Abstract
Growing evidence has implicated members of the genus Enterovirus of the family Picornaviridae in the etiology of some cases of type 1 diabetes (T1D). To contribute to an understanding of the molecular determinants underlying this association, we determined the complete nucleotide sequence of a strain of echovirus 3 (E3), Human enterovirus B (HEV-B) species, isolated from an individual who soon after virus isolation developed autoantibodies characteristic of T1D. The individual has remained positive for over 6 years for tyrosine phosphatase-related IA-2 protein autoantibodies and islet cell autoantibodies, indicating an ongoing autoimmune process, although he has not yet developed clinical T1D. The sequence obtained adds weight to the observation that recent enterovirus isolates differ significantly from prototype strains and provides further evidence of a role for recombination in enterovirus evolution. In common with most HEV-B species members, the isolate exhibits 2C and VP1 sequences suggested as triggers of autoimmunity through molecular mimicry. However, comparisons with the E3 prototype strain and previously reported diabetogenic and nondiabetogenic HEV-B strains do not reveal clear candidates for sequence features of PicoBank/DM1/E3 that could be associated with autoantibody appearance. This is the first time a virus strain isolated at the time of commencement of beta-cell damage has been analyzed and is an invaluable addition to enterovirus strains isolated previously at the onset of T1D in the search for specific molecular features which could be associated with diabetes induction.
Human enteroviruses (HEVs) are important pathogens and include viruses which are capable of causing diverse diseases, such as poliomyelitis, encephalitis, meningitis, conjunctivitis, carditis, respiratory infections, and human hand-foot-and-mouth disease (12). They include polioviruses, echoviruses (E), coxsackieviruses A (CAV) and B (CBV), and the more recently isolated serotypes designated enteroviruses 68 to 71 and 73 to 78. Taxonomically, they are assigned to the genus Enterovirus, the most diverse genus of the family Picornaviridae, and the genus includes five species causing human disease (Poliovirus and Human enterovirus A, B, C, and D [46, 47]). Enteroviruses have a single-stranded, positive-sense RNA genome around 7,400 nucleotides long, which has one open reading frame. This is preceded by a 5′ untranslated region (UTR) (about 750 nucleotides) and is followed by a short 3′ UTR and a poly(A) tract (45).
For more than 100 years, viruses have been suspected to be a factor in the etiology of type 1 diabetes mellitus (T1D) (15), and most reports have concerned the involvement of enteroviruses, frequently the HEV-B species members and CBVs (18). Viruses seem to be involved in the triggering of an autoimmune reaction, which subsequently leads to the destruction of islet cells. Over the past few years, a number of studies have reported an association between enteroviruses and diabetes on the basis of reverse transcription (RT)-PCR amplification from clinical material, using specific sequences from the well-conserved 5′ UTR (1, 8, 10). However, these studies have provided limited information on the precise nature of the enterovirus present, as frequent recombination means that 5′-UTR sequences do not correlate with other regions of the genome, notably, the capsid-encoding region, which determines the serotype and receptor binding properties of the virus (19, 25, 27, 39, 44). Virus propagation from diabetic subjects has been another approach. CBV4 and E9 strains isolated in this way have been studied, and attempts to identify diabetic determinants have been made (21, 37, 48, 52). Other studies have identified sequences in enteroviruses that could be involved in autoimmunity through molecular mimicry (13, 14, 36, 53). Recently, enterovirus RNA has been demonstrated in islet cells of some diabetic patients, and these cells were shown to express several receptors recognized by enteroviruses (55). In addition, work on a strain of CBV4 indicated that changes close to the canyon, the site of receptor binding in enteroviruses, correlated with persistent infection in pancreatic islet cells, emphasizing a potential role for receptor interactions in T1D (54). However, our understanding of the role of enteroviruses in diabetes and potential molecular determinants is still very incomplete.
During a prospective study of T1D, the Finnish Type 1 Diabetes Prediction and Prevention (DIPP) study, an enterovirus was isolated from an individual who soon after virus isolation developed a range of typical autoantibodies seen in diabetic and prediabetic subjects, although to date the patient has not developed clinical T1D. This is the first description of such an enterovirus strain being isolated at the time when the beta-cell damage process starts, and thus, it can provide new information about the role of enteroviruses in these early stages of pathogenesis. The virus proved to be a strain of echovirus 3 (E3), and as there were limited sequence data available for this serotype, we obtained the complete nucleotide sequence of the isolate. Here, we show that the virus differs significantly from the E3 prototype, Morrisey, but is very similar to other recently circulating strains.
MATERIALS AND METHODS
DIPP study.
The Finnish DIPP trial is a prospective study in which serum and stool samples are collected from children at high genetic risk of developing T1D (26). In this study, all newborn infants born at the university hospitals in Oulu, Tampere, and Turku are first screened for HLA-DQB1 risk alleles for type 1 diabetes. Children with increased genetic risk (HLA-DQB1*02/*0302), the *0302/x genotype [x ≠ *02, *0301, or *0602], and also boys with the genotype DQB1*02/y-DQA1*05/z [y ≠ *0301, *0302, *0602, *0603; z ≠ *0201] are observed from birth. The protocol was described in detail previously (24). Serum samples are collected from birth at intervals of 3 to 6 months to test for the possible appearance of diabetes-associated autoantibodies, including islet cell autoantibodies (ICA) and insulin autoantibodies (IAA), as well as autoantibodies (GADA) against the 65-kDa isoform of glutamic acid decarboxylase (GAD65) and protein tyrosine phosphatase-related IA-2 molecule (IA-2A). The clinical symptoms are recorded every visit using a questionnaire. Serum samples are also used for virus analyses. In addition, stool samples are collected every month from the age of 3 months for virus isolation.
Patient and virus isolation.
