Abstract
Tubulins, the protein constituents of the microtubule cytoskeleton, are present in all known eukaryotes but have never been found in the Bacteria or Archaea. Here we report the presence of two tubulin-like genes [bacterial tubulin a (btuba) and bacterial tubulin b (btubb)] in bacteria of the genus Prosthecobacter (Division Verrucomicrobia). In this study, we investigated the organization and expression of these genes and conducted a comparative analysis of the bacterial and eukaryotic protein sequences, focusing on their phylogeny and 3D structures. The btuba and btubb genes are arranged as adjacent loci within the genome along with a kinesin light chain gene homolog. RT-PCR experiments indicate that these three genes are cotranscribed, and a probable promoter was identified upstream of btuba. On the basis of comparative modeling data, we predict that the Prosthecobacter tubulins are monomeric, unlike eukaryotic α and β tubulins, which form dimers and are therefore unlikely to form microtubule-like structures. Phylogenetic analyses indicate that the Prosthecobacter tubulins are quite divergent and do not support recent horizontal transfer of the genes from a eukaryote. The discovery of genes for tubulin in a bacterial genus may offer new insights into the evolution of the cytoskeleton.
It is evident that at some point in their evolution, the Eucarya acquired a structural complexity unrivaled by members of the other two domains of life. One of the major structural features that separates the Eucarya from the Bacteria and the Archaea is the presence of an internal cytoskeleton composed of actin and tubulin. Notably, these cytoskeletal elements are present in all known eukaryotes, even the a-mitochondriate protozoa (1, 2). Furthermore, their acquisition represented an important step in the evolution of eukaryotic cells by facilitating the engulfment of bacterial endosymbionts, which later became chloroplasts and mitochondria (3).
In contrast, there have been no conclusive reports of these cytoskeletal elements in the bacterial or archaeal domains. Over the years, there have been numerous reports of “microtubule-like” structures or “rhapidosomes” in members of both the Bacteria and the Archaea (summarized in ref. 4); however, thus far these observations lack any genetic basis. At present, the leading candidate for an evolutionary precursor of tubulin in the bacterial/archaeal domains is the cell division protein, FtsZ. Although there is strong evidence from their 3D structures that tubulin and FtsZ are homologous proteins (5, 6), they share only very low sequence identity, most of which is confined to the GTP-binding region (7). The strikingly low sequence identity is difficult to reconcile with the fact that tubulins and FtsZs are among the slowest-evolving proteins known and raises the question of whether any more closely related homologs of tubulin exist in members of the Bacteria or Archaea (8, 9).
Reports of microtubule-like structures in bacterial ectosymbionts (“epixenosomes”) of ciliates in the genus Euplotidium present the most compelling structural evidence yet for the existence of tubulin-containing elements in bacteria. These organisms, which belong to the little-studied division, Verrucomicrobia, have been shown to possess tubular structures with diameters of 22 ± 3 nm, the size range of eukaryotic microtubules. These structures crossreact with anti-Paramecium tubulin antibodies and display sensitivity to microtubule-depolymerizing agents (10, 11). On the basis of these observations, we searched the partially sequenced genome of a free-living member of the Verrucomicrobia, Prosthecobacter dejongeii, for genes homologous to those for tubulin. To our knowledge, P. dejongeii is the first member of the division Verrucomicrobia to be subjected to genome-sequencing studies.
Materials and Methods
Bacterial Strains and Culture.
Cultures of P. dejongeii [American Type Culture Collection (ATCC) 27091], Prosthecobacter vanneervenii (ATCC 700199), Prosthecobacter debontii (ATCC 700200), and Prosthecobacter fusiformis (ATCC 25309) were grown aerobically at 28°C in modified medium B broth (12).
DNA Extraction.
DNA was extracted from all four Prosthecobacter strains by using the Instagene matrix (Bio-Rad).
Amplification of btuba and btubb.
