Dabis, and C. Rouzioux. 2005. Transfer and evaluation of an automated, low-cost real-time reverse transcription-PCR test for diagnosis and monitoring of human immunodeficiency virus type 1 Infection in a West African resource-limited setting. J. Clin. Microbiol. 43:2709-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruelle, J., B. K. Mukadi, M. Schutten, and P. Goubau. 2004. Quantitative real-time PCR on Lightcycler for the detection of human immunodeficiency virus type 2 (HIV-2). J. Virol. Methods 117:67-74. [DOI] [PubMed] [Google Scholar]
- 41.Ryncarz, A. J., J. Goddard, A. Wald, M. L. Huang, B. Roizman, and L. Corey. 1999. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J. Clin. Microbiol. 37:1941-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schacker, T., A. J. Ryncarz, J. Goddard, K. Diem, M. Shaughnessy, and L. Corey. 1998. Frequent recovery of HIV-1 from genital herpes simplex virus lesions in HIV-1-infected men. JAMA 280:61-66. [DOI] [PubMed] [Google Scholar]
- 43.Si-Mohamed, A., L. Andreoletti, I. Colombet, M.-P. Carreno, G. Lopez, G. Chatelier, M. D. Kazatchkine, and L. Belec. 2001. Quantitation of human immunodeficiency virus type 1 (HIV-1) RNA in cell-free cervicovaginal secretions: comparison of reverse transcription-PCR amplification (AMPLICOR HIV-1 MONITOR 1.5) with enhanced-sensitivity branched-DNA assay (Quantiplex 3.0). J. Clin. Microbiol. 39:2055-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spinillo, A., F. Zara, B. Gardella, E. Preti, R. Mainini, and R. Maserati. 2005. The effect of vaginal candidiasis on the shedding of human immunodeficiency virus in cervicovaginal secretions. Am. J. Obstet. Gynecol. 192:774-779. [DOI] [PubMed] [Google Scholar]
- 45.Swanson, P., V. Soriano, S. G. Devare, and J. Hackett, Jr. 2001. Comparative performance of three viral load assays on human immunodeficiency virus type 1 (HIV-1) isolates representing group M (subtypes A to G) and group O: LCx HIV RNA quantitative, AMPLICOR HIV-1 MONITOR version 1.5, and Quantiplex HIV-1 RNA version 3.0. J. Clin. Microbiol. 39:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trouplin, V., F. Salvatori, F. Cappello, V. Obry, A. Brelot, N. Heveker, M. Alizon, G. Scarlatti, F. Clavel, and F. Mammano. 2001. Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J. Virol. 75:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viard, J. P., M. Burgard, J. B. Hubert, L. Aaron, C. Rabian, N. Pertuiset, M. Lourenco, C. Rothschild, and C. Rouzioux. 2004. Impact of 5 years of maximally successful highly active antiretroviral therapy on CD4 cell count and HIV-1 DNA level. AIDS 18:45-49. [DOI] [PubMed] [Google Scholar]
- 48.Wald, A., and K. Link. 2002. Risk of human immunodeficiency virus infection in herpes simplex virus type 2-seropositive persons: a meta-analysis. J. Infect. Dis. 185:45-52. [DOI] [PubMed] [Google Scholar]
- 49.Weiss, H. 2004. Epidemiology of herpes simplex virus type 2 infection in the developing world. Herpes 11(Suppl. 1):24A-35A. [PubMed] [Google Scholar]
- 50.World Health Organization and UNAIDS. 2001. Herpes simplex virus type 2. Programmatic and research priorities in developing countries. World Health Organization, Geneva, Switzerland.