The patient was born on 3 February 1998, and he was recruited into the study. He had increased genetic risk of developing T1D, as he carried the DR4/DQ8 haplotype. He had normal diet and weight/height gain and had no chronic diseases, allergies, or first-degree relatives with T1D or other autoimmune diseases. The isolation was done from a stool sample collected at the age of 6.5 months (on 19 August 1998). The child is still in follow-up, the latest examination being at the age of 7 years and 1 month (on 3 March 2005). He has remained consistently positive for diabetes-associated autoantibodies but has not yet developed T1D. Virus in the stool sample, termed PicoBank/DM1/E3, was cultured in CaCo-2 (human colon carcinoma) cells using methods previously described (43).
Measurement of neutralizing antibodies.
Serotype-specific antibodies against the isolated E3 strain (PicoBank/DM1/E3) were measured in the serial serum samples of this child, using the standard plaque neutralization assay as previously described (40). The green monkey kidney (GMK) cell line used was purchased from the American Type Culture Collection. Serum was incubated in two different dilutions (1/16 and 1/64) with the virus (100 PFU) for 1 h at 36°C. Then, the samples were applied to GMK cell monolayers grown in six-well plates (Nunclon; NUNC, Denmark) and further incubated at 36°C for 48 h. The amount of infectious virus was measured by counting the plaques; the reciprocal of the last serum dilution able to block virus infectivity by 80% was taken as the titer of neutralizing antibodies. A fourfold or greater increase in antibody titer between two follow-up samples was considered significant, indicating acute infection.
Extraction of RNA from a cultured clinical sample and RT-PCR.
RNA was extracted from 100 μl of the tissue culture supernatant from infected CaCo-2 cells, using a Nucleospin RNA II extraction kit (Macherey Nagel) according to the manufacturer's instructions. The RNA was resuspended in 60 μl of nuclease-free water, and 5 μl was then subjected to one-step RT-PCR by the use of a SuperScript One-Step RT-PCR kit (Invitrogen) and primers designed specifically for the detection of members of the HEV-B species of enteroviruses (Table 1).
TABLE 1.
Sequences of general primers for the amplification of the genomes of HEV-B members
| Primer name | Position in genome (CAV9 numbering) (6) | Primer sequence (5′ → 3′) |
|---|---|---|
| OL1073 | 1 → 20 | TTAAAACAGCCTGTGGGTTG |
| OL1074 | 1506 ← 1526 | AGGGTTGATCCACTGRTGNGG |
| OL1075 | 1320 → 1339 | GTAGTGTGCGTNCCNGARGC |
| OL1076 | 2375 ← 2398 | CACTGAGAAATCATTRCANGCNGA |
| OL1077 | 2238 → 2260 | CCATGGATCAGCCARACNCAYTA |
| OL1078 | 3774 ← 3796 | CCCTGTTCCATTGCRTCRTCYTC |
| OL998 | 3660 → 3679 | GAGCCAGGGGAYTGTGGNGG |
| OL999 | 5235 ← 5255 | TCTGACGTGCTTYTCNATYTG |
| OL1003 | 5019 → 5038 | ATGTTCAGGGAGTAYAAYCA |
| OL1000 | 6504 ← 6525 | GCCTCATCGCCACNGARTCRTT |
| OL1001 | 6384 → 6406 | AAGGAATGCATGGAYAARTAYGG |
| OL1002 | 7426 ← 7452 | GAGAGAACGCGTTTTTTTTTTTTTTTT |
Purification, cloning, and sequencing of PCR products.
Amplicons were run on 1 to 1.5% (depending on their size) agarose gels (Seakem GTG; Flowgen), excised from the gel with sterile scalpel blades, and then purified by the use of a GFX PCR DNA and Gel Band Purification kit (Amersham Pharmacia) according to the manufacturer's instructions. The DNA (0.25 to 0.35 μg) was ligated into pGEMT-Easy vector (Promega) and transformed into competent Escherichia coli XL1-Blue MRF′ cells. White colonies were analyzed by restriction enzyme digestions, and large-scale plasmid DNA isolations were carried out using a HiSpeed Plasmid midi kit (QIAGEN). The resulting DNA preparation was then sequenced commercially (MRCgeneservice) in both orientations, using either universal primers or custom primers designed on the basis of the newly generated sequences.
Determination and analysis of the complete genome sequence of PicoBank/DM1/E3.
The primer sequences were removed, and the overlapping sequences were assembled into a contiguous sequence. Sequence alignments were carried out by the use of ClustalW (51; http://www.ebi.ac.uk/clustalw/index.html). For phylogenetic analysis, sequences were aligned by using ClustalW and jalview to introduce gaps where necessary. The neighbor-joining algorithm implemented in the ClustalX software (50) was used to generate trees, which were bootstrapped 1,000 times. The trees were drawn using the NJPLOT software (38).
The following complete enterovirus sequences (accession numbers are in parentheses) were used in the construction of phylogenetic trees: CBV1 Japan (M16560), CBV2 Ohio (AF085363), CBV3 Nancy (M88483), CBV4 JVB (X05690), CBV4-D (E2 variant—AF311939) CBV5 Faulkner (AF114383), CBV6 Schmitt (AF105342), swine vesicular disease virus UKG/27/72 (X54521), CAV9 Griggs (D00627), E1 Farouk (AF029859), E2 Cornelis (AY302545), E3 Morrisey (AY302553), E4 Pesacek (AY302557), E5 Noyce (AF083069), E6 Charles (U16283), E6 D'Amori (AY302558), E7 Wallace (AY302559), E9-Hill (X84981), E9-BartyINF (AF524866), E9-DM (AF524867), E11 Gregory (X80059), E11-Kar/87 (AY167104), E12 Travis (X79047), E13 Del Carmen (AY302539), E14 Tow (AY302540), E15 Ch 96-51 (AY302541), E16 Harrington (AY302542), E17 CHHE-29 (AY302543), E18 Metcalf (AF317694), E19 Burke (AY302544), E20 JV-1 (AY302546), E21 Farina (AY302547), E24 DeCamp (AY302548), E25 JV-4 (AY302549), E26 Coronel (AY302550), E27 Bacon (AY302551), E29 JV-10 (AY302552), E30 Bastianni (AF311938), E31 Caldwell (AY302554), E32 PR-10 (AY302555), E33 Toluca-3 (AY302556), EV69 Toluca-1 (AY302560), EV73 CA55-1988 (AF241359), and EV77 CF496-99 (AJ493062). Partial sequences from the following E3 isolates, whose countries of isolation and accession numbers are given in parentheses, were also used: W178-128/99 (France; AY208114), 94CF858 (France; AJ241446), mo/Roma98 (Italy; AJ309243), si/Roma97 (Italy; AJ309242), 810/85 (Sweden; AF295490), 2392-82 (Sweden; AF295453), 1446-81 (Sweden; AF295439), Morrisey (United States; AF081316), and MD86-6393 (United States; AF152264).