The btuba and btubb genes were amplified from the three remaining species of Prosthecobacter via PCR. PCR was optimized by using the PCR Optimizer kit (Invitrogen). Specific PCR primers (5′–3′) were as follows: for btuba, ACGGTTTGCCTTGAGCATGG (forward) and CATGCCTTCGTTGAGATACCA (reverse); and for btubb, AGCATTCACGTCGGACAGTG (forward) and TCATCCACATCCTTCGCCTTC (reverse). PCRs were carried out in a 25-μl volume with one cycle at 96°C, 2 min; 35 cycles of denaturation at 96°C, 30 sec; annealing at 58°C, 30 sec; extension at 72°C, 1 min and one cycle at 72°C, 8 min. Gene walking was performed as described in ref. 13 by using the specific nested primers (5′–3′): ACAGGAAGGGCAGGTGCGGACG and CCAGACCTGGCTCGTGAGACC.
Sequence Analysis.
Gene searches within the P. dejongeii genome were conducted by using the similarity search (blast) function of the ERGO database (http://wit.IntegratedGenomics.com/IGwit). Protein sequences were analyzed for signature motifs by using the program fingerprintscan (www.bioinf.man.ac.uk/fingerPRINTScan). The Neural Network Promoter Prediction software (www.fruitfly.org/seq_tools/promoter.html) was used to identify probable promoter sequences. genedoc (14) was used to align protein sequences and calculate identities.
RNA Extraction and RT-PCR.
RNA was extracted from 500 ml of P. dejongeii culture. Cells were pelleted, washed, and treated with lysozyme and 10% Triton X-100. Cell lysis with achieved with guanidine isothiocyanate and SDS. The cell lysate was loaded onto a cesium chloride cushion and ultracentrifuged for 18 h at 35,000 rpm by using a Beckman L8-M ultracentrifuge with an SW55Ti rotor. RNA was washed with 70% ethanol, resuspended in RNase-free water, and treated with DNase. RT-PCR was performed by using the ProStar HF Single-Tube RT-PCR System (Stratagene).
Comparative Modeling.
Modeling of BtubA and BtubB used the ramp software suite of programs (http://compbio.washington.edu; http://protinfo.compbio.washington.edu). The Protein Data Bank structures -A and -B were used as the templates to construct the initial models by using a minimum perturbation approach that aims to preserve as much information as possible from the template x-ray structure. Variable side chains and main chains were constructed by using a graph-theory clique-finding approach, which explores a variety of possible conformations for the respective side chains and main chains and finds the optimal combination by using an all-atom scoring function. These approaches are described in further detail elsewhere (15, 16).
Phylogenetic Analyses.
Trees were constructed by using aligned protein sequences via parsimony, distance [ProtPars and ProtDist with Neighbor in phylip Ver. 3.6 (17)] and maximum likelihood (protml Ver. 2.3b3; http://bioweb.pasteur.fr/seqanal/interfaces/prot_nucml.html) methods. For parsimony trees, branch lengths were recalculated by using protdist and fitch in phylip. Each analysis used 1,000 bootstrap replications. For all other variables, the default options were used. Trees were either rooted at the midpoint or outgroup rooted with ζ tubulin.
Results and Discussion
Sequence Comparisons.
Two genes with corresponding protein sequences closely matching eukaryotic tubulins were retrieved from the genome sequence of P. dejongeii. Henceforth, these genes will be referred to as btuba (for bacterial tubulin A) and btubb (for bacterial tubulin B). The deduced protein sequences will be referred to as BtubA and BtubB. blast analyses, the calculation of sequence identities and protein sequence comparisons, were carried out to determine the relationship of the P. dejongeii BtubA and BtubB sequences relative to the other members of the tubulin family. BtubA and BtubB had their top 60–100 blast matches with eukaryotic α and β tubulins respectively and were shown to share 31–35% and 34–37% sequence identity with these proteins. In contrast, BtubA and BtubB displayed <25% identity with eukaryotic γ tubulin, <20% with eukaryotic δ and ɛ tubulins, <12% with ζ tubulin, and only 8–11% identity with FtsZ sequences. These identity values indicate that BtubA and BtubB share a specific relationship with eukaryotic α and β of all of the members of the tubulin family. Interestingly, BtubA and BtubB were not found to have a close relationship with the bacterial/archaeal protein FtsZ, which until now was the only member of the tubulin family found in noneukaryotes.