Nucleotide sequence accession data.
The complete nucleotide sequence of PicoBank/DM1/E3 reported in this paper has been deposited in the GenBank database under the accession number AJ849942.
RESULTS
Virus isolate.
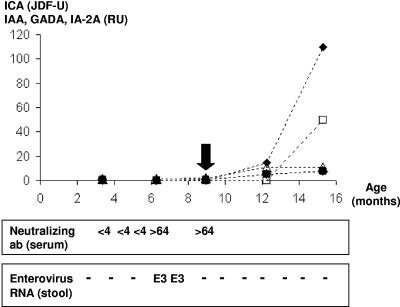
The subject turned positive for IAA for the first time on 2 November 1998 at the age of 9 months (Fig. 1). In the next follow-up sample, at the age of 1 year, he also turned positive for ICA (11 February 1999), and later (12 May 1999) for antibodies against GAD65 and IA-2. Thereafter, he remained constantly positive for both ICA and IA-2A (the last measurements, at the age of 7 years 1 month, were 44 Juvenile Diabetes Foundation units [JDF-U] and 141 relative units [RU], and the peak values observed during follow up were 872 JDF-U and 183 RU, respectively). In contrast, the IAA and GADA levels are currently negative. The clearly elevated levels of IA-2A, the most specific surrogate marker for T1D, suggest that he has an ongoing autoimmune process, even though he has not yet developed clinical T1D. The child's medical management and diet have not changed since he turned positive for autoantibodies. Analysis of stool samples taken at the ages of 6.5 and 7.5 months indicated the presence of enterovirus RNA. Virus isolation was done from the stool sample collected at the age of 6.5 months. The parents did not report any clinical symptoms for the child at the time of virus isolation, even though they reported rhinitis 2 months earlier and 3 months after virus isolation.
FIG. 1.
Autoantibody levels, neutralizing antibody titer against the isolated PicoBank/DM1/E3 strain (top), and viral RNA detected using enterovirus-specific RT-PCR in stool samples during the follow-up of the child participating in the DIPP study. The subject turned autoantibody positive (IAA) at the age of 9 months (marked by a vertical arrow). He had two enterovirus RNA-positive stool samples during the follow-up. The virus isolation was done from a stool sample collected at the age of 6.5 months. E3, PicoBank/DM1/E3 strain. The units of autoantibody levels were as follows: ICA (▴), JDF-U; GADA (•), RU; IAA (▵), RU; and IA-2A (□), RU.
Neutralizing antibodies against PicoBank/DM1/E3.
The isolated virus strain was used in a plaque neutralization assay to determine antibodies against the strain in the serum of the child from whom it was isolated. A rise in antibody titer (0 to >64) was seen in the serum sample taken on 12 August 1998, the same day that the virus was isolated from stools.
Sequence.
Partial sequence analysis of the virus (PicoBank/DM1/E3) propagated from the stool sample suggested that it is a HEV-B species member. By comparing enterovirus sequences, we have developed primers with the potential to amplify the whole genome from all HEV-B members in six overlapping fragments, using RT-PCR (Table 1). Each of these fragments was generated from the isolate, and the fragments were sequenced in both orientations. The derived sequence is 7,427 nucleotides long, including nucleotides 1 to 20, which were used as the priming site for the 5′-UTR PCR primer (and thus not determined), plus the poly(A) tract.
Relationship of PicoBank/DM1/E3 to other enteroviruses.
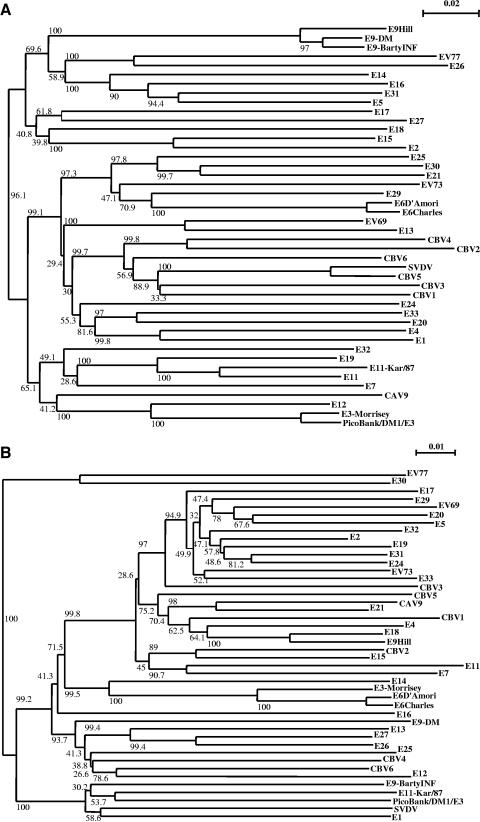
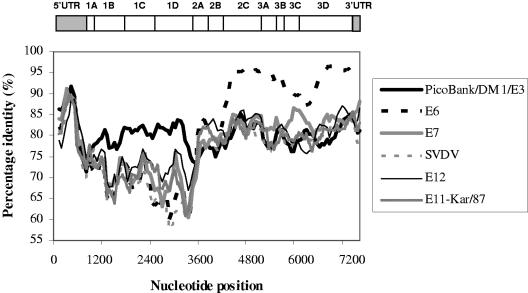
A dendrogram representing the relationship between the amino acid sequences of the P1 regions of all HEV-B members (35) indicates that PicoBank/DM1/E3 clusters with the E3 prototype strain Morrisey (Fig. 2a). It has 95% amino acid identity to E3-Morrisey in VP1 (data not shown), well within the identity range seen in an enterovirus serotype, suggesting strongly that it is a member of the E3 serotype (33, 34). In contrast, the 2C-plus-3CD region, also used regularly for the analysis of picornavirus phylogeny, showed no clear linkage to E3-Morrisey (Fig. 2b). We used the nucleotide sequence for this analysis, as HEV-B amino acid sequences are very similar. In this region, the closest relative of PicoBank/DM1/E3 is Kar/87, an E11 isolate, but it is only marginally more similar to this isolate than to a range of other sequenced enteroviruses. To study this disparity between the P1 and 2C-plus-3CD regions further, graphs of nucleotide sequence identity across the genome, compared with a selection of other enteroviruses, were plotted (Fig. 3). These confirmed that PicoBank/DM1/E3 and E3-Morrisey are closely related throughout the capsid region. However, downstream of the P1/P2 junction, E3-Morrisey is highly similar to the E6 prototype, Charles, while PicoBank/DM1/E3 is much less closely related, providing evidence that at least one of the two E3 strains has undergone a recombination event during divergence. There are also significant differences between the E3 strains in the 5′ UTR (data not shown).
FIG. 2.
Phylogenetic trees expressing the relationship between HEV-B members in different regions of the genome. The trees were derived using the N-J method in the ClustalX software and were bootstrapped 1,000 times before being plotted using the NJPLOT software. Bootstrap values (%) are shown on the branches. (a) Amino acid sequence of P1. (b) Nucleotide sequence of 2C plus 3CD.
FIG. 3.
Nucleotide sequence identities across the genome between E3-Morrisey and a range of enteroviruses, including PicoBank/DM1/E3, together with a schematic representation of the enterovirus genome. The enteroviruses selected are the closest relatives of the two E3 strains. E6, E9, and E12 are prototype strains, and E11-Kar/87 is a recent E11 isolate. SVDV, swine vesicular disease virus. A window of 300 nucleotides and steps of 60 nucleotides were used.
Relationships with other E3 isolates.
A few E3 clinical isolates (28, 30, 31, 32, 34) have been partially sequenced, and these were compared with PicoBank/DM1/E3. A 302-nucleotide region of VP1 (nucleotides 2573 to 2874 in PicoBank/DM1/E3) was chosen, as it lies within most of the available sequences. The results show that PicoBank/DM1/E3 is closely related to other strains circulating in Europe in the 1990s and less so to the E3 prototype, Morrisey (Fig. 4). Indeed, it is 98% identical in this region to W178-128/99, a French isolate (31). The European isolates (W178-128/99, 810/85, 2392-82, 1446-81, mo/Roma98, si/Roma97, and 94CF858) differ significantly from the U.S. isolate MD86-6393.
FIG. 4.
Phylogenetic tree expressing the relationship between PicoBank/DM1/E3 and other E3 isolates (W178-128/99, 94CF858, mo/Roma98, si/Roma97, 810/85, 2392-82, 1446-81, Morrisey, and MD86-6393) based on nucleotide identity in part of VP1 (nucleotides 2573 to 2874). Bootstrap values (%) are shown on the branches.
Previously reported correlates of diabetogenicity.
PicoBank/DM1/E3 has the highly conserved amino acid sequence PEVKEK (amino acids 1147 to 1152 in the polyprotein) found in the 2C protein of HEV-B species members and in GAD65 and proposed to be involved in autoimmunity related to diabetes (4, 22, 53). The VP4/VP2 and VP1 sequences reported to have homology to heat shock protein 60 (HSP60) are very conserved among HEV-B species members, and PicoBank/DM1/E3 and E3-Morrisey are identical to one another at these points (data not shown). They are also very similar to the sequences in CAV9 and CBV4, where reciprocal cross-reactivity with HSP60 and tyrosine phosphatase was demonstrated (13, 14).
Previous analyses of enterovirus sequences (partial 5′ UTR) found in the blood of patients presenting with T1D indicated that they were related to CBV3 and CBV4 (1, 10). Comparisons of PicoBank/DM1/E3 with these sequences (10) indicate no clear grouping (Fig. 5), in common with comparisons with sequences (1) from adult patients (data not shown). Additionally, the PicoBank/DM1/E3 sequence does not add weight to the importance of 5′-UTR residues highlighted by these previous studies on the basis of differences between the prototypes and clinical isolates. Positions 229, 234, and 303, identified in these studies, are identical in E3-Morrisey and PicoBank/DM1/E3. The diabetogenic E9 isolate DM (E9-DM) also does not differ from the E9 prototype, Barty (E9-Barty INF), at these positions (37).
FIG. 5.
Molecular relationships in part of the 5′ UTR (nucleotides 150 to 448 in CBV3-Nancy) between previously reported enterovirus sequences from juvenile diabetic patients (10) and potentially diabetogenic E3, E9, and CBV4 isolates, together with corresponding reference strains. The trees were plotted as for Fig. 2. Bootstrap values (%) are shown on the branches.
Comparison with E3-Morrisey.
E3-Morrisey is the only other completely sequenced E3 isolate and was useful for detailed comparisons with PicoBank/DM1/E3. The 5′ UTRs of these viruses share around 83.8% nucleotide sequence identity. Both conform to the predicted structure, seen in both HEV-A and HEV-B members, and there are no clear structural differences (data not shown). The capsid region P1 of PicoBank/DM1/E3 shows 27 amino acid differences from that of E3-Morrisey (Table 2). However, at several of the VP1 positions where PicoBank/DM1/E3 and E3-Morrisey differ, recent European isolates have the same sequence as PicoBank/DM1/E3, and in this protein, only three differences seem to be specific for PicoBank/DM1/E3 (Table 2). There are 53 amino acid differences between PicoBank/DM1/E3 and E3-Morrisey in the nonstructural region, but no significant patterns emerge. PicoBank/DM1/E3 has the typical predicted 3′-UTR secondary structure (data not show) seen in other HEV-B members, with three domains, X, Y, and Z, and a tertiary interaction between domains X and Y (29). The structure is identical to that of E3-Morrisey, as there are only two nucleotide differences, which are covariant, in the domain Y stem and two in the presumably unpaired domain Y loop. Similarly, there is no significant difference in the structure of the cis-acting replication element (cre), the RNA element necessary for initiation of virus RNA replication through VPg uridylylation (11), between the E3 strains.