To further investigate these relationships, BtubA and BtubB were analyzed for the presence of conserved protein motifs specific to tubulins. BtubA and BtubB were shown to contain each of the nine motifs specific to tubulin but only two to three of the six FtsZ signatures, again indicating a closer relationship to tubulins than FtsZs (Table 1). As shown by the P values, the BtubA and BtubB sequences most closely matched the motif fingerprints of eukaryotic α and β tubulin, respectively. The results are consistent with the blast analyses; however, the missing signature motifs indicate that BtubA and BtubB are quite divergent relative to eukaryotic α and β tubulins.
Table 1.
Tubulin and FtsZ motifs identified in P. dejongeii BtubA and BtubB sequences
| Tubulin subgroup | No. of motifs | P value |
|---|---|---|
| BtubA | ||
| Tubulin | 9 of 9 | 1.4 × 10−62 |
| α tubulin | 7 of 13 | 5.9 × 10−17 |
| ɛ tubulin | 4 of 10 | 1.2 × 10−15 |
| β tubulin | 6 of 13 | 1.4 × 10−14 |
| δ tubulin | 3 of 12 | 1.9 × 1012 |
| γ tubulin | 5 of 8 | 3.3 × 10−11 |
| FtsZ | 2 of 6 | 8.3 × 10−6 |
| BtubB | ||
| Tubulin | 9 of 9 | 1.9 × 10−72 |
| β tubulin | 8 of 13 | 4.2 × 10−26 |
| ɛ tubulin | 6 of 10 | 3.3 × 10−19 |
| α tubulin | 5 of 13 | 2.5 × 10−14 |
| γ tubulin | 4 of 8 | 1.4 × 10−13 |
| FtsZ | 3 of 6 | 1.7 × 10−10 |
| δ tubulin | 2 of 12 | 5.6 × 10−9 |
Probability values based on scoring matches to the motifs.
The discovery of tubulin genes in members of the Prosthecobacter genus is significant, because these cytoskeletal elements were previously thought to be confined to the Eucarya. Although there have been many reports of microtubule-like structures in members of the Bacteria and Archaea (4), genetic evidence for these reports is lacking. Thus, one of the major implications of this study is that the notion that tubulins exist in bacteria now has a molecular basis.
Tubulin Gene Homologs in Prosthecobacter.
Although the P. dejongeii genome is sequenced to only 90% completion, it is notable that btuba and btubb were the only tubulin homologs located in the sequence. In eukaryotes, α and β tubulin are the only tubulins required for microtubule formation. Other members of the tubulin family perform auxiliary functions but are not necessary for microtubule polymerization. Thus, it was hypothesized that the presence of a single α (btuba) and a single β (btubb) tubulin gene homolog in Prosthecobacter may be sufficient for the formation of microtubule-like structures in these organisms. Furthermore, the presence of microtubule-like structures in epixenosome Verrucomicrobia (10, 11) led us to believe that similar structures might be found in Prosthecobacter.
After the discovery of α and β tubulin gene homologs in Prosthecobacter, two major questions were addressed: (i) Are microtubule-like structures being formed in cells of P. dejongeii? and, (ii) What is the evolutionary origin of these genes? We approached the first question by investigating the expression of btuba and btubb, examining thin sections of P. dejongeii for microtubule-like structures and assessing the polymerization potential of BtubA and BtubB. The second question was addressed via phylogenetic analysis of BtubA and BtubB.
Gene Expression and Organization.