TABLE 2.
Amino acid differences between PicoBank/DM1/E3 and E3-Morrisey in capsid regiona
| Location of difference | Difference and its positiona |
|---|---|
| VP4 | T184N |
| VP2 | T372V |
| VP2 | S402N |
| VP2 | G1592E |
| VP2 | V1692A |
| VP2 | E2332K |
| VP3 | V623I |
| VP3 | S643G |
| VP3 | E663D |
| VP3 | H793R |
| VP3 | I1683V |
| VP3 | V1903I |
| VP3 | P2063S |
| VP3 | V2143A |
| VP1 | E81D |
| VP1 | L181M |
| VP1 | I1441L |
| VP1 | S1931T |
| VP1 | N2281S |
| VP1 | L2361V |
| VP1 | S2561C |
| VP1 | V2591T |
| VP1 | T2731D |
| VP1 | M2811Vb |
| VP1 | M2871Tb |
| VP1 | G2881E |
| VP1 | S2911Nb |
The PicoBank/DM1/E3 amino acid is given first and the corresponding E3-Morrisey residue is given after the amino acid position. Amino acid positions are numbered according to the CAV9 sequence, and the final digit refers to the protein, e.g., D81E is position 8 in VP1. At all the VP1 positions, recent E3 isolates have the same amino acid as PicoBank/DM1/E3.
Amino acid differs from that in PicoBank/DM1/E3.
DISCUSSION
Enteroviruses are important causes of acute clinical disease, and there is growing evidence that they may be involved in chronic conditions, including T1D (18). T1D is characterized by autoimmunity leading to destruction of the islet cells (20, 23). The mechanism of enterovirus involvement is not currently understood, although both molecular mimicry and direct beta-cell damage leading to sequestration of proteins and presentation to autoreactive T cells have been suggested (17, 36). If there is an association, it is important to determine if an individual enterovirus serotype or groups of serotypes are involved or whether enteroviruses capable of triggering autoimmunity share specific molecular or biological features. This has important implications for future vaccines and other control strategies. To address this issue, a number of viruses putatively associated with diabetes need to be studied, but few such studies have been reported (21, 37, 48). Here, we present the complete nucleotide sequence of an E3 strain isolated from a fecal sample taken 2.5 months before a patient developed a range of diabetes-associated autoantibodies, which reflect the initiation of the beta-cell-damaging autoimmune process. This is the first time a virus has been isolated from a patient close to the time of appearance of autoantibodies; thus, the strain represents an invaluable resource for studying potential molecular determinants associated with autoantibody induction and T1D.
E3 has been comparatively little studied, and in addition to the reference strain, there are very limited sequence data available for only 11 isolates. Thus, the present study greatly extends the molecular information available on this serotype. It is clear that E3 is a typical HEV-B member in terms of sequence (Fig. 2). It also conforms to the pattern seen in several recent reports of uncoupling of capsid region evolution from that of the 5′ UTR and nonstructural region through frequent recombination (9, 25, 27, 44). E3-Morrisey is closely related to the E6 prototype, Charles, in the nonstructural region and much less so to PicoBank/DM1/E3 in this region (Fig. 3), suggesting recombination since the divergence of the lineages giving rise to E3-Morrisey and PicoBank/DM1/E3. The recombination point between the two E3 lineages seems to be close to the P1/P2 boundary, as seen in a number of other enteroviruses (27), as the lines representing the identity between E3-Morrisey and E6-Charles, and between E3-Morrisey and PicoBank/DM1/E3, cross over around position 3600 (Fig. 3). In nucleotide sequence identity terms (Fig. 3), the closest sequenced relative of PicoBank/DM1/E3 in the nonstructural region is a recent (1988) Russian isolate of E11 (Kar/87), supporting the suggestion that recombination may allow rapid exchange of nonstructural lineages between serotypes (27). In this case, it is likely that a recombination event has occurred in the 3CD region, as the PicoBank/DM1/E3/E11-Kar/87 identity increases sharply there (data not shown).
Previous molecular and serology studies have tended to implicate CBVs in diabetes, and the identification of PicoBank/DM1/E3 as an E3 isolate extends the range of enterovirus serotypes that may be involved in diabetes. A recent report describes the molecular and biological characterization of an E9 strain, E9-DM, shown to be diabetogenic, while patients infected during an E16 outbreak in Cuba were shown to develop autoantibodies characteristic of T1D (5, 37). Thus, there is growing evidence that echoviruses could be an environmental factor in T1D. As yet, no other echoviruses have been identified in the DIPP study, but this may be a reflection of the fact that few viruses have been successfully isolated at the time of autoantibody seroconversion.
The PicoBank/DM1/E3 sequence was examined for previously suggested correlates of diabetogenicity. In common with most other HEV-B members, it has the 2C and VP1 sequences previously implicated in autoimmunity and so could have a similar effect (4, 13, 14, 22, 53). More specific determinants of strains associated with diabetes have also been reported. A study of partial enterovirus 5′-UTR sequences detected in the blood of children presenting with T1D indicated that they were related to CBV3 and CBV4 and clustered together (10). However, neither PicoBank/DM1/E3 nor the diabetogenic E9 isolate E9-DM clusters with these sequences (Fig. 5), and the same is also true when comparisons are made between sequences (1) from adult patients (data not shown). In addition, specific nucleotide differences between the sequences from blood and the CBV3 and CBV4 prototype strains are not seen in either PicoBank/DM1/E3 and E3-Morrisey comparisons or comparisons between E9-DM and the E9 prototype, Barty. It should be noted that the blood samples were geographically and temporally close in origin and that a limited region of the genome was analyzed. It may well be that the clustering seen is due to these factors rather than being a common feature of viruses linked to T1D. The whole-genome sequencing approach adopted here and in the E9 study (37) promises to be more useful in determining such common features.
Comparisons with E3-Morrisey do not reveal any clear candidates for sequence features of PicoBank/DM1/E3 that could be associated with autoantibody appearance. There is also no obvious link with work done on encephalomyocarditis virus, a member of the genus Cardiovirus of the family Picornaviridae, where the critical difference between diabetogenic and nondiabetogenic strains was mapped precisely to VP1 position 167 (3), as PicoBank/DM1/E3 shows no significant differences from E3-Morrisey and the recent European isolates in the corresponding region of VP1. However, when the capsid differences are positioned on a three-dimensional structure predicted using CAV9, another HEV-B member, as a model (16), several differences cluster around the threefold axis (data not shown). These include a difference immediately at the threefold axis, in a location analogous to one that brings about altered cell tropism in CAV9 (Ç. H. Williams and G. Stanway, unpublished data), and a difference at this position could influence the tropism of PicoBank/DM1/E3.
The patient from whom PicoBank/DM1/E3 was isolated has remained positive for IA-2A and ICA for 6 years, indicating an ongoing autoimmune process, although he has not yet developed clinical T1D. This virus strain, isolated immediately prior to the appearance of autoantibodies, is particularly interesting, as it may be involved in the initiation of the beta-cell-damaging process. This is the first time that such a strain has been isolated, and further biological studies should give important information. Mouse models and human islets have been widely used in previous studies and are logical approaches for future work, although both have limitations. It is not clear how well animal models reflect the diabetogenicity of enteroviruses in humans, as there are significant differences in pathology (2). In addition, there is no established mouse model for echovirus 3. The use of human islet cells is complicated by the observation that all enteroviruses seem to infect these cells, causing a wide range of alterations (apoptosis, necrosis, or changes in insulin secretion without clear damage), and which of these correlate with diabetogenicity is not clear (7, 41, 42, 49, 54). Thus, a range of models and additional enterovirus strains isolated during prospective studies and at the time of onset are required to further address the role of enteroviruses in T1D. The identification of consistent molecular features in such strains would be additional strong evidence of a role for enteroviruses in T1D and would provide a basis for strategies, such as vaccination, aimed at preventing the disease.
Acknowledgments
This work was made possible by funding from the EC (VIRDIAB Project, Quality of Life Programme, KA2, contract number QLK 2-CT-2001-01910). The virus studied was isolated in the DIPP study, and the work has been funded by the Juvenile Diabetes Research Foundation, the Foundation for Diabetes Research in Finland, and the Academy of Finland.
C.H.W., S.O., H.H., and G.S. are members of the VIRDIAB study group, whose other members provided helpful discussions. Other members of the VIRDIAB study group include Kaisa Anturamäki (University of Tampere, Tampere, Finland), Didier Hober (Service de Virologie CHRU Lille and UPRES EA, Université Lille, Lille, France), Bernadette Lucas (Service de Virologie CHRU Lille and UPRES EA, Université Lille, Lille, France), Peter Muir (King's College London and Health Protection Agency South West, United Kingdom), Keith Taylor (Queen Mary and Westfield College, London, United Kingdom), Christos S. Bartsocas (University of Athens, Athens, Greece), Andriani Gerasimidou-Vazeou (University of Athens, Athens, Greece), Evangelos Bozas (University of Athens, Athens, Greece), and Johnny Ludvigsson (University of Linköping, Linköping, Sweden).
REFERENCES
- 1.Andréoletti, L., D. Hober, C. Hober-Vandenberghe, S. Belaich, M. C. Vantyghem, J. Lefebvre, and P. Wattre. 1997. Detection of coxsackie B virus RNA sequences in whole blood samples from adult patients at the onset of type I diabetes mellitus. J. Med. Virol. 52:121-127. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson, M. A., and E. H. Leiter. 1999. The NOD mouse model of type 1 diabetes: as good as it gets? Nat. Med. 5:601-604. [DOI] [PubMed] [Google Scholar]
- 3.Bae, Y.-S., H.-M. Eun, and J.-W. Yoon. 1989. Genomic differences between the diabetogenic and nondiabetogenic variants of Encephalomyocarditis Virus. Virology 170:282-287. [DOI] [PubMed] [Google Scholar]
- 4.Baekkeskov, S., H. J. Aanstoot, S. Christgau, A. Reetz, M. Solimena, M. Cascalho, F. Folli, H. Richterolesen, and P. D. Camilli. 1990. Identification of the 64K autoantigen in insulin-dependent diabetes as the GABA-synthesising enzyme glutamic acid decarboxylase. Nature 347:151-156. [DOI] [PubMed] [Google Scholar]
- 5.Cabrera-Rode, E., L. Sarmiento, C. Tiberti, G. Molina, J. Barrios, D. Hernandez, O. Diaz-Horta, and U. Di Mario. 2003. Type 1 diabetes islet associated antibodies in subjects infected by echovirus 16. Diabetologia 46:1348-1353. [DOI] [PubMed] [Google Scholar]
- 6.Chang, K. H., P. Auvinen, T. Hyypiä, and G. Stanway. 1989. The nucleotide sequence of coxsackievirus A9—implication for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269-3280. [DOI] [PubMed] [Google Scholar]