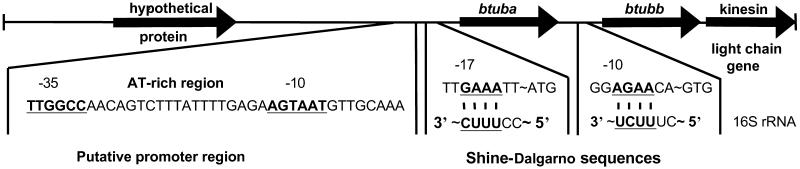
The organization and expression of the P. dejongeii tubulin genes was investigated via gene walking and RT-PCR. btuba and btubb were found to exist as adjacent loci on the genome sequence with the btuba sequence located upstream of btubb (Fig. 1). This organization is similar to that seen for many eukaryotic α and β tubulins, whereby the genes are arranged within the genome as pairs or clusters (18). Interestingly, gene walking revealed a third ORF downstream of btubb (Fig. 1), which was shown by blast analysis to be homologous to the light chain of kinesin (E value: 1 × 10−11), kinesin being the microtubule motor protein of eukaryotes. RT-PCR results indicated that btuba and btubb are expressed (Fig. 2) and are cotranscribed with the kinesin light chain homolog (data not shown), indicating that these three genes form an operon. A probable promoter sequence for the operon was identified upstream of btuba, with the consensus sequences AGTAAT (−10) and TTGGCC (−35).
Fig 1.
Organization of the P. dejongeii tubulin genes in the genome. A probable promoter sequence upstream of btuba is marked. Consensus sequences and Shine–Dalgarno sequences are underscored.
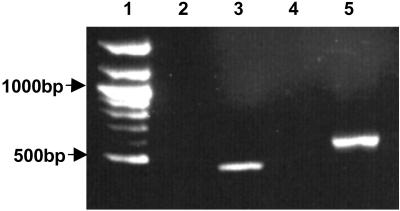
Fig 2.
Agarose gel showing RT-PCR products with and without the addition of RT. Lane 1, 100-bp ladder (New England Biolabs); lane 2, BtubA primers − RT; lane 3, BtubA primers + RT; lane 4, BtubB primers − RT; lane 5, BtubB primers + RT.
Because BtubA and BtubB did not crossreact with commercially available antibovine tubulin antibodies, we were unable to test directly for the presence of these proteins in Prosthecobacter. However, the presence of Shine–Dalgarno sequences upstream of the btuba and btubb start codons (Fig. 1) indicates that the tubulin RNAs may be translated into protein. Evidence for the expression of btuba and btubb and the presence and coexpression of a kinesin light chain gene homolog was consistent with the idea that these genes may play a role in the formation of microtubule-like structures.
Electron Microscopy.
To determine whether the btuba and btubb genes are involved in the formation of microtubule-like structures, we examined thin sections of P. dejongeii by using techniques described for epixenosomes (11). Despite extensive examination of cell sections, we were unable to locate any such structures (data not shown). Although this suggested that microtubule-like structures might not be present in Prosthecobacter, the visualization of these structures depends on numerous factors, including the availability of favorable sections and the fixation technique used. Therefore, these results were not regarded as definitive evidence for the absence of microtubule-like structures in Prosthecobacter.
Comparative Modeling.
Eukaryotic α and β tubulins are known to interact to form dimers, which then polymerize during microtubule formation. We directly addressed the question of whether BtubA and BtubB would be predicted to interact in a similar manner to α and β tubulin via comparative modeling of these protein sequences.
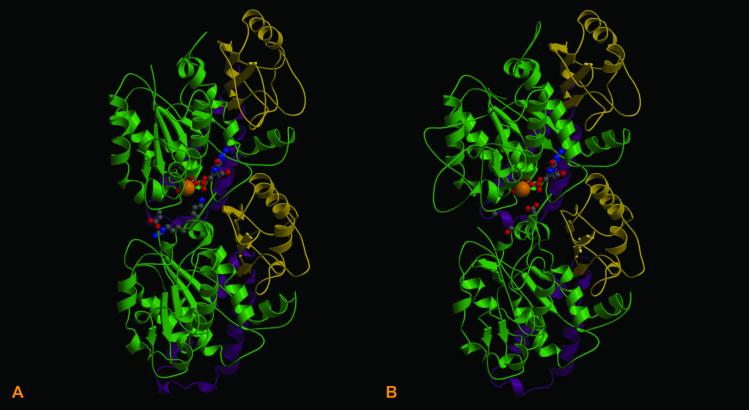
To ascertain the polymerization potential of BtubA and BtubB, the 3D structures of these proteins were reconstructed by using the refined crystal structure of the bovine αβ tubulin dimer (19) as a template. Similarities between the Prosthecobacter models and the eukaryotic tubulin structures include the presence of a hydrophobic loop formed by two C-terminal helices, which in eukaryotic protofilaments are involved in contacting adjacent tubulin monomers, and the presence of a Rossmann-fold characteristic of GTP-binding proteins (5) (Fig. 3 A and B).