- 7.Chehadeh, W., J. Kerr-Conte, F. Pattou, G. Alm, J. Lefebvre, P. Wattre, and D. Hober. 2000. Persistent infection of human pancreatic islets by coxsackievirus B is associated with alpha interferon synthesis in beta cells. J. Virol. 74:10153-10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chehadeh, W., J. Weill, M. C. Vantyghem, G. Alm, J. Lefebvre, P. Wattre, and D. Hober. 2000. Increased level of interferon-alpha in blood of patients with insulin-dependent diabetes mellitus: relationship with coxsackievirus B infection. J. Infect. Dis. 181:1929-1939. [DOI] [PubMed] [Google Scholar]
- 9.Chevaliez, S., A. Szendröi, V. Caro, J. Balanant, S. Guillot, G. Berencsi, and F. Delpeyroux. 2004. Molecular comparison of echovirus 11 strains circulating in Europe during an epidemic of multisystem hemorrhagic disease of infants indicates that evolution generally occurs by recombination. Virology 325:56-70. [DOI] [PubMed] [Google Scholar]
- 10.Clements, G. B., D. N. Galbraith, and K. W. Taylor. 1995. Coxsackie B virus infection and onset of childhood diabetes. Lancet 346:221-223. [DOI] [PubMed] [Google Scholar]
- 11.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grist, N. R., E. J. Bell, and F. Assaad. 1978. Enteroviruses in human disease. Prog. Med. Virol. 24:114-157. [PubMed] [Google Scholar]
- 13.Härkönen, T., H. Lankinen, B. Davydova, T. Hovi, and M. Roivainen. 2002. Enterovirus infection can induce immune responses that cross-react with β-cell autoantigen tyrosine phosphatase IA-2/IAR. J. Med. Virol. 66:340-350. [DOI] [PubMed] [Google Scholar]
- 14.Härkönen, T., M. Puolakkainen, M. Sarvas, U. Airaksinen, T. Hovi, and M. Roivainen. 2000. Picornavirus proteins share antigenic determinants with Heat Shock Proteins 60/65. J. Med. Virol. 62:383-391. [DOI] [PubMed] [Google Scholar]
- 15.Harris, H. F. 1898. A case of diabetes mellitus quickly following mumps. Boston Med. Surg. J. 140:465-469. [Google Scholar]
- 16.Hendry, E., H. Hatanaka, E. Fry, M. Smyth, J. Tate, G. Stanway, J. Santti, M. Maaronen, T. Hyypiä, and D. Stuart. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7:1527-1538. [DOI] [PubMed] [Google Scholar]
- 17.Horwitz, M. S., A. Ilic, C. Fine, E. Rodriguez, and N. Sarvetnick. 2002. Presented antigen from damaged pancreatic β cells activates autoreactive T cells in virus-mediated autoimmune diabetes. J. Clin. Investig. 109:79-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyöty, H., and K. W. Taylor. 2002. The role of viruses in human diabetes. Diabetologia 45:1353-1361. [DOI] [PubMed] [Google Scholar]
- 19.Hyypiä, T., T. Hovi, N. J. Knowles, and G. Stanway. 1997. Classification of enteroviruses based on molecular and biological properties. J. Gen. Virol. 78:1-11. [DOI] [PubMed] [Google Scholar]
- 20.Jenson, A. B., H. S. Rosenberg, and A. L. Notkins. 1980. Pancreatic islet-cell damage in children with fatal viral infections. Lancet ii:354-358. [PubMed] [Google Scholar]
- 21.Kang, Y., N. K. Chatterjee, M. J. Nodwell, and J.-W. Yoon. 1994. Complete nucleotide sequence of a strain of Coxsackie B4 virus of human origin that induces diabetes in mice and its comparison with nondiabetogenic Coxsackie B4 JBV strain. J. Med. Virol. 44:353-361. [DOI] [PubMed] [Google Scholar]
- 22.Kaufman, D. L., M. G. Erlander, M. Claresalzler, M. A. Atkinson, N. K. Maclaren, and A. J. Tobin. 1992. Autoimmunity to 2 forms of glutamate-decarboxylase in insulin-dependent diabetes-mellitus. J. Clin. Investig. 89:283-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolb, H., V. Kolbbachofen, and B. O. Roep. 1995. Autoimmune versus inflammatory type-I diabetes—a controversy. Immunol. Today 16:170-172. [DOI] [PubMed] [Google Scholar]
- 24.Kupila, A., P. Muona, T. Simell, P. Arvilommi, H. Savolainen, A.-M. Hämäläinen, S. Korhonen, T. Kimpimaki, M. Sjoroos, J. Ilonen, M. Knip, O. Simell, and the Juvenile Diabetes Research Foundation Centre for the Prevention of Type I Diabetes in Finland. 2001. Feasibility of genetic and immunological prediction of type 1 diabetes in a population-based cohort. Diabetologia 44:290-297. [DOI] [PubMed] [Google Scholar]
- 25.Lindberg, A. M., P. Andersson, C. Savolainen, M. N. Mulders, and T. Hovi. 2003. Evolution of the genome of human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species J. Gen. Virol. 84:1223-1235. [DOI] [PubMed] [Google Scholar]
- 26.Lönnrot, M., K. Korpela, M. Knip, J. Ilonen, O. Simell, S. Korhonen, K. Savola, P. Muona, T. Simell, P. Koskela, and H. Hyöty. 2000. Enterovirus infection as a risk factor for beta-cell autoimmunity in a prospectively observed birth cohort: the Finnish Diabetes Prediction and Prevention Study. Diabetes 49:1314-1318. [DOI] [PubMed] [Google Scholar]
- 27.Lukashev, A. N., V. A. Lashkevich, O. E. Ivanova, G. A. Koroleva, A. E. Hinkkanen, and J. Ilonen. 2003. Recombination in circulating enteroviruses. J. Virol. 77:10423-10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manzara, S., M. Muscillo, G. La Rosa, C. Marianelli, P. Cattani, and G. Fadda. 2002. Molecular identification and typing of enteroviruses isolated from RT clinical specimens. J. Clin. Microbiol. 40:4554-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirmomeni, M., P. J. Hughes, and G. Stanway. 1997. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J. Virol. 71:2363-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norder, H., L. Bjerregaard, and L. O. Magnius. 2001. Homotypic echoviruses share aminoterminal VP1 sequence homology applicable RT for typing. J. Med. Virol. 63:35-44. [PubMed] [Google Scholar]