Fig 3.
Comparison of the refined crystal structure of the bovine αβ tubulin dimer (A) with the modeled P. dejongeii BtubA/BtubB structures (B). Each structure indicates the position of GTP and a Mg2+ ion at the intradimer active site (N-site). The hydrophobic C-terminal loop and helices are marked in magenta. Rossmann folds are marked in green.
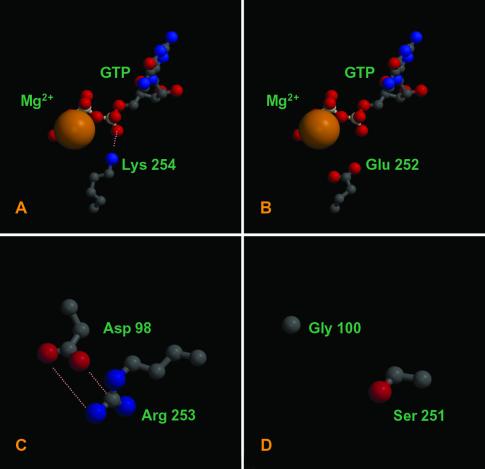
In contrast, at the intradimer interface several important differences between the eukaryote and P. dejongeii structures were observed. In eukaryotic tubulins, the exposed negative charges of the GTP molecule at the intradimer interface (N-site) are neutralized by a magnesium ion and a highly conserved lysine residue in the β subunit (K254) (19) (Fig. 4A). In BtubB, a glutamic acid residue substitutes lysine at the corresponding alignment position (E252). Rather than satisfying the negative charge of the N-site GTP γ-phosphate group, the glutamic acid residue adds a further negative charge to the active site (Fig. 4B). Furthermore, a second substitution in BtubB (S251 instead of R253) combined with a substitution in BtubA (G100 instead of D98) results in the loss of a salt bridge between the monomers (20) (Fig. 4 C and D). Importantly, these substitutions were found in the tubulin sequences of all four Prosthecobacter species but not in any eukaryotic tubulin sequences (20).
Fig 4.
Several notable changes are present at the P. dejongeii tubulin intradimer interface. Lysine residue 254, which neutralizes the N-site GTP phosphate groups (A) is highly conserved in eukaryotic β tubulin but is substituted for glutamic acid in BtubB (B). A key salt bridge at the interface of eukaryotic tubulins (C) is absent in the P. dejongeii tubulin dimer (D). BtubA and BtubB are >35% identical to bovine α and β tubulin. Models ranging from 1.5 to 2.5 Å rms deviation for this degree of sequence relationship are routinely produced using these methods (15, 16); thus, our models can be considered highly reliable. Figures were generated with MOLSCRIPT (21) and RASTER 3D (22).
To determine whether these variations in the Prosthecobacter tubulin structures would be predicted to have a negative effect on dimerization, the conditional probability score (23) of each model was calculated before and after introducing theoretical mutations at the substituted sites. The conditional probability score indicates the probability of the model being correct given a set of interatomic contacts (23).
For eukaryotic tubulin, the probability score for the dimer is more favorable (lower value) than the sum of the scores of the individual monomers indicating that they form a stable structure. In contrast, before the amino acid substitutions were introduced, the Prosthecobacter dimer has a less favorable score than the sum of the monomer scores, indicating that dimerization is not favored (Table 2).
Table 2.
Conditional probability scores of the bovine αβ dimer and the modeled Prosthecobacter BtubA/BtubB dimer
| Structure | Conditional probability score |
|---|---|
| Eukaryotic tubulin | |
| α tubulin (monomer) | −10,296 |
| β tubulin (monomer) | −11,132 |
| αβ tubulin (dimer) | −21,756 |
| Prosthecobacter tubulin | |
| BtubA (monomer) | −9,970 |
| BtubB (monomer) | −10,884 |
| BtubA/BtubB (dimer) | −20,139 |
| Prosthecobacter tubulins with computational substitutions | |
| BtubA (G100D)/BtubB dimer | −20,206 |
| BtubA/BtubB (S251R) dimer | −20,158 |
| BtubA (G100D)/BtubB (S251R) dimer | −20,264 |
| BtubA/BtubB (E252K) dimer | −20,118 |
Scores are for the refined crystal structures presented in ref. 19.
The computational mutations introduced into the Prosthecobacter sequence correspond to the eukaryotic residues at the same alignment position: in BtubA, G100D, and in BtubB, S251R and E252K. In the case of the G100D and S251R mutations, corresponding to the two residues involved in formation of a salt bridge in eukaryotic tubulin (20), the conditional probability score of the mutated dimers was significantly more favorable than in the unmutated structures (Table 2). These scores indicate that the BtubA/BtubB dimer is significantly less stable due to the absence of the salt bridge at the intradimer interface. Substituting the BtubB residue E252K at the interface of the active site appears to have a slight destabilizing effect on the BtubA/BtubB dimer. We predict that this mutation would confer a stabilizing effect once GTP is bound, due to neutralization of the negative charges by the lysine residue (20). That the residue neutralizing GTP in eukaryotic tubulin is substituted in Prosthecobacter tubulins, combined with the absence of a key salt bridge at the BtubA/BtubB interface, suggests that the dimer is not the native form of the Prosthecobacter tubulins. Although the hydrophobic site of interaction formed by the C-terminal loops is conserved, the conditional probability scores indicate that this alone is insufficient to stabilize the interface.
That BtubA and BtubB are not predicted to dimerize suggests they do not play a role in the formation of microtubule-like structures. These results are consistent with the electron microscopy data, which did not demonstrate the presence of these structures in Prosthecobacter. These findings leave open the question of the function of the btuba and btubb genes in Prosthecobacter. This question can be more effectively addressed via mutational analyses. Genetic tools are not currently available for any members of the Verrucomicrobia, therefore this would be a worthwhile direction for future studies on bacterial tubulins.
Phylogeny of btuba and btubb.
To assess the distribution of the btuba and btubb genes within the genus Prosthecobacter, specific PCR primers were used to amplify the gene sequences from the three remaining Prosthecobacter species, P. vanneervenii, P. debontii, and P. fusiformis. All sequences have been submitted to GenBank.
The Prosthecobacter BtubA and BtubB sequences were aligned with sequences from each tubulin subgroup as well as several FtsZs. Phylogenetic assessment showed that the BtubA sequences from the four Prosthecobacter species form a monophyletic group, as do the BtubB sequences (Fig. 5A). Trees constructed using the tubulin/FtsZ alignments demonstrated that the Prosthecobacter proteins clustered with the tubulins and not with the FtsZ sequences (data not shown). In most trees constructed by using the Prosthecobacter tubulins and members of each eukaryotic tubulin subgroup, BtubA was shown to branch at the base of the α tubulin cluster, although this relationship was less robust when all of the tubulin subgroups were analyzed simultaneously. The branching pattern of the BtubB sequences was less stable and depended on which tubulin subgroups were included in the analysis. Trees constructed by using only α and β tubulins showed the BtubB sequences to cluster at the base of the β tubulin subgroup (Fig. 5A). However, the inclusion of the other tubulin subgroups (γ, δ, ɛ, and ζ) caused the BtubB sequences to fall out of the β subgroup and form a separate cluster, as in Fig. 5B. Sometimes the BtubB sequences clustered at the base of one of the other tubulin subgroups (data not shown).
Fig 5.
Phylogenetic trees showing the relationship of BtubA and BtubB relative to eukaryotic α and β tubulins (A) and other members of the tubulin family (B). Trees presented are parsimony trees rooted at the midpoint in A and outgroup rooted with ζ tubulin in B. Circles indicate bootstrap values. Nodes supported at >75% in the majority of analyses are indicated by the filled circles. Nodes supported at 50–74% in most analyses are indicated by the open circles. Unsupported nodes (<50%) have no circle. (Bar = 0.1 substitutions per site.)