- 31.Norder, H., L. Bjerregaard, L. O. Magnius, B. Lina, M. Aymard, and J. J. Chomel. 2003. Sequencing of ‘untypable’ enteroviruses reveals two new types, EV-77 and EV-78, within human enterovirus type B and substitutions in the BC loop of the VP1 protein for known types. J. Gen. Virol. 84:827-836. [DOI] [PubMed] [Google Scholar]
- 32.Oberste, M. S., K. Maher, M. R. Flemister, G. Marchetti, D. R. Kilpatrick, and M. A. Pallansch. 2000. Comparison of classic and molecular approaches for the identification of RT untypeable enteroviruses. J. Clin. Microbiol. 38:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberste, M. S., K. Maher, D. R. Kilpatrick, M. R. Flemister, B. A. Brown, and M. A. Pallansch. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oberste, M. S., K. Maher, and M. A. Pallansch. 2004. Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J. Virol. 78:855-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oldstone, M. B. 1987. Molecular mimicry and autoimmune diseases. Cell 50:819-820. [DOI] [PubMed] [Google Scholar]
- 37.Paananen, A., P. Ylipaasto, E. Rieder, T. Hovi, J. Galama, and M. Roivainen. 2003. Molecular and biological analysis of Echovirus 9 strain isolated from a diabetic child. J. Med. Virol. 69:529-537. [DOI] [PubMed] [Google Scholar]
- 38.Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 39.Pöyry, T., L. Kinnunen, T. Hyypiä, B. Brown, C. Horsnell, T. Hovi, and G. Stanway. 1996. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 77:1699-1717. [DOI] [PubMed] [Google Scholar]
- 40.Roivainen, M., M. Knip, H. Hyöty, P. Kulmala, M. Hiltunen, P. Vähäsalo, T. Hovi, and H. K. Åkerblom. 1998. Several different enterovirus serotypes can be associated with prediabetic autoimmune episodes and onset of overt IDDM. J. Med. Virol. 56:74-78. [DOI] [PubMed] [Google Scholar]
- 41.Roivainen, M., S. Rasilainen, P. Ylipaasto, R. Nissinen, J. Ustinov, L. Bouwens, D. L. Eizirik, T. Hovi, and T. Otonkoski. 2000. Mechanisms of coxsackievirus-induced damage to human pancreatic beta-cells. J. Clin. Endo. Metab. 85:432-440. [DOI] [PubMed] [Google Scholar]
- 42.Roivainen, M., P. Ylipaasto, C. Savolainen, J. Galama, T. Hovi, and T. Otonkoski. 2002. Functional impairment and killing of human beta cells by enteroviruses: the capacity is shared by a wide range of serotypes, but the extent is a characteristic of individual virus strains. Diabetologia 45:693-702. [DOI] [PubMed] [Google Scholar]
- 43.Salminen, K. K., T. Vuorinen, S. Oikarinen, M. Helminen, S. Simell, M. Knip, J. Ilonen, O. Simell, and H. Hyöty. 2004. Isolation of enterovirus strains from children with preclinical type 1 diabetes. Diabet. Med. 21:156-164. [DOI] [PubMed] [Google Scholar]
- 44.Santti, J., T. Hyypiä, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanway, G. 1990. Structure, function and evolution of picornaviruses. J. Gen. Virol. 71:2483-2501. [DOI] [PubMed] [Google Scholar]
- 46.Stanway, G., F. Brown, P. Christian, T. Hovi, T. Hyypiä, A. M. Q. King, N. J. Knowles, S. M. Lemon, P. D. Minor, M. A. Pallansch, A. C. Palmenberg, and T. Skern. 2004. Picornaviridae, p. 757-778. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.). Virus taxonomy, VIIIth Report of the ICTV. Elsevier, London, United Kingdom.
- 47.Stanway, G., T. Hyypiä, N. J. Knowles, and T. Hovi. 2002. Biological and molecular basis of picornavirus classification, p. 17-24. In B. L. Semler, and E. Wimmer (ed.), Molecular biology of picornaviruses. American Society for Microbiology, Washington, D.C.
- 48.Szopa, T. M., P. A. Titchener, N. D. Portwood, and K. W. Taylor. 1993. Diabetes due to viruses—some recent developments. Diabetologia 36:687-695. [DOI] [PubMed] [Google Scholar]
- 49.Szopa, T. M., T. Ward, D. M. Dronfield, N. D. Portwood, and K. W. Taylor. 1990. Coxsackie-b4 viruses with the potential to damage beta-cells of the islets are present in clinical isolates. Diabetologia 33:325-328. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Titchener, P. A., O. Jenkins, T. M. Szopa, K. W. Taylor, and J. W. Almond. 1994. Complete nucleotide-sequence of a beta-cell tropic variant of coxsackievirus-B4. J. Med. Virol. 42:369-373. [DOI] [PubMed] [Google Scholar]
- 53.Vreugdenhil, G. R., A. Geluk, T. H. M. Ottenhoff, W. J. G. Melchers, B. O. Roep, and J. M. D. Galama. 1998. Molecular mimicry in diabetes mellitus: the homologous domain in coxsackie B virus protein 2C and islet autoantigen GAD65 is highly conserved in the coxsackie B-like enteroviruses and binds to the diabetes associated HLA-DR3 molecule. Diabetologia 41:40-46. [DOI] [PubMed] [Google Scholar]
- 54.Yin, H., A. K. Berg, J. Westman, C. Hellerstrom, and G. Frisk. 2002. Complete nucleotide sequence of a Coxsackievirus B-4 strain capable of establishing persistent infection in human pancreatic islet cells: effects on insulin release, proinsulin synthesis, and cell morphology. J. Med. Virol. 68:544-557. [DOI] [PubMed] [Google Scholar]
- 55.Ylipaasto, P., K. Klingel, A. M. Lindberg, T. Otonkoski, R. Kandolf, T. Hovi, and M. Roivainen. 2004. Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia 47:225-239. [DOI] [PubMed] [Google Scholar]