Unfortunately, these phylogenies cannot be considered reliable given the variations in branch position of these sequences. The instability in branch position is not surprising given the level of sequence divergence of the bacterial tubulins and the tendency for long branch groups to be attracted toward more distant outgroups (24). It is interesting to note, however, that at no stage was a relationship between the bacterial tubulins and those from a specific eukaryote lineage observed.
Evolutionary Origin of Prosthecobacter Tubulin Genes.
A significant question raised by this study relates to the evolutionary origin of the Prosthecobacter tubulin genes and may be summarized as two main hypotheses. First, the genes arose via a horizontal gene transfer from a eukaryote, and second, that the bacterial tubulins are ancestral to eukaryotic tubulins.
Relationships between the Prosthecobacter tubulins and a specific eukaryotic lineage, which would implicate a recent gene transfer, were never observed regardless of the sequence representatives, alignment subset, or mode of analysis used. Furthermore, btuba and btubb genes are present in all four species of the Prosthecobacter genus, suggesting that the genes were acquired before the divergence of this lineage. Thus, if the Prosthecobacter tubulin genes arose via horizontal transfer from a eukaryote, it was not during the recent history of the lineage.
The second hypothesis, that the bacterial tubulin genes are ancestral to eukaryotic tubulin genes, could be explained in terms of a shared ancestry between the two groups or a gene transfer from an ancestor of the Verrucomicrobia to a protoeukaryotic organism, before the radiation of extant eukaryotes. A gene transfer between the groups could also encompass a fusion event between an ancestor of the Verrucomicrobia and another organism, such as an archaeon (25). The phylogenetic analyses superficially support this hypothesis, in that the bacterial tubulin sequences were always seen to branch more deeply than eukaryotic α and β tubulin; however, this relies on the assumption that α and β tubulins were the first members of the tubulin family to arise. Even if this assumption is correct, caution is required in the interpretation of the analyses, given that the level of sequence divergence in the bacterial sequences may cause them to migrate to the base of the tree artifactually (24). The various evolutionary models for the origin of tubulins that are implied by these hypotheses are to be discussed in detail elsewhere.
Although the current evidence does not allow an effective distinction between the two hypotheses presented here, further indications as to the origin of the Prosthecobacter tubulin genes may be facilitated by determining the distribution of the genes within the division Verrucomicrobia. If the genes were present in members of several subdivisions of the Verrucomicrobia, this would suggest that the genes have been in these organisms for a long time. Furthermore, closer examination of the P. dejongeii genome, such as searching for other genes unique to eukaryotes, may aid in determining whether a large transfer event or a fusion occurred between members of the Verrucomicrobia and eukaryotes.
If it were true that the bacterial tubulins are ancestral to eukaryotic tubulins, it would have a significant impact on our understanding of eukaryote cell evolution. Although FtsZ is a homolog of tubulin, the evolutionary distance between the two proteins is substantial. Indeed, it has been suggested several times that a more immediate evolutionary precursor of tubulin may reside in some as-yet-undiscovered bacterial or archaeal lineage (26) or was acquired from an extinct lineage (25, 27) or “chronocyte” (2). Whether the Prosthecobacter tubulins satisfy this role as evolutionary intermediate between FtsZ and eukaryotic tubulin remains to be seen.
Acknowledgments
We thank Carl Woese for advice during the course of this research and for providing useful comments on the manuscript. We are indebted to Kendall Gray, Jeremy Dodsworth, and Heather Bouzek, who contributed to related aspects of this work, and to Rose Ann Cattolico, Frank Harold, and Sujatha Srinivasan for providing helpful comments on the manuscript. Likewise, we appreciate the generosity of Integrated Genomics for its support of basic science research. We are also grateful to the University of Washington's National Science Foundation Integrated Graduate Education Research Traineeship doctoral program, Astrobiology: Life in and Beyond Earth's Solar System (NSF DGE-9870713); the Royalty Research Fund at the University of Washington; and the University of Washington National Aeronautics and Space Administration Astrobiology Institute program. The modeling portion of this work was supported by a Searle Scholar Award to R.S.
References
- 1.Keeling P. J. & Doolittle, W. F. (1996) Mol. Biol. Evol. 13, 1297-1305. [DOI] [PubMed] [Google Scholar]
- 2.Hartman H. & Fedorov, A. (2002) Proc. Natl. Acad. Sci. USA 99, 1420-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanier R. Y. (1970) Symp. Soc. Gen. Microbiol. 20, 1-38. [Google Scholar]
- 4.Bermudes D., Hinkel, F. & Margulis, L. (1994) Microbiol. Rev. 58, 387-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nogales E., Wolf, S. G. & Downing, K. H. (1998) Nature 391, 199-203. [DOI] [PubMed] [Google Scholar]
- 6.Löwe J. & Amos, L. A. (1998) Nature 391, 203-206. [DOI] [PubMed] [Google Scholar]
- 7.de Boer P., Crossley, R. & Rothfield, L. (1992) Nature 359, 254-256. [DOI] [PubMed] [Google Scholar]
- 8.Doolittle R. F. (1995) Philos. Trans. R. Soc. London 349, 235-240. [DOI] [PubMed] [Google Scholar]
- 9.Dacks J. B. & Doolittle, W. F. (2001) Cell 107, 419-425. [DOI] [PubMed] [Google Scholar]
- 10.Petroni G., Spring, S., Schleifer, K.-H., Verni, F. & Rosati, G. (2000) Proc. Natl. Acad. Sci. USA 97, 1813-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosati G., Lenzi, P. & Verni, F. (1993) Micron 24, 465-471. [Google Scholar]
- 12.Staley J. T. & Mandel, M. (1973) Int. J. Syst. Bacteriol. 23, 271-273. [Google Scholar]
- 13.Siebert P. D., Chenchik, A., Kellogg, D. E., Lukyanov, K. A. & Lukyanov, S. A. (1995) Nucleic Acids Res. 6, 1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholas K. B., Nicholas, H. B., Jr. & Deerfield, D. W., II (1997) EMBNEW.NEWS 4, 14. [Google Scholar]
- 15.Samudrala R. & Moult, J. (1997) Proteins 29S, 43-49. [DOI] [PubMed] [Google Scholar]
- 16.Samudrala R. & Levitt, M. A. (2002) BMC Struct. Biol. 2, 3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. (1989) Cladistics 5, 164-166. [Google Scholar]
- 18.Vaughan S., Attwood, T., Navarro, M., Scott, V., McKean, P. & Gull, K. (2000) Curr. Biol. 10, R258-R259. [DOI] [PubMed] [Google Scholar]
- 19.Löwe J., Li, H., Downing, K. H. & Nogales, E. (2001) J. Mol. Biol. 313, 1045-1057. [DOI] [PubMed] [Google Scholar]
- 20.Nogales E., Whittaker, M., Milligan, R. A. & Downing, K. H. (1999) Cell 96, 79-88. [DOI] [PubMed] [Google Scholar]
- 21.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 22.Merritt A. E. & Bacon, D. J. (1997) Methods Enzymol. 277, 505-524. [DOI] [PubMed] [Google Scholar]
- 23.Samudrala R. & Moult, J. (1998) J. Mol. Biol. 275, 895-916. [DOI] [PubMed] [Google Scholar]
- 24.Philippe H. & Laurent, J. (1998) Curr. Opin. Genet. Dev. 8, 616-623. [DOI] [PubMed] [Google Scholar]
- 25.Sogin M. L. (1991) Curr. Opin. Genet. Dev. 1, 457-463. [DOI] [PubMed] [Google Scholar]
- 26.Erickson H. P. (1995) Cell 80, 367-370. [DOI] [PubMed] [Google Scholar]
- 27.Faguy D. M. & Doolittle, W. F. (1998) Curr. Biol. 8, R338-R341. [DOI] [PubMed] [Google Scholar]